Abstract
Often used as a proxy for the transition to reproduction, flowering time (FT) is an integrative trait of two successive biological processes, i.e. bolting time (BT) and the interval between bolting and flowering time (INT). In this study, we aimed to identify candidate genes associated with these composite traits in Arabidopsis thaliana using a field experiment. Genome-wide association (GWA) mapping was performed on BT, INT and FT based on a sample of 179 worldwide natural accessions genotyped for 216,509 SNPs. The high resolution conferred by GWA mapping indicates that FT is an integrative trait at the genetic level, with distinct genetics for BT and INT. BT is shaped largely by genes involved in the circadian clock whereas INT is shaped by genes involved in both the hormone pathways and cold acclimation. Finally, the florigen TSF appears to be the main integrator of environmental and internal signals in ecologically realistic conditions. Based on FT scored in a previous field experiment, we also studied the genetics underlying reaction norms across two years. Only four genes were common to both years, emphasizing the need to repeat field experiments. The gene regulation model appeared as the main genetic model for genotype × year interactions.
Keywords: phenology, Arabidopsis thaliana, association mapping, QTL, common garden, phenotypic plasticity
Introduction
The transition from a vegetative to reproductive stage is a key component of the plant life cycle. In order to maximize fitness, plants must synchronize this transition with the period most favorable for reproduction (Koornneef, Alonso-Blanco and Vreugdenhil 2004; Engelmann and Purugganan 2006; Brachi et al. 2012). Natural variation in the timing of this transition is related to latitude in many plant species (Stinchcombe et al. 2004; Uga et al. 2007; Van Dijk and Hautekèete 2007; Ducrocq et al. 2008; Jones et al. 2008), suggesting adaptation to large-scale environmental factors such as photoperiod, temperature or precipitations.
Synchronization of the reproductive transition with optimal environmental conditions is realized through the integration of environmental and internal signals (see Roux et al. 2006 for a review). In the model species Arabidopsis thaliana, a complex genetic network regulates the transition from vegetative to reproductive stages (Komeda 2004; Roux et al. 2006). The photoperiod and vernalization pathways integrate photoperiod and cold period requirements, while the autonomous pathway and the gibberellin pathway integrate internal signals.
Most knowledge about the genetics of the transition to flowering in A. thaliana comes from forward genetic approaches (Alonso and Ecker 2006). An increasing number of studies now aim to explore the genetics underlying natural variation in flowering time as observed across the species range. Two approaches can be considered. The first is traditional linkage mapping, based on crosses of two or more natural accessions (see for example Simon et al. 2008; Balasubramanian et al. 2009; Bentsink et al. 2010). More recently association mapping, first applied in human genetics (Stein and Elston 2009), was introduced in plant genetics (Rafalski 2010). By exploiting the numerous recombination events along a species history and providing a high density of markers, association mapping has great resolution for dissecting the genetics of a complex trait (Aranzana et al. 2005; Zhao, Nordborg, and Marjoram 2007; Ehrenreich et al. 2009; Atwell et al. 2010). Association studies can be carried out on either candidate genes or at the genome scale. However, the candidate gene approach requires detailed knowledge of the biochemistry and genetics of a trait (Bergelson and Roux 2010; Rafalski 2010), and may even be difficult to apply to well described complex traits like flowering time (Ehrenreich et al. 2009). In contrast, the genome-wide strategy allows one to search blindly for genomic regions associated with a trait of interest (Atwell et al. 2010). The performance of Genome-Wide Association (GWA) mapping has been demonstrated for A. thaliana by enrichment in a priori candidate genes for 16 floral transition related traits; this method also facilitates the identification of new regions with no a priori candidate genes (Atwell et al. 2010).
We recently performed a field experiment in Northern France (September 2007 to April 2008), with the aim of identifying the genetic bases of natural variation in flowering time in A. thaliana grown under field conditions (Brachi et al. 2010). In this study, we combined GWA mapping based on 184 worldwide natural accessions genotyped for 216,509 SNPs, and traditional linkage mapping based on 13 RIL families, representing a total of 4,366 RILs. Two main conclusions were drawn from this field experiment. First, our dual mapping strategy clearly reduces both the rate of false positives, i.e. spurious associations due to population structure, and the rate of false negatives, i.e. causative SNPs whose signal is lost after correction for population structure because their distribution of allele frequencies overlaps with population structure. As a consequence, enrichment in a priori candidate genes was increased when SNPs identified through GWAs mapping also overlapped with QTL confidence intervals (Brachi et al. 2010). Second, in contrast to greenhouse conditions, genes involved in the regulation of the plant circadian clock were prevalent among the genetic determinants of flowering time expressed in natural conditions. This result stresses the importance of estimating natural variation in ecologically realistic conditions (Bergelson and Roux 2010).
Because climatic conditions present yearly stochastic variation at a specific geographic location, we re-grew 179 out of 184 worldwide natural accessions genotyped for 216,509 SNPs in a second field experiment at the exact same location in Northern France, from September 2008 to April 2009. The first objective of this new field experiment was to measure the reproducibility of flowering time scored in 2007-2008. Because flowering time may result from sensing light and temperature cues in A. thaliana, expressing flowering time in photothermal units (PTU) was shown to improve the reproducibility of flowering time measurement among years (Brachi et al. 2010).
The second objective of this new field experiment was to identify the genetics underlying individual components of flowering time in natural conditions. Flowering time is an integrative trait of two successive phenological processes, i.e. bolting time and the interval between bolting and flowering time. Bolting time, which corresponds to the onset of elongation of the internodes of the leaf zone (Pouteau and Albertini 2009) is related to latitude in A. thaliana (Stinchcombe et al. 2004). This suggests a global selective cline in the transition to the reproductive phase of the life cycle. The interval between bolting and flowering is a potential target of selection, a shorter interval improving the fitness of individuals in various light and competition conditions (Dorn, Hammond Pyle and Schmitt 2000). Here, we sought to identify the genetic bases of bolting time, flowering time and the interval between these developmental events using GWA mapping of 179 natural accessions phenotyped under ecologically realistic conditions. Based on a priori candidate genes list for floral transition, GWA mapping allowed us to detect SNPs underlying natural variation common to these three traits, as well as SNPs associated with each trait alone. Our results present flowering time as a composite character of two successive components, each with distinct genetics.
Interestingly, GWA mapping performed on flowering time scored in this second field experiment only identified a subset of the candidate genes associated with flowering time in the first field experiment (data from Brachi et al. 2010 without considering QTL overlapping). This led us to study the genetic bases of flowering time reaction norms across the two successive years. We suggest that GWA mapping might be precise enough to distinguish between gene regulation vs. allelic sensitivity as the main model for the genetic of phenotypic plasticity (Via et al. 1995).
Materials and methods
Plant material and field experimental design
This study is based on phenotypes measured for a worldwide collection of 192 A. thaliana natural accessions in a field experiment performed at the University of Lille from September 2008 to April 2009. The field experiment design is fully described elsewhere (Brachi et al. 2010). Briefly, a total of 1188 plants were organized in three blocks, each block being an independent randomization of 2 replicates of each of the 192 natural accessions. Bolting date was scored as the number of days between germination and the date the inflorescence was distinguishable from the leaves at a size < 5mm. Flowering date was scored as the number of days between germination and the appearance of the first open flower. The interval between bolting time and flowering time was measured as the difference between the bolting time and flowering time.
Flowering time (FT) data scored in this study from September 2008 to April 2009 were previously used in Brachi et al. 2010 in order to study the reproducibility of flowering time measurement obtained in a field experiment that took place from September 2007 to April 2008. These 2009 flowering time data have not previously been subjected to GWAs analysis.
Having found differences in the genetic bases of flowering time in this study, and our previous 2008 experiment (Brachi et al. 2010), we additionally used the 2008 data in order to calculate reaction norms for flowering time calculated as the flowering time difference between the two years (DIFF), based on flowering time genotypic means (least-square means, see below) estimated for each accession in each year.
Data analysis
In this study, bolting time (BT), flowering time (FT) and the interval between bolting and flowering (INT) were converted to photothermal units (PTU) as previously described in Brachi et al. (2010).
BT, FT and INT were analyzed with the general linear model (GLM) procedure in SAS9.1 (SAS Institute Inc., Cary, North Carolina, USA) according to a model previously described in Brachi et al. (2010). Data were not transformed as the residuals from the model were close to normality for all traits.
Pearson genetic correlations among phenological traits were calculated based on the least-square (LS) means obtained for each natural accession from the GLM described above.
Multiple linear regressions were used to study the relationships between phenological trait variation (LS means) with latitude and longitude of the location from which natural accessions were originally sampled (see Dataset S1, Table 1 in Brachi et al. 2010).
GWA mapping and enrichment for a priori candidate genes
GWA analyses were based on a subset of the 192 natural accessions included in the experiment for which we had genotypic data. One hundred and seventy-nine natural accessions were genotyped for 216, 509 SNPs evenly spaced across the genome (i.e. on average, 1 SNP every 500bp; see Dataset S1, Table 1 in Brachi et al. 2010). The design of the Affymetrix genotyping array and genotyping protocols have been detailed elsewhere (Atwell et al. 2010).
When testing the association between phenotypes and genotypes for each marker, we first used a naive approach not taking into account the potential effect of population structure, i.e. Wilcoxon rank-sum tests. To control for population structure, we then ran EMMA, based on a mixed model that includes a matrix of genotype similarities among accessions (Kang et al. 2008).
For each trait, we focused on the SNPs presenting the highest association (top SNPs) for each GWA mapping analysis (Wilcoxon and EMMA). An association peak was defined as a group of top SNPs that co-occur within a physical distance less than 20kb.
Based on an a prior list of 282 candidate genes for floral transition (Brachi et al. 2010), computation of enrichment for a priori candidate genes and with respect to linkage disequilibrium has been fully described elsewhere (Atwell et al. 2010, Brachi et al. 2010).
All GWA analyses and enrichment computations were performed using the software R (R Development Core Team 2009; Brachi et al. 2010).
Detection of a priori candidate genes
Following our literature searches, candidate genes detected within 20 kb of a top SNP were assigned to one of seven flowering time related genetic pathways. “Circadian clock” refers to genes implicated in the regulation and entrainment of the circadian rhythm; “Cold acclimation and vernalization” refers to genes implicated to the triggering of flowering in response to cold-induced stress or to a cold period; “Hormones” refers to genes related to the gibberellins and auxins; “Autonomous” refers to genes involved in the autonomous pathway; “Light quality and photoperiod” refers to genes implicated in light perception; “Integrators” refers to genes triggering flowering by integrating signals from the other pathways; “Meristem identity and flower formation” refers to genes specifically expressed in the meristem and responsible for formation of floral organs. An 8th class consisted of genes that could not be addressed to any other classes, their function being generally unclear.
Comparison of FT genetics across two successive years
The comparison of the identity of candidate genes detected in both years was based on the 200 top EMMA SNPs for FT in the 2007/2008 experiment without considering QTL overlapping (data from Brachi et al. 2010) and FT in the 2008/2009 experiment.
Results
Natural variation in semi-natural conditions
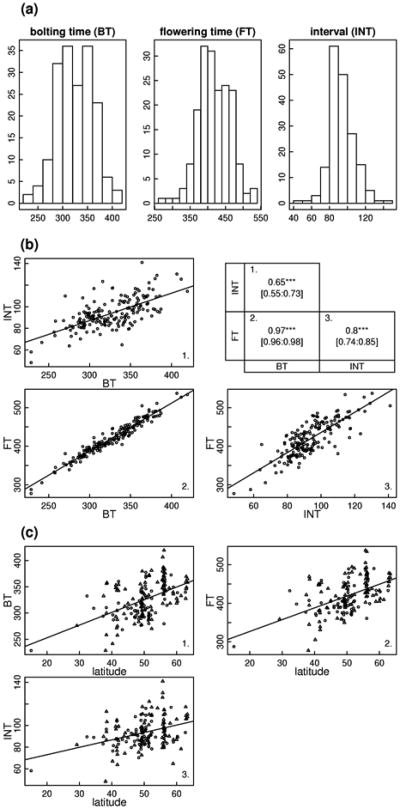
The first bolting occurred on the 17th of November 2008 (Pa-1 from Sicilia, Italy), and the last accession flowered on the 15th of April 2009 (Lund from Sweden). Highly significant variation was observed among natural accessions for bolting time (BT), flowering time (FT) and the interval between bolting and flowering (INT) expressed in PTU (Supplementary Table 1). BT ranged from 228.5 to 419.4 PTU (i.e. 61.7 to 175.5 Julian days), FT ranged from 277.3 to 537 PTU (i.e. 116.1 to 192.8 Julian days), and INT ranged from 48.3 to 141 PTU (i.e. 13.2 to 77 Julian days) (Figure 1a). Bolting time and flowering time, as expected, were highly correlated (Pearson's correlation coefficient= 0.97, P < 0.001). The correlations between INT and either BT or FT were much weaker, although significant (Figure 1b).
Figure 1.
Natural variation of floral transition related traits. (a) Distributions of bolting time (BT), flowering time (FT) and the interval between bolting and flowering (INT), expressed in PTU. (b) Genetic correlations among traits. 1. INT vs. BT, 2. FT vs. BT, and 3. FT vs. INT. The table gives the Pearson's correlation coefficients corresponding to each pairwise combination. The confidence intervals of the correlation coefficients are given in brackets. (c) Relationship of each trait with latitude. 1. BT, 2. FT and 3. INT. ***: P < 0.001; ns: non-significant.
Strong and significant latitudinal clines were detected for BT and FT, while a weaker but significant latitudinal cline was detected for INT (Table 1). To avoid outliers in the geographical distribution, latitudinal and longitudinal clines were also investigated at the European scale (latitude > 33°, -50° < longitude < 50°). The latitude regression coefficients for BT and FT are even stronger when only considering the European accessions. Because the functionality of the FRIGIDA (FRI) flowering time gene was previously shown to influence the relationship between the timing of floral transition and the latitude (Stinchcombe et al. 2004), latitudinal and longitudinal clines were tested in this study according to the FRI functionality. For each trait, the latitudinal cline in Europe was strong for accessions bearing a functional FRI allele (Table 1). For accessions bearing a non functional FRI allele in Europe, latitudinal clines were weaker but still significant for BT and FT, while non-significant for INT. In this sample, only weak or non-significant longitudinal clines were detected for BT, FT and INT (Table 1).
Table 1.
Relationships of phenological variation scored in the second field experiment with latitude and longitude.
| Worldwidea | Europeb | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Trait | Groupc | Latitude | Longitude | Latitude | Longitude |
| BT | All | 2.65 ± 0.34 *** | -0.11 ± 0.07 ns | 3.99 ± 0.46 *** | -0.07 ± 0.29 ns |
| F | 2.89 ± 0.46 ** | -0.15 ± 0.09 ns | 4.62 ± 0.55 *** | -0.96 ± 0.40 * | |
| NF | 1.75 ± 0.48 *** | -0.06 ± 0.11 ns | 2.07 ± 0.76 ** | 0.86 ± 0.41 * | |
|
| |||||
| INT | All | 0.72 ± 0.14 *** | -0.01 ± 0.03 ns | 1.03 ± 0.21 *** | 0.04 ± 0.14 ns |
| F | 0.76 ± 0.21 *** | -0.01 ± 0.04 ns | 1.30 ± 0.28 *** | -0.20 ± 0.20 ns | |
| NF | 0.47 ± 0.18 * | -0.02 ± 0.04 ns | 0.22 ± 0.31 ns | 0.28 ± 0.17 ns | |
|
| |||||
| FT 2008/2009 | All | 3.89 ± 0.46 *** | -0.21 ± 0.09 * | 5.69 ± 0.61 *** | 0.13 ± 0.39 ns |
| F | 4.15 ± 0.63 *** | -0.26 ± 0.12 * | 6.66 ± 0.75 *** | -0.88 ± 0.54 ns | |
| NF | 2.78 ± 0.62 *** | -0.14 ± 0.14 ns | 2.81 ± 0.90 ** | 1.25 ± 0.50 * | |
|
| |||||
| DIFF | All | 0.57 ± 0.16 *** | -0.09 ± 0.03 ** | 0.67 ± 0.23 ** | 0.17 ± 0.15 ns |
| F | 0.52 ± 0.21 * | -0.10 ± 0.04 * | 0.75 ± 0.28 ** | 0.27 ± 0.20ns | |
| NF | 0.63 ± 0.25 * | -0.07 ± 0.05 ns | 0.52 ± 0.42 ns | 0.11 ± 0.23 ns | |
Multiple linear regression performed on worldwide accessions with available latitude and longitude data.
Multiple linear regression performed on European accessions (latitude > 33°, -50° < longitude < 50°) with available latitude and longitude data.
All: all accessions (Worldwide: n = 179; Europe: n = 154), F: only accessions bearing a functional FRI allele (Worldwide: n = 113; Europe: n = 96), NF: only accessions bearing a non-functional FRI allele (Worldwide: n = 66; Europe: n = 58).
P < 0.001;
non significant
GWA mapping
Effect of population structure and Minor Allele Relative Frequency
To assess the influence of population structure on GWA mapping results, the distribution of p-values was compared to a uniform (expected) distribution of p-values (Supplementary Figure 1). The Wilcoxon tests led to a strong departure from the null hypothesis of a uniform distribution for BT, FT, and to a lesser extent, for INT. As previously shown by Atwell et al. (2010), EMMA appears to efficiently correct for population structure for all traits. Distributions of p-values obtained with EMMA were much closer to a uniform distribution except for the strongest associations, i.e. the lowest p-values. We should note, however, that EMMA may also introduce false negatives (see discussion).
Minor Allele Relative Frequencies (MARF) also seemed to have an impact on the distribution of p-values. SNPs with MARF < 10% show an excess of high p-values when analyzed using EMMA and a lack of high p-values when analyzed using the Wilcoxon test. Although less dramatic than the effect of population structure, we followed Kang et al. (2008) and only considered SNPs with MARF over 10% for candidate genes detection. The impact of retaining SNPs with MARF lower than 10% was nevertheless considered when calculating enrichment ratios (see below).
Shared vs. specific association peaks across the genome
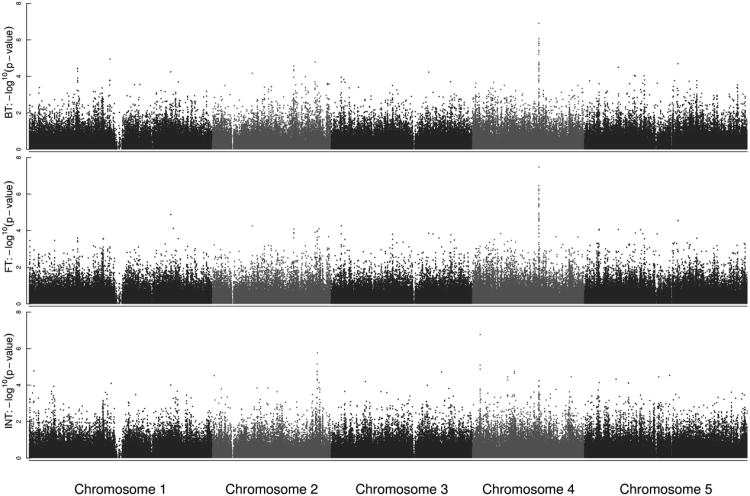
GWA revealed common regions detected for BT and FT (Figure 2). For both traits, a clear peak in the middle of chromosome 4 is detected with EMMA (Figure 2), with -log10(p-values) reaching 7.12 and 7.46 for BT and FT, respectively. Although less obvious, this peak also appeared when population structure was not taken into account (Supplementary Figure 2). For INT, two regions present clear association peaks, one at the end of chromosome 2 and one at the beginning of chromosome 4, with -log10(p-values) reaching 5.82 and 6.8 with EMMA, respectively (Figure 2). These regions are much less associated with BT and FT.
Figure 2.
Manhattan plots of the genome-wide association mapping (EMMA) results for BT, FT and INT. The x-axis indicates the physical position along each chromosome. The five chromosomes are presented in a row along the x-axis in different degrees of grey. The y-axis gives the −log10(p-value) obtained by EMMA for each SNP.
Enrichment ratios
Enrichment ratios were calculated for all traits based on a list of 282 a priori candidate genes for floral transition. When analyzed with EMMA, enrichment for BT was significant only when considering SNPs with MARF exceeding 10% and restricting attention to the 200 top, or fewer, SNPs (Supplementary Figure 3). For FT, enrichment was significant when considering up to 100 top SNPs overall or up to 200 top SNPs with MARF over 10%, (Supplementary Figure 3). For INT, enrichment was generally much weaker but significant for up to the 1000 top SNPs and MARF did not seem to have an effect on enrichment (Supplementary Figure 3). When population structure was not accounted for, no significant enrichment was detected for BT, FT or INT (Supplementary Figure 4).
Shared vs. specific genetics
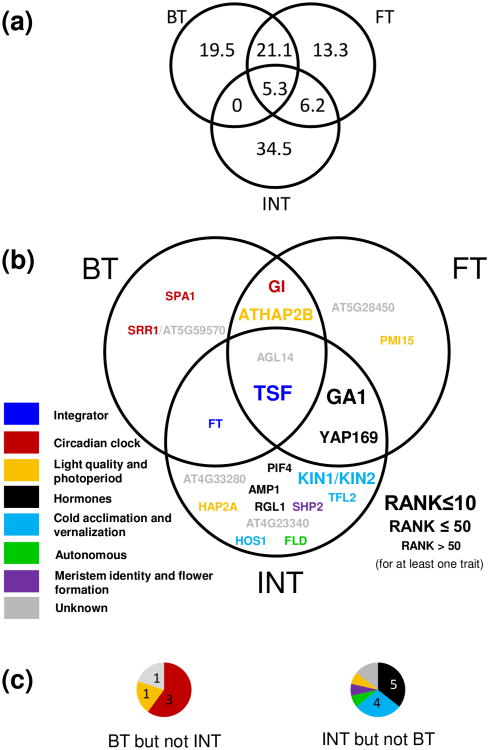
We chose the 200 top SNPs as a common threshold for all traits as it captures the most information in the GWA signal without including too much noise. The 200 top SNPs for the three traits analyzed with EMMA were then checked for being specific to a trait or shared between two or three traits. These three lists of 200 SNPs included a total of 435 different SNPs (Figure 3a), 67.3% of which were detected specifically for one trait; i.e. 19.5, 13.3 and 34.5% being specific to BT, FT and INT, respectively. More than 27% of the top SNPs were detected for two traits. BT and FT shared 21.1% of the top SNPs, while FT and INT only shared 6.2%. Interestingly, BT and INT did not uniquely share any top SNPs. Only 5.3% top SNPs were shared by the three floral transition related traits.
Figure 3.
Identification of candidate genes associated with phenological variation. (a) Venn diagram presenting the partitioning (in %) of the 435 different SNPs detected among the lists of 200 top SNPs for BT, FT and INT. (b) Venn diagram showing the partitioning of genes detected for BT, FT and INT. The size of the gene names indicates the highest rank of the SNP within 20kb of that candidate gene, across all traits for which it was detected. Colors indicate the genetic pathway to which the genes were assigned. (c) Pie charts illustrate the partitioning of genes into the different pathways (colors) for genes detected for BT and not for INT (left), and for INT but not BT (right).
All 435 top SNPs were checked for being within 20 kb of any candidate gene for flowering time. Twenty-four out of the 282 a priori candidate genes appeared to be associated with at least one of the traits. Among those genes, 17 were detected for one trait, 5 were common to two traits and 2 were common to all traits (Figure 3b). From our literature search, 20 of these 24 genes could be assigned to a genetic pathway involved in floral transition (Table 2 and Figure 3b). Shared SNPs showed significantly better association with candidate genes than SNPs detected for only one trait (Kruskall-Wallis test: Chi square = 4.82, P = 0.028).
Table 2.
Candidate genes detected for bolting time (BT), flowering time (FT), and interval between bolting and flowering time (INT).
| Genes ID | Genes (names) | BTa | FTa | INTa | Pathway | Reference |
|---|---|---|---|---|---|---|
| AT2G46340 | SPA1 | 1 (69) | . | . | circadian clock | (Ishikawa, Kiba and Chua 2006) |
| AT5G59560/AT5G59570 | SRR1/AT5G59570 | 1 (199) | . | . | circadian clock/. | (Staiger et al. 2003) |
| AT5G38150 | PMI15 | . | 1 (96) | . | blue light perception | (Luesse, DeBlasio, and Hangarter. 2006) |
| AT5G28450 | AT5G28450 | . | 1 (181) | . | . | . |
| AT5G15960/AT5G15970 | KIN1/KIN2 | . | . | 1 (19) | cold acclimation | (Kurkela and Borg-Franck 1992) |
| AT4G33280 | AT4G33280 | . | . | 2 (56) | . | . |
| AT2G42830 | SHP2 | . | . | 1 (57) | flower formation | . |
| AT3G54720 | AMP1 | . | . | 1 (96) | hormone | (Vidaurre et al. 2007) |
| AT5G17690 | TFL2 | . | . | 1 (98) | vernalization | (Sung et al. 2006) |
| AT1G66350 | RGL1 | . | . | 3 (150) | gibberelin | (Wen and Chang 2002) |
| AT4G23340 | AT4G23340 | . | . | 1 (154) | . | . |
| AT5G12840 | HAP2A | . | . | 1 (158) | photoperiod | (Wenkel et al. 2006) |
| AT2G39810 | HOS1 | . | . | 1 (169) | cold acclimation and vernalization | (Lee et al. 2001) |
| AT3G10390 | FLD | . | . | 1 (178) | autonomous | (Jin et al. 2008) |
| AT2G43010 | PIF4 | . | . | 1 (193) | light and gibberellin signaling | (Lucyshyn and Wigge 2009) |
| AT3G05690 | ATHAP2B | 5 (43) | 6 (27) | . | photoperiod | (Wenkel et al. 2006) |
| AT1G22770 | GI | 1 (45) | 1 (179) | . | circadian clock | (Mizoguchi et al. 2005) |
| AT4G02780 | GA1 | . | 1 (132) | 7 (1) | gibberelin | (Sun and Kamiya 1994) |
| AT5G07200 | YAP169 | . | 1 (94) | 1 (49) | gibberelin | (Phillips et al. 1995) |
| AT1G65480 | FT | 2 (154) | . | 2 (114) | integrator | (King et al. 2008) |
| AT4G20370 | TSF | 24 (1) | 25 (1) | 17 (25) | integrator | (Michaels et al. 2005) |
| AT4G11880 | AGL14 | 1 (130) | 1 (86) | 1 (159) | . | . |
BT, FT and INT indicate the number of top SNPs detected by EMMA within 20 kb of the candidate gene. The number in brackets is the corresponding rank of the most associated SNP. For example, for BT, 24 (1) for the TSF gene indicates that 24 out of the 200 top SNPs were detected within 20 kb of TSF and that the rank of the most associated SNP of these 24 is 1. “.” indicates that no top SNP was found within 20 kb of the candidate gene. When top SNPs were within 20kb of two candidate genes, genes ID, genes names, genes pathways (if different) and references are separated by “/”.
Genes detected for FT mainly overlapped with genes detected for BT and/or INT. Only two genes specific to FT were detected, one of unknown function (AT5G28450), the second related to the photoperiod pathway (PMI15). Three out of five genes detected for BT but not for INT have been implicated in the circadian clock (GI, SPA1 and SRR1) (Table 2, Figures 3b and 3c). Of the 14 genes detected for INT but not for BT, five were related to hormones (GA1, YAP169, AMP1, PIF4 and RGL1) and four to cold acclimation and vernalization (KIN1, KIN2, TFL2 and HOS1) (Table 2, Figures 3b and 3c). Three genes were detected for both BT and INT, two of these are known floral integrators (FT and TSF) (Table 2; Figure 3b, c).
TSF and GA1 correspond to the peaks in the middle of chromosome 4 detected for BT and FT and the peak at the beginning of chromosome 4 detected for INT, respectively (Figure 2 and Supplementary Figure 5). The peak in the middle of chromosome 4 is located approximately 11 kb on the 3′ side of TSF (Supplementary Figure 5), close to a retro-transposon copia element (AT4G20365) shown to play a role in the regulation of TSF (Yang et al. 2010). At the beginning of chromosome 4, the SNP showing the lowest p-value for INT is located 10.8 kb from the 5′ end of the GA1 (Supplementary Figure 5). The peak at the beginning of chromosome 2 does not seem to be in the vicinity of any candidate genes from the list (Supplementary Figure 5). Further search of the TAIR9 database for this region did not reveal any other likely candidate.
Comparison of FT genetics across two successive years
Surprisingly, only 4 candidate genes (TSF, YAP169, ATHAP2B, PMI15) identified for flowering time in this field experiment (2008/2009) had already been identified for flowering time during the 2007/2008 experiment (Table 3). This prompted us to study the genetic bases of flowering time plasticity across the two successive years. GWA mapping was performed on flowering time difference between the two years (DIFF), based on flowering time genotypic means (LS means) estimated for each accession in each year.
Table 3.
Candidate genes associated with flowering time reaction norms.
| Genes ID | Genes names | FT2007/2008a | FT 2008/2009 a | DIFFa |
|---|---|---|---|---|
| AT4G20370 | TSF | 19 (1) | 25 (1) | . |
| AT3G05690 | ATHAP2B | 5 (16) | 6 (27) | . |
| AT5G07200 | YAP169 | 1 (154) | 1 (94) | . |
| AT5G38150 | PMI15 | 1 (53) | 1 (96) | . |
| AT4G11880 | AGL14 | . | 1 (86) | . |
| AT4G02780 | GA1 | . | 1 (132) | . |
| AT1G22770 | GI | . | 1 (179) | . |
| AT5G28450 | AT5G28450 | . | 1 (181) | . |
| AT4G02560 | LD | 7 (29) | . | 2 (147) |
| AT1G69120 | AP1 | 1 (38) | . | . |
| AT5G62640 | ELF5 | 1 (50) | . | . |
| AT4G32980 | ATH1 | 2 (111) | . | 2 (23) |
| AT4G01060 | ETC3 | 1 (163) | . | . |
| AT1G63030 | DDF2 | 1 (184) | . | . |
| AT5G39660 | CDF2 | . | . | 2 (22) |
| AT4G27060 | TOR1 | . | . | 2 (74) |
| AT3G24440 | VRN5 | . | . | 3 (79) |
| AT1G18450 | ATARP4 | . | . | 1 (108) |
| AT5G51810/ AT1G78050 | AT2353/PGM | . | . | 1 (109) |
| AT4G22140 | EBS | . | . | 1 (118) |
| AT3G46640 | PCL1 | . | . | 1 (134) |
| AT1G50960 | ATGA2OX7 | . | . | 2 (153) |
| AT5G11530 | EMF1 | . | . | 1 (154) |
| AT5G28490 | LSH1 | . | . | 1 (160) |
| AT5G14920 | AT5G14920 | . | . | 1 (179) |
| AT5G12840 | HAP2A | . | . | 1 (185) |
FT 2007/2008, , FT 2008/2009, and DIFF indicate the number of top SNPs detected by EMMA within 20 kb of the candidate gene. The number in brackets is the corresponding rank of the most associated SNP. For example, for FT 2008/2009, 25(1) for the TSF gene indicates that 25 out of the 200 top SNPs were detected within 20 kb of TSF and that the rank of the most associated SNP of these 25 is 1. “.” indicates that no top SNP was found within 20 kb of the candidate gene. When top SNPs were within 20kb of two candidate genes, genes ID and genes names are separated by “/”.
The mean DIFF among natural accessions (38.2 PTU ± 14.8) represented 12 % of the flowering time variation observed among accessions in the 2007/2008 field experiment (Brachi et al. 2010). DIFF ranged from negative values with accessions that flowered with more PTU in the 2008/2009 field experiment (Pro-0, Spain, DIFF = - 20.6 PTU) to positive values up to 79.5 PTU (Spr1-6, Sweden), to accessions displaying low plasticity (LL-0, Spain, DIFF = 1.88 PTU). Multiple regression on latitude and longitude revealed a weak but significant latitudinal cline for DIFF, at both worldwide and European scales (worldwide: latitude regression coefficient ± SE = 0.57 ± 0.16, P < 0.001; Europe: latitude regression coefficient ± SE = 0.67 ± 0.23, P < 0.01). A very weak longitudinal cline was detected at the worldwide scale (longitude regression coefficient ± SE = -0.09 ± 0.03, P < 0.01) but no longer significant at the European scale (longitude regression coefficient ± SE = 0.17 ± 0.15, P > 0.05).
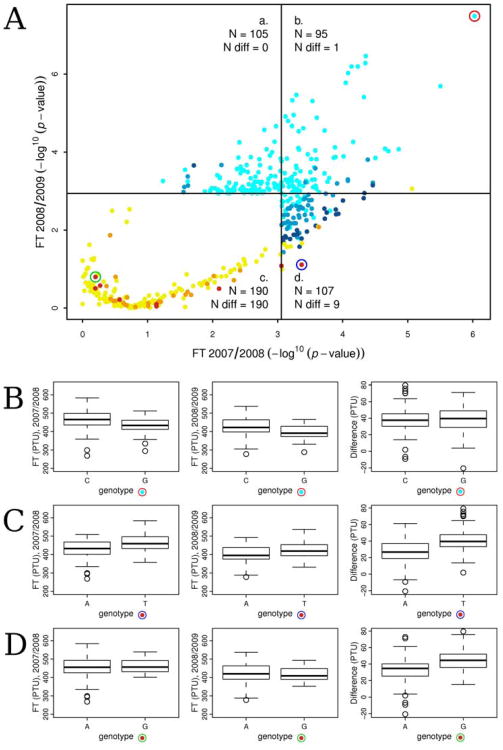
In order to study the genetics of phenotypic plasticity, we focused on the 200 top EMMA SNPs for FT in the 2007/2008 experiment (data from Brachi et al. 2010 without considering QTL overlapping), FT in the 2008/2009 experiment and DIFF. The resulting 600 SNPs corresponded to 497 non-redundant top SNPs (Figure 4a). The genetics underlying flowering time plasticity displayed two main features. First, among all the top SNPs identified for FT across both field experiments (i.e. 307 SNPs; Figure 4a, categories a, b and c), only 31 % were associated with FT in both years (i.e. 95 SNPs; Figure 4a, category a). Second, 95 % of top SNPs associated with DIFF were not associated with FT in either field experiments (Figure 4a, category d).
Figure 4.
Genetics of phenotypic plasticity. A. The x- and y-axes indicate the –log10(p-value) from the association tests for FT in the 2007/2008 and the 2008/2009 field experiments, respectively. After sorting the 200 top SNPs for each trait (FT in 2007/2008, FT in 2008/2009 and the difference between the two years), i.e. 600 SNPs, 497 non redundant SNPs were identified. Each point correspond to one of these 497 SNPs. Horizontal and vertical black lines indicate the lowest association score observed among the 200 best associations for FT in the 2007/2008 and the 2008/2009 field experiments, respectively. The color of each point is indicative of the association score (−log10(p-value)) with the difference in flowering time between the two years (DIFF): light blue < 1 < blue < 2 < dark blue < threshold < yellow < 4 < orange < 5 < dark red, ‘threshold’ corresponding to the lowest association score observed among the 200 best associations for DIFF, i.e. ∼ 3.05. ‘N’ indicates the number of SNPs in each of the four categories (a, b, c and d) defined by the lowest association score observed among the 200 best associations for FT in the 2007/2008 and the 2008/2009 field experiments. ‘N diff’ indicates the number of SNPs with association scores over the 3.05 threshold for DIFF. B, C and D. Three contrasting cases of genetics underlying flowering time reaction norms. The box-plots are representative of phenotypic distributions according to the genotype at three top SNPs circled in red, blue and green in the A panel, respectively.
Among the 497 non-redundant top SNPs, we identified three types of SNPs illustrating different genetic controls of phenotypic plasticity. First, the SNP with the highest association with FT in both years (chromosome 4, position 10,989,741 bp), in the vicinity of the florigen TSF, was not associated with DIFF (Figure 4b). Second, the SNP with the highest association with DIFF (chromosome 4, position 11,691,632 bp) was associated with FT in the 2007/2008 field experiment but not with FT in the 2008/2009 field experiment (Figure 4c). Third, the second best associated SNP for DIFF (chromosome 5, position 9,816,674 bp) was not detected among the 200 top SNPs for FT in either field experiments (Figure 4d).
Based on a list of 282 a priori candidate genes for flowering time, candidate genes detected for DIFF are given in Table 3. In agreement with the patterns observed for the 497 non-redundant top SNPs, candidate genes detected for DIFF were not associated with FT in either field experiments, except LD and ATH1. Twelve a priori candidate genes were specifically detected for DIFF, but no obvious candidate genes were found in the vicinity of the 20 top SNPs, even after a close inspection of the gene annotation of the genome.
Discussion
Natural variation of floral transition related traits
Highly significant variation was observed among natural accessions for three traits related to floral transition, i.e. bolting time (BT), flowering time (FT) and the interval between bolting and flowering (INT). The relationship with latitude was strong for BT and FT and was consistent with previous studies (Stinchcombe et al. 2004; Brachi et al. 2010). As previously suggested (Stinchcombe et al. 2004; Van Dijk and Hautekèete 2007), the strong relationship with latitude found for BT suggests a global selective cline in the transition to the reproductive phase of the life cycle, linked to globally varying environmental factors like photoperiod, temperature or precipitations. The latitudinal cline for INT was much weaker, although significant. This suggests that selection on INT may act at a more local scale, perhaps as a consequence of local competition. This is plausible in that fast growth of the shoot is important for maximizing access to light of the cauline leaves in dense stands (Franklin 2008). Indeed, the interval between bolting and flowering has previously been shown to be under directional selection when A. thaliana plants experience competition (Dorn, Hammond Pyle and Schmitt 2000; Brachi et al. 2012). The stronger relationship with latitude for BT than INT might suggest that they are under selective pressures operating at contrasting geographical scales, and variation for FT could be the consequence of a complex combination of selective pressures.
The genetics of the integrative flowering time trait
Our results suggest that flowering time is an integrative trait of at least two underlying biological processes. Bolting time relates to the transition of the apical meristem activity, shifting from producing vegetative tissue to reproductive structures (Pouteau and Albertini 2009). The interval between bolting and flowering relates to shoot growth and the beginning of offspring production.
Genes detected for bolting time were mostly implicated in the circadian clock. This finding is consistent with the fact that fitness is enhanced when the clock period tracks the environment (Michael et al. 2003; Dodd et al. 2005). Circadian rhythms are determined by daily variation of light quality and temperature and follow a latitudinal cline. Together with the significant relationship between bolting time and latitude, the detection of circadian clock related genes strengthens the inference that the transition to reproduction varies at the scale of the species, following a latitudinal cline.
Genes associated with the interval between bolting and opening of the first flower were related to both the hormone pathways (particularly gibberellins) and cold acclimation. Interestingly, both pathways seem to be involved in ‘the shade avoidance syndrome’ (SAS) (Franklin 2008). Vegetational shading leads to a decrease in the ratio of photon irradiance in the red (R) region of the spectrum to that in the far-red (FR) region (termed R: FR ratio). To increase light-foraging capacity, dicotyledonous plants may elevate leaves within the canopy or show increased elongation rates of stems and leaves (Franklin 2008). A hypothetical model depicting the molecular mechanism underlying SAS that includes many hormone related candidate genes detected in this study has been proposed (Franklin 2008). PIF4 is thought to induce production of gibberellins (GA1 and YAP169) (Sun and Kamiya 1994; Phillips et al. 1995) in low R: FR ratio by overriding the repression of gibberellins by DELLA proteins like RGL1 (reviewed in Hussain and Peng 2003). Given the high density of plants in our study (576 plants/m2), we cannot rule out a shade avoidance response induced by neighbors. However, SAS results from the overlapping of the plants' leaves, modifying the light quality and intensity. In our experiment, only 2.6 % of plants had a rosette diameter at bolting greater than four cm, suggesting that very few plants were covered by leaves from neighbouring individuals before bolting. We therefore believe that plants experienced a small (or negligible) reduction in the R: FR ratio and in photosynthetically active radiation induced by neighbors. In addition, PIF4 was previously found to be associated with the interval between bolting and flowering in the absence of competition through a candidate gene approach (Brock, Maloof and Weining 2010). Altogether, our results may rather suggest that the hormone related genes detected for INT may underlie natural variation of “constitutive shade avoidance” (Franklin 2008). In our study, genes involved in cold acclimation, KIN1 or KIN2 (Kurkela and Borg-Franck 1992), were also detected. Cold acclimation related genes were previously shown to interact with shade avoidance signaling (Franklin 2008). Their detection in this study warrants further investigation to better understand their interaction with other signaling pathways.
A causal polymorphism in the promoter region of the florigen FLOWERING LOCUS T (FT) mediates natural variation in flowering time measured in greenhouse conditions (Schwartz et al. 2009). In the ecologically realistic conditions of our second field experiment, a role of FT was suggested for BT and INT. The florigen TWIN SISTER OF FT (TSF) appears as the major integrator of environmental and internal signals triggering flowering in A. thaliana. The causal polymorphism might not be located in TSF itself but rather in a regulatory region located nearby. Indeed, the association peak detected in our study is located 10kb downstream of the 3′ end of TSF. A heterochromatic copia retro-element AT4G20635 is located closer to the association peak, and influences the chromatin environment for the TSF locus (Yang et al. 2010). The observation that different genes appears to govern variation in flowering time in the field versus greenhouse demonstrates the need to study natural variation in naturally complex environments (Bergelson and Roux 2010), and highlights the importance of floral integrators in the timing of reproductive transition in A. thaliana.
The genetics of flowering time reaction norms across two successive years
A high correlation coefficient was detected between flowering time (PTU) measured in the two successive field experiments (Brachi et al. 2010). This correlation was however different from 1, suggesting a fine tuning of flowering time through genotype × environment interactions.
Two main models for the genetics of phenotypic plasticity have been considered in the literature (Scheiner 1993; Via et al. 1995; Pigliucci 2005). The allelic sensitivity model, i.e. pleiotropic model, suggests that phenotypic plasticity results from genes having different effects on phenotypic variation depending on the environment. The gene regulation model, i.e. epistatic model, suggests that two sets of genes interact epistatically to affect the two components of the reaction norms; the first determines the difference in the average phenotype across environments; the second determines the slope of the reaction norms. Among the 200 top SNPs associated to DIFF, 95 % were not associated with flowering time in either field experiments, therefore favouring the gene regulation model. This result contributes to growing support for the gene regulation model in the literature (reviewed in Lacaze, Hayes and Korol 2009).
Interestingly, the only four candidate genes (including TSF) associated with flowering time in both years were not associated with genotype × year interactions, suggesting that the effect of these genes may not be influenced by the different environmental cues encountered by plants between the two years. As previously reported in Brachi et al. (2010), the second field experiment was much colder than the first field experiment, leading to an almost two-fold accumulation of chilling degrees during the second field experiment. In this study, four candidate genes associated with flowering time reactions norms across two successive years interact with FLC, a major floral repressor which is down regulated by vernalization (Michaels and Amasino 1999). The EMBRYONIC FLOWER 1(EMF1) and LUMINIDEPENDENS (LD) genes repress FLC expression (Michaels and Amasino 1999; Kim, Zhu and Sung 2010), while the ARABIDOPSIS THALIANA HOMEOBOX 1 (ATH1) gene acts as a general activator of FLC expression (Proveniers et al. 2007). VERNALIZATION 5(VRN5) is thought to maintain the vernalization-mediated chromatin modifications of FLC after exposure of plants to a cold period (Greb et al. 2007; Amasino 2010). Other candidate genes associated with flowering time reactions norms in this study may be related to other environmental factors known to affect flowering time, but not measured in this study, like precipitations, light quality or pathogens prevalence.
Power of GWA mapping in A. thaliana
Genome wide association mapping was previously shown to be an effective way to finely map complex traits in plants (Atwell et al. 2010). Here, we confirm this observation with a significant candidate gene enrichment detected for the three floral transition related traits. FT shows a stronger enrichment than BT and INT, probably because the candidate genes list was designed based on forward genetics studies scoring flowering time as a proxy of floral transition. Enrichment ratio calculations for different numbers of top SNPs suggest that 200 top SNPs retained most of the information about the genetics of BT and FT.
While it is necessary to correct for population structure in order to reduce the false positive rate, this correction likely introduces false negatives; that is, causative SNPs that are not detected because their patterns of allele frequencies overlap with population structure. The challenge of avoiding both false positives and false negatives can be resolved by combining GWA mapping and traditional linkage mapping (Zhao et al. 2007). The power of this dual mapping strategy was illustrated in a study on A. thaliana flowering time in outdoor conditions in which it estimated that GWA analysis alone led to a false positive rate of 40% and a false negative rate of 24% (Brachi et al. 2010). In the second field experiment reported here, the gene LKP2, related to photoperiod perception and the circadian clock, was only detected by GWA mapping when not correcting for population structure (data not shown) yet was previously validated by a QTL in the first field experiment (Brachi et al. 2010).
In addition, the coarse resolution of traditional linkage mapping usually does not allow easy discrimination between the two main models for the genetics of phenotypic plasticity (Lacaze, Hayes and Korol 2009). In this study, the high resolution of GWA mapping suggests that it may be possible to discriminate between the allelic sensitivity and gene regulation models.
Conclusion
All together, the finding of few common candidate genes across two successive years emphasizes the need to repeat the estimation of flowering time variation in natural conditions over multiple years. In order to better understand the genetic determinism of flowering time plasticity, the predominance of the gene regulation model for flowering time plasticity needs to be confirmed by further experiments, with a special emphasis on environmental factors that differed among years and/or locations.
Supplementary Material
Supplementary Figure 1. Quantile-Quantile plot of –log10 p-values in genome-wide scans for the three reproductive transition related traits, from left to right, BT,FT and INT.
Supplementary Figure 2. Manhattan plots of the genome-wide association mapping (Wilcoxon) results for BT, FT and INT.
Supplementary Figure 3. Enrichment ratios as a function of the number of top SNPs chosen in the GWA mapping results using the EMMA method.
Supplementary Figure 4. Enrichment ratios as a function of the number of top SNPs chosen in the GWA mapping results using a Wilcoxon test.
Supplementary Figure 5. Peaks of association in the regions spanning 20kb on each side of the top SNP near TSF (left), GA1 (middle), and at the beginning of chromosome 2 (right).
Supplementary Table 1. Natural variation for bolting time, flowering time and the interval between bolting and flowering scored in nature.
Acknowledgments
We are grateful to Angélique Bourceaux, Marie Carpentier, Clarisse Galliez and Cédric Glorieux for their assistance during the field experiment. This study was supported by a BQR grant to FR from the University of Lille 1, NSF DEB 0519961 and NIH R01 GM083068 grants to JB, and a PhD fellowship from the French Research Ministry and a mobility grant from the Collège Doctoral Européen to BB.
References
- Alonso JM, Ecker JR. Moving forward in reverse: Genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nature Reviews Genetics. 2006;7:524–536. doi: 10.1038/nrg1893. [DOI] [PubMed] [Google Scholar]
- Amasino R. Seasonal and developmental timing of flowering. The Plant Journal. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, Toomajian C, Traw B, Zheng H, Bergelson J, Dean C, Marjoram P, Nordborg M. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genetics. 2005;1, no. 5:e60. doi: 10.1371/journal.pgen.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang Y, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone A, Hu TT, Jiang R, Muliyati NW, Zhang X, Amer MA, Baxter I, Brachi B, Chory J, Dean C, Debieu M, de Meaux J, Ecker JR, Faure N, Kniskern JM, Jones JDG, Michael T, Nemri A, Roux F, Salt DE, Tang C, Todesco M, Traw MB, Weigel D, Marjoram P, Borevitz J, Bergelson J, Nordborg M. Genome-wide association study of 107 phenotypes in a common set of Arabidopsis thaliana inbred lines. Nature. 2010;465:632–636. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Schwartz C, Singh A, Warthmann N, Kim MC, Maloof JN, Loudet O, Trainer GT, Dabi T, Borevitz JO, Chory J, Weigel D. QTL mapping in new Arabidopsis thaliana advanced intercross-recombinant inbred lines. PLoS One. 2009;4, no. 2:e4318. doi: 10.1371/journal.pone.0004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy ME, Alonso-Blanco C, Teresa de Andrés M, Reymond M, van Eeuwijk F, Smeekens S, Koornneef M. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proceedings of the National Academy of Sciences, USA. 2010;107(9):4264–4269. doi: 10.1073/pnas.1000410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nature Reviews Genetics. 2010;11:867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- Brachi B, Aimé C, Glorieux C, Cuguen J, Roux F. Adaptive value of phenological traits in stressful environments: predictions based on seed production and laboratory natural selection. PLoS One. 2012;7, no. 3:e32069. doi: 10.1371/journal.pone.0032069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Nathalie F, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genetics. 2010;6, no. 5:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MT, Maloof JN, Weinig C. Genes underlying quantitative variation on ecologically important traits: PIF4 (PHYTOCHROME INTERACTING FACTOR 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana. Molecular Ecology. 2010;19:1187–1199. doi: 10.1111/j.1365-294X.2010.04538.x. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Dorn LA, Hammond Pyle E, Schmitt J. Plasticity to light cues and resources in Arabidopsis thaliana: Testing for adaptive value and cost. Evolution. 2000;54(6):1982–1994. doi: 10.1111/j.0014-3820.2000.tb01242.x. [DOI] [PubMed] [Google Scholar]
- Ducrocq S, Veyrieras JB, Camus-Kulandaivelu L, Kloiber-Maitz M, Presterl T, Ouzunova M, Manicacci D, Charcosset A. Key impact of vgt1 on flowering time adaptation in maize: Evidence from association mapping and ecogeographical information. Genetics. 2008;178, no. 4:2433–2437. doi: 10.1534/genetics.107.084830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I, Hanzawa Y, Chou L, Roe J, Kover PX, Purugganan MD. Candidate gene association mapping of Arabidopsis flowering time. Genetics. 2009:109.105189. doi: 10.1534/genetics.109.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann K, Purugganan MD. The molecular evolutionary ecology of plant development: flowering time in Arabidopsis thaliana. Advances in Botanical Research. 2006;44:507–526. [Google Scholar]
- Franklin KA. Shade avoidance. New Phytologist. 2008;179:930. doi: 10.1111/j.1469-8137.2008.02507.x. [DOI] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Current Biology. 2007;17(1):73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Hussain A, Peng J. DELLA proteins and GA signalling in Arabidopsis. Journal of Plant Growth Regulation. 2003;22:134–140. [Google Scholar]
- Ishikawa M, Kiba T, Chua NH. The Arabidopsis SPA1 gene is required for circadian clock function and photoperiodic flowering. The Plant Journal. 2006;46, no. 5:736–746. doi: 10.1111/j.1365-313X.2006.02737.x. [DOI] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. The Plant Journal. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H, Leigh FJ, Mackay I, Bower MA, Smith LMJ, Charles MP, Jones G, Jones MK, Brown TA, Powell W. Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the fertile crescent. Molecular Biology and Evolution. 2008;25, no. 10:2211–2219. doi: 10.1093/molbev/msn167. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178, no. 3:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Zhu T, Sung ZR. Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiology. 2010;152, no. 2:516–528. doi: 10.1104/pp.109.143495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Hisamatsu T, Goldschmidt EE, Blundell C. The nature of floral signals in Arabidopsis. I. Photosynthesis and far-red photoresponse independently regulate flowering by increasing expression of FLOWERING LOCUS T (FT) Journal of Experimental Botany. 2008;59, no. 14:3811–3832. doi: 10.1093/jxb/ern231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Genetic regulation of time to flower in Arabidopsis thaliana. Annual Review of Plant Biology. 2004;55, no. 1:521–535. doi: 10.1146/annurev.arplant.55.031903.141644. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occuring genetic variation in Arabidopsis thaliana. Annual Review of Plant Biology. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Kurkela S, Borg-Franck M. Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Molecular Biology. 1992;19:689–692. doi: 10.1007/BF00026794. [DOI] [PubMed] [Google Scholar]
- Lacaze X, Hayes PM, Korol A. Genetics of the phenotypic plasticity: QTL analysis in barley, Hordeum vulgare. Heredity. 2009;102:163–173. doi: 10.1038/hdy.2008.76. [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Zhizhong G, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a ring finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Gene & Development. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucyshyn D, Wigge PA. Plant development: PIF4 integrates diverse environmental signals. Current Biology. 2009;19, no. 6:R265–R266. doi: 10.1016/j.cub.2009.01.051. [DOI] [PubMed] [Google Scholar]
- Luesse DR, DeBlasio SL, Hangarter RP. Plastid movement impaired 2, a new gene involved in normal blue-ligth-induced chloroplast movement in Arabidopsis. Plant Physiology. 2006;141:1328–1337. doi: 10.1104/pp.106.080333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. Enhanced fitness conferred by natural occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell. 1999;11(5):949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiology. 2005;137:149–156. doi: 10.1104/pp.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, Coupland G. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. The Plant Cell. 2005;17:2255–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Ukness S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiology. 1995;108, no. 3:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M. Evolution of phenotypic plasticity: Where are we going now? Trends in Ecology & Evolution. 2005;20, no. 9:481–486. doi: 10.1016/j.tree.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Pouteau S, Albertini C. The significance of bolting and floral transitions as indicators of reproductive phase change in Arabidopsis. Journal of Experimental Botany. 2009;60, no. 12:3367–3377. doi: 10.1093/jxb/erp173. [DOI] [PubMed] [Google Scholar]
- Proveniers M, Rutjens B, Brand M, Smeekens S. The Arabidopsis TALE homeobox gene ATH1 controls floral competency through positive regulation of FLC. The Plant Journal. 2007;52, no. 5:899–913. doi: 10.1111/j.1365-313X.2007.03285.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Rafalski JA. Association genetics on crop improvement. Current Opinion in Plant Biology. 2010;13:1–7. doi: 10.1016/j.pbi.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Roux F, Touzet P, Cuguen J, Le Corre V. How to be early flowering: An evolutionary perspective. Trends in Plant Science. 2006;11, no. 8:375–381. doi: 10.1016/j.tplants.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Scheiner SM. Genetics and evolution of phenotypic plasticity. Annual Review of Ecology and Systematics. 1993;24:35–68. [Google Scholar]
- Schwartz C, Balasubramanian S, Warthmann N, Michael TP, Lempe J, Sureshkumar S, Kobayashi Y, Maloof JN, Borevitz JO, Chory J, Weigel D. Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering response of Arabidopsis thaliana. Genetics. 2009;183:723–732. doi: 10.1534/genetics.109.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Loudet O, Durand S, Berard A, Brunel D D, Sennesal FX, Durand-Tardif M, Pelletier G G, Camilleri C. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics. 2008;178, no. 4:2253–2264. doi: 10.1534/genetics.107.083899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, Chory J, Fankhauser C. The Arabidopsis SRR1 gene mediates phyb signaling and is required for normal circadian clock function. Genes and Development. 2003;17:256–268. doi: 10.1101/gad.244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CM, Elston RC. Finding genes underlying human disease. Clinical Genetics. 2009;75:101–106. doi: 10.1111/j.1399-0004.2008.01083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proceedings of the National Academy of Sciences, USA. 2004;101, no. 13:4712–4717. doi: 10.1073/pnas.0306401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase a of gibberellin biosynthesis. The Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires like heterochromatin protein 1. Nature Genetics. 2006;38, no. 6:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- Uga Y, Nonoue Y, Liang Z, Lin H, Yamamoto S, Yamanouchi U, Yano M. Accumulation of additive effects generates a strong photoperiod sensitivity in the extremely late-heading rice cultivar ‘Nona Bokra’. Theoretical and Applied Genetics. 2007;114, no. 8:1457–1466. doi: 10.1007/s00122-007-0534-0. [DOI] [PubMed] [Google Scholar]
- Van Dijk H, Hautekèete NC. Long day plants and the response to global warming: Rapid evolutionary change in day length sensitivity is possible in wild beet. Journal of Evolutionary Biology. 2007;20, no. 1:349–357. doi: 10.1111/j.1420-9101.2006.01192.x. [DOI] [PubMed] [Google Scholar]
- Via S, Gomulkiewicz R, de Jong G, Scheiner SM, Schlichting CD, van Tienderen PH. Adaptive phenotypic plasticity - consensus and controversy. Trends in Ecology & Evolution. 1995;110:212–217. doi: 10.1016/s0169-5347(00)89061-8. [DOI] [PubMed] [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T. AMP1and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development. 2007;134:2561–2567. doi: 10.1242/dev.006759. [DOI] [PubMed] [Google Scholar]
- Wen CK, Chang C. Arabidopsis RGL1 encoldes a negative regulator of gibberellin responses. The Plant Cell. 2002;14:87–100. doi: 10.1105/tpc.010325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Jiang D, Jiang J, He Y. A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. The Plant Journal. 2010;62, no. 4:663–673. doi: 10.1111/j.1365-313X.2010.04182.x. [DOI] [PubMed] [Google Scholar]
- Zhao K, Aranzana M, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M. An Arabidopsis example of association mapping in structured samples. PLoS Genetics. 2007;3, no. 1:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Nordborg M, Marjoram P. Genome-wide association mapping using mixed-models: Application to GAW15 Problem 3. BMC Proceedings. 2007;1(1):S164. doi: 10.1186/1753-6561-1-s1-s164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Quantile-Quantile plot of –log10 p-values in genome-wide scans for the three reproductive transition related traits, from left to right, BT,FT and INT.
Supplementary Figure 2. Manhattan plots of the genome-wide association mapping (Wilcoxon) results for BT, FT and INT.
Supplementary Figure 3. Enrichment ratios as a function of the number of top SNPs chosen in the GWA mapping results using the EMMA method.
Supplementary Figure 4. Enrichment ratios as a function of the number of top SNPs chosen in the GWA mapping results using a Wilcoxon test.
Supplementary Figure 5. Peaks of association in the regions spanning 20kb on each side of the top SNP near TSF (left), GA1 (middle), and at the beginning of chromosome 2 (right).
Supplementary Table 1. Natural variation for bolting time, flowering time and the interval between bolting and flowering scored in nature.