Summary
The inner ear of mammals uses neurosensory cells derived from the embryonic ear for mechanoelectric transduction of vestibular and auditory stimuli (the hair cells) and conducts this information to the brain via sensory neurons. As with most other neurons of mammals, lost hair cells and sensory neurons are not spontaneously replaced and result instead in age-dependent progressive hearing loss. We review the molecular basis of neurosensory development in the mouse ear to provide a blueprint for possible enhancement of therapeutically useful transformation of stem cells into lost neurosensory cells. We identify several readily available adult sources of stem cells that express, like the ectoderm-derived ear, genes known to be essential for ear development. Use of these stem cells combined with molecular insights into neurosensory cell specification and proliferation regulation of the ear, might allow for neurosensory regeneration of mammalian ears in the near future.
Introduction
Development of the vertebrate ear is a coordinated molecular transformation of a set of epidermal cells (the otic placode) into the fully developed ear with its neurosensory component, necessary for signal extraction and transmission, and the non-sensory component, forming the labyrinth necessary for directing sensory stimuli to specific sensory epithelia (Fig. 1). Three developmental steps ensure that (1) the ectoderm is transformed to otic ectoderm, including neurosensory precursor cells, (2) neurosensory precursor cells generate neurons, and (3) sensor precursor cells form hair cells and supporting cells in the designated area of sensory epithelia (Fig. 1). As with other developing systems, differentiation of the epidermal cells into the four major cell types of the ear (sensory neurons, hair cells, supporting cells and non-sensory epithelial cells) occurs through molecular fate specification followed by clonal expansion of committed precursors to produce the final number of a specific cell type in embryos. These neurosensory cells have a limited life span that is further truncated by numerous environmental insults (loud sound, ototoxic substances such as cysplatin or aminoglycoside antibiotics) and genetic predisposition (numerous genes related to hearing loss). Combined with the increased longevity of humans, genetic predisposition and cumulative insults lead to an increasing likelihood of neurosensory hearing loss with age, thus depriving half of people age 70 and older from one of the most important aspect of communication as well as negatively affecting their sense of balance.
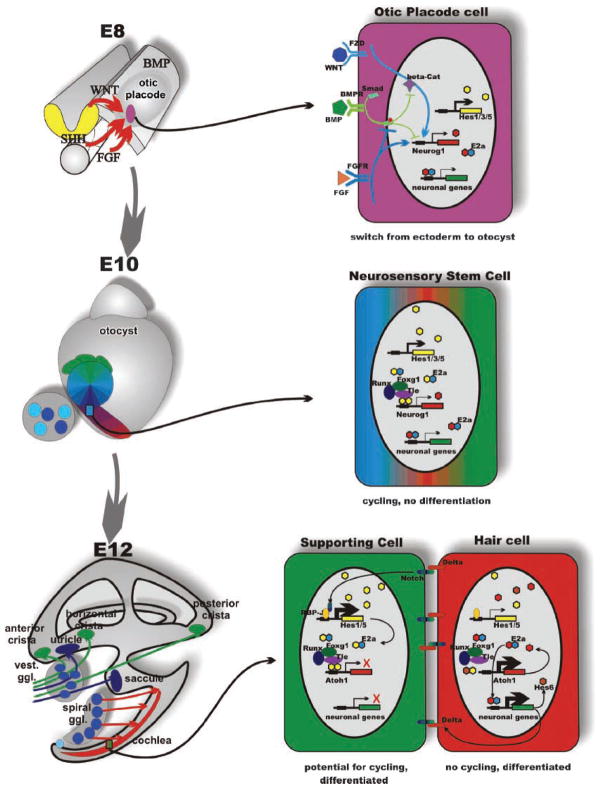
Figure 1.
Organ, cell and molecular interactions in ear development. The morphogenesis (left) and some molecular interactions underlying proliferation and cell fate decision (right) are depicted in this scheme. Morphogenesis transforms a small patch of ectoderm between embryonic days 8 and 12 into a complex labyrinth of ducts and recesses that harbors the six sensory epithelia of the mammalian ear in strategic positions for extraction of epithelia-specific energy. Delamination of sensory neurons generates the vestibular and cochlear sensory neurons that connect specific sensory epithelia of the ear to specific targets in the hindbrain. One of the earliest steps in this process is the selection of otic placode cells through the interaction of several diffusible factors; in particular, FGF and WNT signaling upregulates both inhibitory and activating bHLH genes to switch the cell fate through downregulation of BMP signaling, specifying the position and size of the otic placode (top right). These stem cells will, through the interaction of activator- and inhibitor-type bHLH genes remain in cycling phase without differentiation resulting in clonal expansion. As cells progress through the cycles, they will change their fate determination, giving rise to neurosensory stem cells (middle right) that form by asymmetric divisions all sensory neurons of the ear. Some neurosensory stem cells as well as independently arising cells of the otic placode turn into sensory epithelia precursor cells (SNP). These cells will give rise by asymmetric divisions to hair cells and supporting cells (bottom right). Exit from the cell cycle, combined with proper cell fate specification to, eg hair cell and supporting cell, will be mediated in part by the NOTCH-reinforced switch to either explosive upregulation of proneuronal bHLH genes (Atoh1 in the case of hair cells) or of inhibitory bHLH genes (such as Hes1 or Hes5) by the γ-secretase-cleaved Notch fragment that binds to RBPSUH (formerly Rbp-J). The action of HES homodimers on N-boxes to turn on proneuronal genes is enhanced through interaction with the TLE, RUNX, FOXG and genes. Consequently, eliminating for example Foxg1 results in diminished efficacy of HES signaling resulting in premature cell cycle exit and differentiation. Shortly after E14, all proliferative activity in the PNP progenitors stops and no new sensory neurons or hair cells will form. Modified after Refs 37,38.
Much like with the adult human brain,(1) there is only limited evidence for the presence of neurosensory stem cells in the mammalian ear that seem to be able to proliferate only under certain circumstances in vitro.(2,3) Consequently, loss of any differentiated neurosensory cell will potentially diminish hearing. In contrast to other vertebrates (like bony fish or chickens), there is no evidence for spontaneous regeneration of lost neurosensory cells in the mammalian cochlea in vivo. Because of the difficulties in accessing these stem cells in the adult human ear without disrupting the very organ that requires regeneration, other sources of stem cells and strategies are being explored that may ultimately provide replacements for lost neurosensory cells or restore hearing:
An already existing therapy is to use remaining sensory neurons in combination with a cochlear implant (an electric device that transforms sound into electric stimuli) to bypass the missing hair cells by directly stimulating nerve fibers, bringing sound information via the sensory neurons to the brain. The viability of this approach rests on the long-term survival of sensory neurons that depend on neurotrophic support from the lost hair cells and the dedifferentiating supporting cells for their survival.(4,5) To maximize the viability of sensory neurons, several strategies are being explored using neurotrophin infusions.(6–8)
Attempts are being made to understand the molecular mechanism that shuts down neurosensory proliferation in the ear through regulation of cyclin-dependent kinase inhibitor expression(9) in analogy to other systems.(10) Conceptually, it seems possible to translate those insights directly into reactivation of the dormant replacement capacity of mammals, comparable to the injury-induced regeneration of chicken hair cells. Recent work has demonstrated that cell cycle reentry is possible in neonatal mammals(11) but manipulation of this pathway is not without risks(9,12) requiring a more sophisticated manipulation of this pathway than simply knocking out cyclin-dependent kinase inhibitor genes.
Proliferation of postmitotic neurosensory cells can be forced through targeted deletion of S-phase entry control genes such as the retinoblastoma gene.(13,14) While such approaches lead to the transient formation of more hair cells and can potentially be initiated via siRNA therapy, such cells ultimately die necessitating further refinement of this approach before it can be therapeutically useful.
Transdifferentiation of the supporting cells of the sensory epithelium into hair cells can be enforced through overexpression of regulatory genes.(15) The problem with such a gene therapy approach is that it will deplete the existing supporting cells, thus leaving the sensory epithelia in an unusual organization with limited functionality of the organ of Corti which, in part, depends on supporting cells.(16)
Stem cells of various tissues are being investigated and some have been successfully incorporated into the developing chicken ear, providing proof of principle for a stem cell approach.(17,18) However, only a limited set of stem cell sources have been investigated. Thus far, the easily accessible stem cells derived from hair follicles(19–22) have not been explored for ear regeneration.
The purpose of this review is to analyze molecular steps that specify the cell fate of neurosensory hair cells out of epidermal cells and that regulate the clonal expansion of those precursors and their differentiation into sensory neurons and hair cells. After presenting these developmental steps, we will discuss the potential use of skin-derived stem cells to generate neurosensory precursors useful for ear implantation.
Turning embryonic ectoderm cells into otic neurosensory cells: the molecular basis for otic neurosensory induction
Induction of the ear requires both mesodermal and neuroectodermal signals.(23) This basic decision is essentially identical to the induction of the neural plate(24,25) and olfactory system.(26) Similarly to these neural inductions, ear induction is based on FGFR signaling, possibly combined with inhibition of BMP signaling (Fig. 1a). Molecularly, these inductions require diffusible signals that cause graded responses in the target cells. Four such diffusible signals have been characterized in mammalian ear development: SHH from the floor plate and notochord,(27) FGF8, FGF10 and FGF3 from mesoderm and neuroectoderm,(28) WNTs from the hindbrain(29,30) and BMP4 from general ectoderm as well as from the ear.(31) The combined action of these signals change the fate of ectodermal cells to acquire an otic placode phenotype instead (Fig. 1a). Within the otic placode, the acquisition of a neurosensory phenotype is consolidated with the upregulation of the proneuronal gene neurogenin 1 (Neurog1). Upregulation of Neurog1 was detected as early as E8.75 in the mouse in a few cells(32) and is thus not unlike the sensory organ precursor cell known to initiate formation of mechanosensors in insects.(33–36) In contrast to most insect mechanosensory organs, the mammalian ear undergoes many more cell cycles to expand first the precursor population followed by a coordinated cell cycle exit of, in order, sensory neurons, hair cells and supporting cells.(37) The adult mouse ear contains approximately 10,000 hair cells and 11,000 sensory neurons. Between embryonic day 8.75 (first expression of the bHLH gene Neurog1) and E13.75 (when all cochlear and most vestibular hair cells and neurons have exited the cell cycle) ear precursors will undergo approximately 16 cell cycles of about 8.5 hours each.(37) Assuming only symmetric divisions, only two initial cells would be needed to generate 32,000 neurosensory cells of the adult mouse ear in only 15 rounds of division. Selecting the right number of cells that express Neurog1 is therefore a crucial final step of otic placode induction.
Neurog1 is not only one of the earliest genes to identify cells of the otic placode but it also has an essential functional role in ear development: Neurog1 is necessary for all sensory neuron formation.(32) However, Neurog1 also affects other aspects of ear development, including development of sensory epithelia and hair cells.(38,39) Misexpression of Neurog1 in frog skin demonstrates that it is not only necessary but also sufficient to induce neuronal transformation of epithelial cells.(40) Understanding otic induction requires therefore a mechanistic understanding of how the four above outlined diffusible factors (SHH, WNTs, BMPs and FGFs) interact at a cellular level to change ectodermal cells to otic cells and eventually to a neurosensory precursor fate by upregulating Neurog1. The ubiquitous use of these factors in neuronal and non-neuronal systems alike suggests that they are necessary but not sufficient to achieve this epithelial transformation. Other transcription factors possibly important for the epithelial–neurosensory transition are also early expressed in the placode such as Gata3,(41) Pax2/8,(42,43) Tbx1,(44) Foxg1,(45,46) Foxi1,(47) Eya1/Six1(48) and Oct4.(49) In particular, the unique overlapping expression of Pax2/8, Gata3, Foxg1, Foxi and Eya1/Six1 may provide a necessary context for inner ear neurosensory development that is dramatically altered in their absence.
How do all these factors interact with each other to achieve epithelial-to-otic transformation? A central cellular event in many cells to induce cell fate changes is regulation of transcription factors via modifying BMP signaling. BMPs signal through dimerized BMP receptors to phosphorylate SMADs(50) which then enter the nucleus to regulate over 500 genes. Entry of SMADs to the nucleus and binding to promoters is tightly regulated by numerous interactions with other signaling pathways, notably the FGF- and EGF-related receptor tyrosine kinase (RTK) signaling pathways (Fig. 1). Activation of the RTK pathway will block SMAD entry to the nucleus.(50) GATA3 can form complexes with SMADS and thereby change binding specificity.(51) Combined with its role in hair follicle stem cells,(20) the early expression and massive reduction of ear development in Gata3 null mice(41) shows that this gene plays an important role in setting up the proliferation capacity of the otocyst through interactions with SMADs(51) and FGFs.(52) Some evidence for PAX signaling affecting SMADS exists for thyroid development,(53) but this has not been demonstrated for the ear. However, an absence of sensory neurons has been claimed for Pax2 null mice,(42) a claim that needs to be reexamined with more sophisticated techniques. FOXG1 has recently been shown to interfere with the SMAD–FOXO complex and thus can alter SMAD-mediated gene regulation(54) and neurosensory development is altered in Foxg1 null mice.(46) WNT signaling through β-catenin is known to act directly on SMAD-mediated gene activation,(50) but other interactions of WNTs and BMPs are known and which of these pathways is active in the ear requires further research. Wnt signaling clearly effects otic placode formation(30) and later ear development,(29) but the effects of β-catenin on SMAD signaling have not been investigated in the ear. Oct4 null zebrafish show no expression of Neurog1 in the otic placode, suggesting that OCT4 regulates Neurog1.(49) Such an epistatic effect of Oct4 has recently been demonstrated in mammalian stem cells(55) but has net yet been shown for the mammalian ear. In the brain, NEUROG1 inhibits SMAD1-mediated signaling by sequestering the SMAD1 complex away from glia-specific promoters, thereby enhancing a neuronal phenotype.(50) Thus, NEU-ROG1, once expressed, could further downregulate SMAD signaling in the otic placode, enhancing the commitment toward neuronal development. Tbx1 is known to suppress Neurog1(44) and thus would remove a proneural signal and reinstall unmitigated SMAD1 signaling, thereby converting neurosensory fate back to epithelial fate. Finally, SHH is known to upregulate bHLH genes in somites and there is neither Neurog1 upregulation nor sensory neuron formation in Shh null mice.(27) Thus, SHH could affect SMAD1 phosphorylation indirectly through expression of Neurog1, possibly allowing Neurog1 expression only in cells with a specific concentration of BMP and SHH signaling, like in the spinal cord.(56) Indeed, recent in vitro data on embryonic stem cells show that treatment with SHH can bias toward hair cell differentiation, albeit at a very low yield.(57) Whether SHH’s effects in vitro are accomplished via regulation of SMAD signaling through Neurog1 expression requires further research. In addition, for a therapeutically useful yield of cells, the propensity for neurosensory differentiation must be increased.
Taken together, these data suggest that several otic transcription factors expressed early in development and diffusible morphogens co-operate to modify BMP–SMAD signaling thereby altering epithelial fate toward neurosensory otic placode fate. While SMADs undoubtedly play a role in ear development, exactly when and where Smad’s are expressed and phosphorylated in mammalian ear development requires further analysis. Presently we only know that, in zebrafish, Smad1 is expressed in the sensory neurons of the ear(58) consistent with our hypothesis that SMAD regulation by various means may be a crucial first step in ectodermal–otic transition. It needs to be noted that most of the molecules thus far identified are used in many other developing systems, suggesting that specific otic identification is achieved through a unique combination of genes and not through a single gene unique to the otic placode. Independent of this uncertainty, the final step in otic neurosensory commitment is the upregulation of Neurog1, consolidating the switch from epidermal to pro-neurosensory fate and initiating pro-neurosensory clonal expansion. Therefore, we will next review the molecular basis that makes this clonal expansion possible and turns a small set of otic placode cells into the several thousand neurosensory cells of the adult ear.
The molecular basis of inner ear neurosensory cell generation
In this section, we will explore the developmental pathways utilized in the formation of neurons of the ear. While the presence of Neurog1-expressing precursors is obvious at E8.75,(32) neither the entire fate of these proneuronal precursors nor the distribution of prosensory precursors is fully known. The first identification of sensory patches that will give rise to hair cells and supporting cells is only possible around E10.5. At this stage or later, several genes highlight to various degrees those prosensory areas, notably the neurotrophins BDNF and Ntf3,(59,60) Bmp4 and Lnfg,(31) Sox2,(61) Islet1(62) and Fgf10.(63) Several of these genes are expressed both in the otocyst wall in likely sensory epithelial precursors as well as in delaminated, proliferating neuronal precursors,(59,62,63) suggesting a possible common precursor for both sensory epithelia and neurons.
This common expression in the otocyst wall and delaminated neuronal precursors is also true for Neurod1,(64,65) a bHLH gene that is regulated in the ear by Neurog1.(32) However, whereas Neurog1 null mice have a severe reduction in hair cells, notably in the saccule and cochlea,(38) there is only a limited shortening of the cochlea in Neurod1 null mice.(66) This suggests that some precursors that express Neurog1 are also forming hair cells and supporting cells of sensory epithelia whereas precursors that express Neurod1 are already committed to the neuronal lineage. Recently, it was shown that some sensory precursors switch their fate in the absence of Neurog1 and differentiate into hair cells.(39) In addition, using sensitive markers, it was shown that some sensory neurons express the otherwise hair-cell-specific bHLH gene Atoh1, a gene essential for hair cell differentiation.(67,68) These indirect suggestions for a clonal relationship between some sensory neurons and hair cells was confirmed with lineage tracing in chicken.(69) Combined, these data suggest that at E10.5 the neurosensory precursors may be composed of three populations: (1) neuronal precursors that form only neurons, (2) neurosensory precursors that form only hair cells (and supporting cells) and (3) precursors that form both neurons and hair cells (Fig. 2). How the selection of these precursors and the determination of their relative size are regulated and whether or not there is a coordinated transition of one precursor into another as in brain development(70) remains unclear. But the existence of a population that can generate both hair cells and neurons from a single line of clonally related cells has therapeutic potential: it would allow for the transformation of neuronal stem cells that give rise to both neurons and hair cells out of the same stem cell. Indeed, recent in vitro data suggest that the yield of hair cells out of bone marrow stem cells can be enhanced when stem cells are selected that express neuronal markers before they are switched to a hair cell differentiation pathway (Heller et al, unpublished data).
Figure 2.
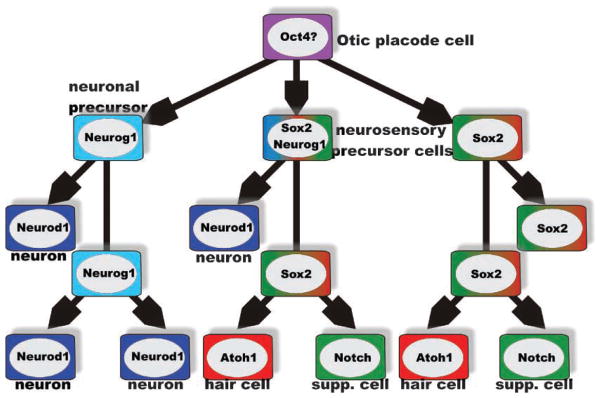
Cell-type-specific and overlapping precursors. Analysis of several null mutations suggest that there is an initial formation of two, partially overlapping, precursor populations, a neuronal precursor characterized by Neurog1 expression and a neurosensory precursor, characterized by Sox2 expression. The 40–80% reduction of hair cell and supporting cell formation in Neurog1 null mice suggests that the size of the common neuronal/neurosensory precursor population varies in different sensory epithelia. The later-expressed bHLH gene Neurod1 does not show this massive effect on hair cells and appears to be exclusively expressed in differentiating neurons. Absence of hair cell differentiation in Sox2 and Atoh1 null mice suggests that these genes are essential for hair cell formation, no matter what origin. Supporting cells depend on the hair-cell-mediated upregulation of Notch (and Hes) for their differentiation and will turn into hair cells in the absence of proper Notch/Hes signaling. Modified after Refs 39,49,61,64,120.
Still, the question remains: what is the function of two or more, instead of one bHLH gene in the neuronal development of the ear? Our understanding of the development of the olfactory system provides clues to begin to answer this question. In the olfactory system, transient amplifying precursors are initially specified by Mash1. The Mash1-expressing precursor gives rise to a transient amplifying precursor population, the immediate neuronal precursor (INP), which expresses Neurog1. INP cells divide, exit the cell cycle accompanied by Neurod1 expression and differentiate into olfactory receptor neurons.(71,72) Both Fgfs and Bmps play a role in specifying the transition from one cell type to the next and hence the degree of clonal expansion(73,74) and allocation to various clones giving rise to olfactory neurons and cells of the olfactory system.(72) As in muscle cell proliferation, an antagonistic interaction between GDF11 and follistatin determine the expression level of the cyclin-dependent kinase inhibitor 1b (Cdkn1b; formerly p27 kip) and thus determine the cell cycle exit.(75)
Comparable to the olfactory system, the ear shows various progenitor populations able to produce either hair cells, supporting cells, and even sensory neurons or hair cells and supporting cells.(69) Cell cycle exit in these progenitors is regulated by cyclin-dependent kinase inhibitors.(9) However, the regulation of the cyclin-dependent kinase inhibitors by GDF 11/follistatin remains to be shown for the ear. Nevertheless, it appears that, in neurosensory development of the ear and olfactory epithelium, we can distinguish a phase of early clonal expansion with limited, if any expression of cyclin-dependent kinase inhibitors followed by a phase of progressive upregulation of these inhibitors to tightly regulate the final number of neurosensory cells.(9,11) The molecular basis of this final phase of progenitor cell cycle regulation and differentiation into distinct cell types is well understood in the ear.(11,13,39,76,77) We will therefore concentrate next on the molecular basis of clonal expansion of neurosensory precursors to provide the right number that can then be regulated to divide and terminally differentiate through these molecularly known pathways.
Molecular basis of otic neuronal stem cell maintenance and expansion: the ear relates closely to other systems?
Recent years have revealed the molecular basis of stem cells in general, which involves the genes Oct4, Nanog and Sox2,(55) and of neuronal stem cells in particular, involving certain bHLH genes.(78) Not surprisingly, WNT and SHH signals seem to interact with bHLH genes to ensure clonal expansion of neuronal stem cells.(79) Not all the details are clear yet for the ear, but several important aspects are known that suggest a rough parallelism to this general principle with ear-specific molecular players. SHH and WNT1/3A are diffusible signals that influence ear histogenesis and morphogenesis from sources outside the ear.(27,29,30) In addition, FGF’s likely signaling through FGFR2B(80) affect morphogenesis and neurosensory formation.(52,63,81,82) How signals generated by these diffusible factors combine with local signals such as EYA1(48) to maintain and alter bHLH-gene-mediated neuronal progenitor specification and proliferation is unclear. Based on the limited data and expanding general principles validated in other systems, the following tentative conclusions can be drawn: in general, neuronal stem cells express both glial and neuronal markers such as GFAP and Nestin(79) but also the activator and repressor-type bHLH genes.(78) Eliminating the repressor-type bHLH gene signaling initiates premature neuronal differentiation combined with limited clonal expansion.(78) This can either be achieved by eliminating Hes genes, Notch genes or the intracellular partners that regulate Hes expression (RBPSUH, formerly RBP-J), or by changing the ability of HES to form homodimers that bind to N-boxes using the WRPW domain (Figs 1,3). An excellent example of the latter is the reduced clonal expansion and premature neuronal differentiation in the forebrain of Foxg1 null mice,(83) in part mediated by alteration in DNA binding of HES homodimers interacting with TLE and RUNX.(84)
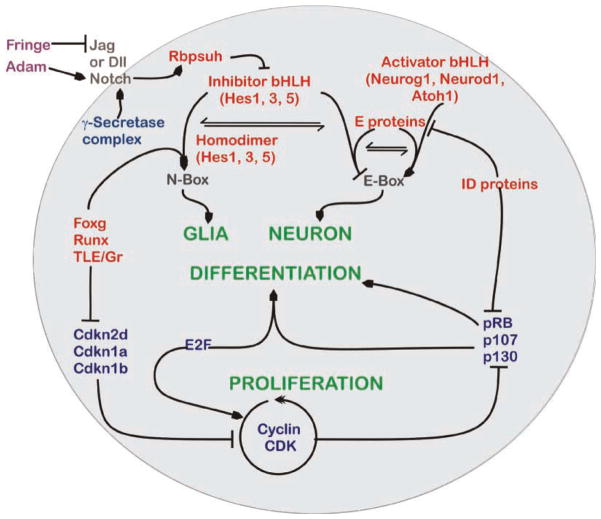
Figure 3.
Signaling pathways for inner ear proliferation and differentiation. This schematic diagram represents an overview of the known and presumed interactive pathways for proliferation and differentiation of the neurosensory cells in the inner ear. Signaling of the membrane-bound (brown) Notch receptors by binding to their ligands, Delta and Jagged, can be influenced by the extracellular (purple) Fringe and ADAM enzymes. Fringe inhibits (blocked line) the Notch binding of Jagged, while Adam cleaves the Notch receptor to potentate its activation (lined arrow). The cleavage of intercellular domain fragment of Notch is done by the cytoplasmic (dark blue) γ-secretase complex which then activates the nuclear protein (red) RBPSUH. Inactivated RBPSUH blocks transcription of the Hes genes whereas activation enhances transcription. Homodimers of HES proteins can bind to N-boxes to initiate differentiation (green) of glial precursors. N-box binding of HES homodimers is regulated further by a FOXG, RUNX and TLE promoter complex. Heterodimers between HES and E proteins bind and competitively block usage of E-box-binding sites. Activation of E-box promoter sequences is through the combined E-protein and the activator bHLH heterodimers and this permits neuronal differentiation. To do this, the activator bHLH proteins compete with HES proteins for the E-protein-binding partners. E proteins can also be inactivated from DNA binding through interaction with the inhibitor of DNA-binding (ID) proteins, which also suppress the cell cycle (blue) retinoblastoma isoforms. The pRB isoforms alone or in combination E2F proteins cause cell differentiation. Cell proliferation (green) is mediated through the proteins of cyclin CDK pathway that phosphorylate Rb to allow E2F proteins to initiate the S-phase entry. The cyclin CDK proteins can also inhibit differentiation via pRB phosphorylation, whereas cyclin-dependent kinase inhibitors (Cdkn) prevent proliferation. Expression of the CDKNs is blocked by the FOXG, RUNX and TLE complex, allowing differentiation of glia cells through enhanced action of HES homodimers on the N-Box. Modified after Refs 11,46,78,85.
Neurog1 drives the upregulation of several genes relevant for the maintenance of neuronal stem cells. The expression of the NOTCH ligand DELTA 1 is delayed in Neurog1 null mice, showing that Neurog1 is epistatic to DELTA 1.(32) Consistent with other developing mammalian neuronal systems,(78) initial upregulation of Neurog1 is not ubiquitous but occurs in a few cells only. Nevertheless, eliminating RBPSUH and thus the NOTCH signaling pathway (Figs. 1,3) results in expansion of Neurog1-expressing areas of the ear.(32) These data show that NEUROG1 signaling affects Notch signaling and may indeed be effective at this early time. Despite the known presence of Notch and several ligands as early as E8.5(85,86) and the known effects of deletions of Notch ligands on ear development,(87–89) there is no direct evidence suggesting expression of any Hes genes in the ear prior to E12.5.(85) Given that activated NOTCH signals through de-repression of Rbpsuh and thus upregulation of Hes1 and Hes5, the expression data are bound to be incomplete and further studies using more sensitive techniques such as green-fluorescent-protein-expressing reporter systems(90) are needed to reveal the spatial and temporal pattern of Hes distribution in the developing otocyst. Thus, at the moment, the role of Hes signaling in neuronal and early neurosensory stem cells of the ear remains unclear (Fig. 1).
Altering the balance between Hes and activator-type bHLH genes determines how long a stem cell cycles and whether they differentiate toward a neuronal or a glial cell type.(78) Eliminating all activator-type bHLH genes can result in phenotypic switch to a glial phenotype.(72) Such switches in phenotype combined with truncation of later formed cells such as hair cells or supporting cells have been described in Neurog1 null mice.(38,39) Most interestingly, Neurod1, a bHLH gene that is immediately downstream of Neurog1 and depends on Neurog1 for early expression,(32) shows a profound upregulation in hair cells that exit the cell cycle prematurely in Neurog1 null mice.(39) Likewise, altering NOTCH signaling, either at the level of ligand/receptor,(87,88) the intracellular effectors Hes1 and Hes5,(91) or a co-factor for binding to the N-box,(46) results in aberrations of hair cell organization. Combined, these data show that proper bHLH signaling is essential for normal neurosensory development of the ear and requires the interaction of both activator and inhibitor-type bHLH genes for transit amplification of precursors. The ear is in this respect essentially identical to other developing neuronal systems,(72,78,79) although it uses a unique combination of players.
Forming the right number of hair cells: complex regulations of a simple outcome
In addition to the above-outlined molecular interactions that result in the formation of sensory neurons and neurosensory precursors, a partially overlapping set of genes regulates the neurosensory and supporting cellular components of the inner ear sensory epithelia development (Fig. 1c). These regulations involve the bHLH network of the neuronal activator gene Atoh1,(39,67) and the repressor genes, Hes1 and Hes5,(91,92) in combination with Notch1, and the delta and jagged/serrate ligands, Dll1, Jag1 and Jag2.(85,88) These two networks are directly linked (Fig. 3) through the expression regulation of and interactions with the Hes genes.(78)
The bHLH network functions through the DNA targeting and binding affinities of a combinatorial complex of proteins(78) that involve bHLH dimers,(93) transducin-like enhancer of split (Tle, groucho in fly), runt-related transcription factor (Runx), and forkhead box G1 protein (Foxg1).(84,94–96) The TLE protein is the central component with binding sites for HES, runt and forkhead proteins and forms the repressor complex that, in general, prevents neurogenesis (Figs 1,3). HESs also exert an additional effect by competing with the activator bHLH proteins for the ubiquitously expressed class I bHLH activator-binding partner (E protein), Tcfe2a (Figs 1,3). TCFE2A functions by facilitating the formation of heterodimers with activator-type bHLH genes (NEUROG1, ATOH1 and certain HESs) that permit binding to the E-box (5-CANNTG-3). Homodimers of activator bHLH proteins either have low E-box-binding affinities or are inactive.(97) HES homodimers bind N-box response elements (5-CCGGAA-3). HES-mediated repression are largely through the Orange and WRPW protein domains. The Orange domain confers specificity for homodimerization among the HES family members and the WRPW domain interacts with the co-repressor TLE protein for enhanced binding to N-boxes. A second class of repressor bHLHs are represented by the inhibitor of DNA-binding (Id) bHLH genes that function as a dominant-negative protein due to the absence of the DNA-binding motif.(77) Strength of activation or repression can be further fine-tuned by qualitative and quantitative ratios of these proteins and paralogue usage.(98–100) HES6 differs in that it can function as a positive- feedback loop in neurogenesis by forming heterodimers with other HESs, inhibiting their repressor activity.(101–103)
This intracellular signaling network is tied into an inter-cellular signaling network that refines fate assignment of hair cells and supporting cells in the sensory epithelia through NOTCH signaling (Figs 1,3). NOTCH signaling contributes to proliferation, apoptosis, stem cell self- renewal and regulation via lateral inhibition between neighboring cells.(85,104) In vertebrates, Notch receptors all share similar functional domains, where the extracellular domain has epidermal growth factor and Lin-Notch repeats (LNR) and the intracellular domain has a RBPSUH-associated motif (RAM). Homomerical oligomerization of the NOTCH receptors and subsequent differential proteolytic cleavage of the intracellular domain (ICD) are modulated by two classes of ligands that induce (Serrate/Jagged) or inhibit (Delta) signaling. The presence of extracellular Fringe modifies NOTCH to signal only with Delta proteins, whereas unmodified NOTCH is responsive to Jagged.(105) Upon binding a ligand, intracellular cleavage by a variable γ-secretase complex containing presinilin related molecules leads to a NOTCH fragment that interacts with RBPSUH to regulate Hes expression (Figs 1,3).
Examination of the inner ear phenotype of mutants for many of these pathway component genes reveals several levels of severity. The least severe are those that alter the cell numbers and rows in the organ of Corti. These include Cdkn1a ( formerly p21), Cdkn2d (formerly p19Ink4d ), Hes6, Hes1, Hes5, Notch1 and Jag2 (9,86,91,101,106,107) with more severe changes in the organ of Corti being observed in the Neurog1, Foxg1, Jag1 and Cdkn1b (formerly p27 ) mutants.(38,39,46,87,108) In contrast to the limited addition of hair cells in Cdkn null mice,(9,11,106) conditional null of the Rb1 gene causes a preferential expansion of the hair cell population leading to cochlear tumors.(13,14)
Beyond these readily understandable effects on inner ear differentiation are less obvious effects that require a deeper insight into the molecular interactions to appreciate them. Some of these effects require the additional interaction of activator-type bHLH genes, more specifically of Neurog1 and Atoh1. In Atoh1-deficient mice, only the differentiation of hair cells is affected with no effect on morphogenesis or formation of undifferentiated precursors in specific sensory epithelia (Fig. 4). In contrast, in the Neurog1 null mice, all inner ear ganglion neurons are absent(13,39) and there are morphogenetic effects such as a reduction of hair cells by 40–80%, depending on the sensory epithelium (Fig. 4). This suggests that the proliferative capacity of neurosensory precursors is also being affected in these activator bHLH-deficient mice. Recently, an interactive network of activator- and inhibitor-type bHLH genes has been described that tightly regulates the proliferation and differentiation of retinal ganglion cells.(109) Specifically, this interaction is mediated with paralogs of two inner ear bHLH genes, Neurog2 and Atoh7 (formerly Math5). It appears that Atoh7 is more profoundly affected by high levels of Hes, possibly through an inhibitory action of Hes homodimers on N-boxes in its promoter region (Fig. 5). In contrast, Neurog2 is compatible with high levels of Hes and promotes continuous cycling of the precursors. Through as yet unclear extracellular signals, possibly mediated by the Delta–Notch system, Hes expression is downregulated, thereby decreasing inhibition of Atoh7 expression. Once ATOH7 protein has reached a critical level, most E proteins will form heterodimers with ATOH7, reducing NEUROG2/E-protein heterodimer signaling. These phases were shown to neatly correlate with clonal expansion (high levels of Neurog2 and Hes), cell cycle exit (equal level of Neurog2 and Atoh7, reduction in Hes) and differentiation (reduced presence of Hes and Neurog2, high expression of Atoh7) of retinal ganglion cells (Fig. 5). While impressive in the technical achievements of single cell quantitative PCR, even this work leaves open the questions open of protein–protein interactions and the half-life of bHLH proteins. Nevertheless, it stresses that technical advances are needed to close the gap between the most-sensitive tissue-based detection systems and the more-sensitive non-tissue-based detection.
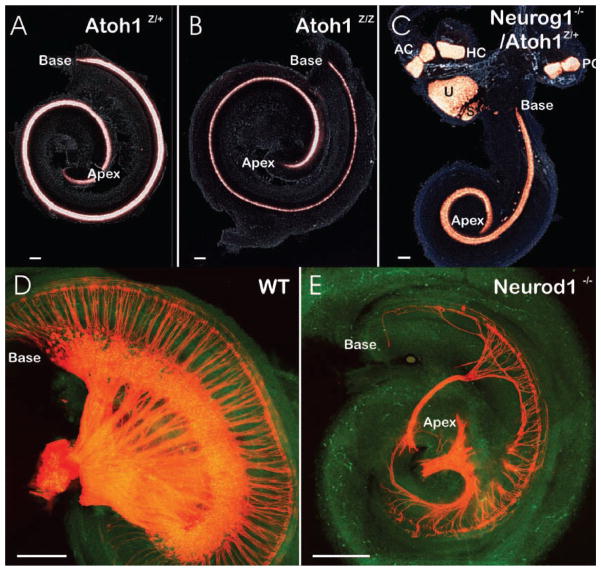
Figure 4.
Examples of gene effects on histogenesis and morphogenesis. A,B,D,E: Flat-mounted cochlea or C: entire ears show the effects of targeted deletion of an activator-type bHLH gene (Atoh1, B; Neurog1, C; Neurod1, E) on the presence of hair cells (revealed by Atoh1–lac Z expression in A–C) or innervation (revealed by lipophilic dye tracing in D,E). Note that both the distribution of Atoh1–lac Z-positive cells as well as the overall length of the cochlea (base and apex are indicated) show little difference in Atoh1–lac Z heterozygote and null mutants, despite the fact that no hair cells differentiate in Atoh1 null mice. This suggests that the late upregulation of a bHLH gene in cells destined to exit the cell cycle is of little consequence for morphogenesis and cellular patterning in the ear. In contrast, earlier upregulated bHLH genes such as Neurog1 (C) or Neurod1 (E) have a more profound morphogenetic effect such as shortening of the cochlea (C,E) or almost complete loss of sensory epithelia (saccule in E). Additional effects are displaced development of some hair cells outside the typical sensory epithelia (C) or loss of a large fraction of sensory neurons combined with an alteration in the pattern of innervation. Modified after Refs 39,64,68 AC, anterior crista; HC, horizontal crista; PC, posterior crista, S, saccule; U, utricle. Bar indicates 100 μm.
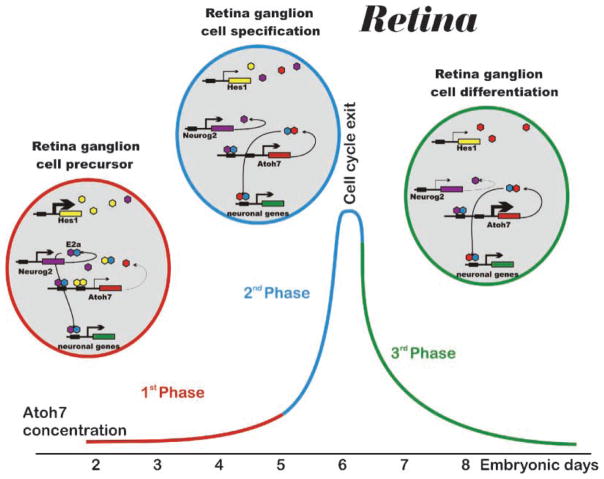
Figure 5.
bHLH gene interactions in retinal ganglion cell specification. The most-detailed single-cell quantitative PCR analysis shows that relative concentrations of bHLH transcripts vary systematically during chicken retina ganglion cell formation. In the first phase (red line), Hes1 transcript exceeds that of Neurog2 and very much that of Atoh7. This dominance of inhibitory bHLH gene expression will result in homodimers on N-boxes (yellow hexagons) as well as few heterodimers of Neurog2 with E2a on E-boxes (lilac/blue hexagons). In phase 2 (blue lines) Hes1 is downregulated allowing Atoh7 transcript to become as prominent as Neurog2 and to form heterodimers with E2a proteins to bind to specific E-boxes (red and blue hexagons). In the third phase (green line) Atoh7 is further upregulated to drive ganglion cell differentiation as well as preventing the developing neuron from reentering the cell cycle. Modified after Refs 109.
A similar regulation is conceivable in the ear, involving instead Aoth1 and Neurog1 and may also play a role in neuronal differentiation of the ear (Neurog1 and Neurod1) and the olfactory system (Mash1, Neurog1, Neurod1). In this context, it is important not only that Neurog1 absence has been shown to reduce formation of hair cells and also to result in loss of sensory neurons, but also that Atoh1 upregulation was recently shown much earlier in the ear using more-sensitive detection systems and some sensory neurons were found to express Atoh1.(39) These dataQ1 support the idea that at least some hair cells are clonally related to sensory neurons and this precursor population may be larger in the mammalian ear compared to the limited clonal relationship thus far found in chicken development.(69) Consistent with the comparatively late upregulation of CDK inhibitors in the ear,(9,106) these data suggest that the initial clonal expansion of neurosensory precursors in the ear may be predominantly regulated via bHLH gene interactions and their effect on cell cycle progression with only limited input from the Delta-Notch system (Figs 3,5). How genes that define sensory epithelia and may be upstream to Atoh1 regulation such as Sox2(61) affect this intracellular signaling remains at the moment unclear as Sox2 might not be the only factor driving upregulation of Atoh1. In summary, these data show a complex intracellular signaling for neurosensory precursor regulation that requires transition between several activator-type bHLH genes that provide the molecular basis for transit-amplification and precursor specification. Some extracellular signals that regulate the expression of activator-type bHLH genes are known in the ear (Sox2, Oct4, Tbx1) and the role of the Delta/Notch regulation in refining hair cell and supporting cell development is becoming clear. How regulation of Cdk inhibitors ultimately causes the irreversible arrest of cell cycle re-entry remains as unclear in the ear(11) as in other non-proliferating systems.(10)
Using easily accessible skin and olfactory precursors to regenerate hair cells of the ear: are we there already?
In this essay, we have outlined our current understanding of the molecular basis for ear neurosensory specification and proliferation. Clearly, this involves the transformation of ectodermal cells into neurosensory cells through the selective expression of a reasonably well understood sequence of gene activations. Interestingly, several of the genes found to be important in ear neurosensory development are also important for skin stem cells. For example, Gata3 is needed (together with other genes) for hair follicle stem cell determination.(20) GATA3 acts with Lef-1/Wnts to define the inner root sheath versus the hair shaft cell fate decision in hair follicle morphogenesis.(20) GATA3 is essential for early ear development and is expressed already in the invaginating ectodermal placode.(41) Thus, isolating hair follicle precursors from skin would provide progenitor cells that have already one of the crucial genes for ear formation expressed. Expression of other crucial genes such as Neurog1, Foxg1, Foxi1 and Pax2/8 in these cells in tissue culture could transform those cells into ear neurosensory precursors able to differentiate into neurons, as already demonstrated with the ectopic expression of Neurog1 in frog ectoderm.(40) Experiments are underway in our laboratories to test this possibility.
Another source of neural-crest-derived stem cells was recently identified in sensory hair roots.(21) These cells have the capacity to express neuronal markers if implanted into the spinal cord.(22) It is likely that these cells are related to the neural-crest-derived Merkel cells,(110) a population of cells that express two genes essential for hair cell development, Pou4f3(111–114) and Atoh1.(67,115) These cells seem to retain their gene expression profile while proliferating. If so, these cells might readily differentiate into hair cells if implanted into ears; this appears to be possible with other stem-cell-derived precursors that are equally characterized by Atoh1 and Pou4f3 expression.(17)
Most importantly in this context, recent work has molecularly characterized the only source of continuously proliferating neuronal stem cells in mammals, the olfactory epithelium.(72) This epithelium is surgically easily accessible and some precursors are characterized by the expression of the same bHLH genes known for ear neuronal development, Neurog1 and Neurod1(116) and Foxg1.(46,74) Isolation of Neurog1-positive precursors and forced expression of other ear-related genes such as Gata3,(41) Foxi1, Pax2/8(43) or Fgf10(63) might help drive such cells in tissue culture towards ear neurosensory development. Clearly, other genes expressed in both the ear and olfactory epithelium, such as Sox2 or Foxg1, would not redirect the fate of these cells beyond olfactory specification.
These approaches might provide sufficient adult cellular stem cell material to restore lost hair cells and sensory neurons of the ear combined with limited surgical intervention to obtain adult stem cells to repopulate the ear. If these simple approaches have too low a yield of cells with ear-specific gene expression, the known steps of neurosensory development in the ear as outlined above can provide appropriate guidance to achieve this goal through additional manipulations. Such manipulations may include, but are not limited to, selective upregulation of miRNA. miRNAs are generally known to be important in cell fate determination and proliferation regulation(117) through regulation of large sets of target genes. Some miRNAs were recently shown to be selectively expressed in hair cells(118) and may be important in consolidating cell cycle exit and maintaining differentiation of neurosensory aspects of the ear but such functions require ear-specific conditional mutations of enzymes necessary for miRNA processing.(119) All the progress towards the molecular basis of ear development during the last five years, combined with recent advances in isolation and molecular manipulation of stem cells from various sources, raises the hope that hearing loss will soon be correctable via stem cell therapy before the baby boom generation will have suffered untreatable neurosensory hearing loss.
Acknowledgments
Funding agency: NIH; Grant numbers: RO1 DC005590, DC005009; Funding agency: NASA; Grant number: NAG 2-1611; Funding agency: COBRE; Grant numbers: 1P20RR018788-01, LB692; Funding agency: Bellucci Fund.
This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number 1 C06 RR17417-01 from the National Center for Research Resources, National Institutes of Health. We acknowledge the use of the confocal microscope facility of the NCCB, supported by EPSCoR EPS-0346476 (CFD 47.076).
References
- 1.Rakic P. Neuroscience. No more cortical neurons for you. Science. 2006;313:928–929. doi: 10.1126/science.1131713. [DOI] [PubMed] [Google Scholar]
- 2.Rask-Andersen H, Bostrom M, Gerdin B, Kinnefors A, Nyberg G, et al. Regeneration of human auditory nerve. In vitro/in video demonstration of neural progenitor cells in adult human and guinea pig spiral ganglion. Hear Res. 2005;203:180–191. doi: 10.1016/j.heares.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Liu H, Heller S. Pluripotent stem cells from the adult mouse inner ear. Nat Med. 2003;9:1293–1299. doi: 10.1038/nm925. [DOI] [PubMed] [Google Scholar]
- 4.Fritzsch B, Tessarollo L, Coppola E, Reichardt LF. Neurotrophins in the ear: their roles in sensory neuron survival and fiber guidance. Prog Brain Res. 2004;146:265–278. doi: 10.1016/S0079-6123(03)46017-2. [DOI] [PubMed] [Google Scholar]
- 5.Sugawara M, Corfas G, Liberman MC. Influence of supporting cells on neuronal degeneration after hair cell loss. J Assoc Res Otolaryngol. 2005;6:136–147. doi: 10.1007/s10162-004-5050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roehm PC, Hansen MR. Strategies to preserve or regenerate spiral ganglion neurons. Curr Opin Otolaryngol Head Neck Surg. 2005;13:294–300. doi: 10.1097/01.moo.0000180919.68812.b9. [DOI] [PubMed] [Google Scholar]
- 7.Gillespie LN, Shepherd RK. Clinical application of neurotrophic factors: the potential for primary auditory neuron protection. Eur J Neurosci. 2005;22:2123–2133. doi: 10.1111/j.1460-9568.2005.04430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staecker H, Brough DE, Praetorius M, Baker K. Drug delivery to the inner ear using gene therapy. Otolaryngol Clin North Am. 2004;37:1091–1108. doi: 10.1016/j.otc.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen P, Zindy F, Abdala C, Liu F, Li X, Roussel MF, Segil N. Progressive hearing loss in mice lacking the cyclin-dependent kinase inhibitor Ink4d. Nat Cell Biol. 2003;5:422–426. doi: 10.1038/ncb976. [DOI] [PubMed] [Google Scholar]
- 10.Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, et al. Increasing p16(INK4a) expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006 doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441:984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- 12.Kanzaki S, Beyer LA, Swiderski DL, Izumikawa M, Stover T, et al. p27(Kip1) deficiency causes organ of Corti pathology and hearing loss. Hear Res. 2006;214:28–36. doi: 10.1016/j.heares.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Mantela J, Jiang Z, Ylikoski J, Fritzsch B, Zacksenhaus E, Pirvola U. The retinoblastoma gene pathway regulates the postmitotic state of hair cells of the mouse inner ear. Development. 2005;132:2377–2388. doi: 10.1242/dev.01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sage C, Huang M, Vollrath MA, Brown MC, Hinds PW, et al. Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc Natl Acad Sci USA. 2006;103:7345–7350. doi: 10.1073/pnas.0510631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- 16.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 17.Rivolta MN, Li H, Heller S. Generation of inner ear cell types from embryonic stem cells. Methods Mol Biol. 2006;330:71–92. doi: 10.1385/1-59745-036-7:71. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Corrales CE, Edge A, Heller S. Stem cells as therapy for hearing loss. Trends Mol Med. 2004;10:309–315. doi: 10.1016/j.molmed.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman CK, Zhou P, Pasolli HA, Rendl M, Bolotin D, et al. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieber-Blum M, Grim M, Hu YF, Szeder V. Pluripotent neural crest stem cells in the adult hair follicle. Dev Dyn. 2004;231:258–269. doi: 10.1002/dvdy.20129. [DOI] [PubMed] [Google Scholar]
- 22.Sieber-Blum M, Schnell L, Grim M, Hu YF, Schneider R, Schwab ME. Characterization of epidermal neural crest stem cell (EPI-NCSC) grafts in the lesioned spinal cord. Mol Cell Neurosci. 2006;32:67–81. doi: 10.1016/j.mcn.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Groves AK. The induction of the otic placode. In: Kelley MW, Wu DK, Popper AN, Fay RR, editors. Development of the inner ear. New York: Springer Verlag; 2005. pp. 10–42. [Google Scholar]
- 24.Lemaire P, Bertrand V, Hudson C. Early steps in the formation of neural tissue in ascidian embryos. Dev Biol. 2002;252:151–169. doi: 10.1006/dbio.2002.0861. [DOI] [PubMed] [Google Scholar]
- 25.Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- 26.Shou J, Rim PC, Calof AL. BMPs inhibit neurogenesis by a mechanism involving degradation of a transcription factor. Nat Neurosci. 1999;2:339–345. doi: 10.1038/7251. [DOI] [PubMed] [Google Scholar]
- 27.Riccomagno MM, Martinu L, Mulheisen M, Wu DK, Epstein DJ. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright TJ, Hatch EP, Karabagli H, Karabagli P, Schoenwolf GC, Mansour SL. Expression of mouse fibroblast growth factor and fibroblast growth factor receptor genes during early inner ear development. Dev Dyn. 2003;228:267–272. doi: 10.1002/dvdy.10362. [DOI] [PubMed] [Google Scholar]
- 29.Riccomagno MM, Takada S, Epstein DJ. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19:1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohyama T, Mohamed OA, Taketo MM, Dufort D, Groves AK. Wnt signals mediate a fate decision between otic placode and epidermis. Development. 2006;133:865–875. doi: 10.1242/dev.02271. [DOI] [PubMed] [Google Scholar]
- 31.Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 33.Fritzsch B, Beisel KW, Bermingham NA. Developmental evolutionary biology of the vertebrate ear: conserving mechanoelectric transduction and developmental pathways in diverging morphologies. Neuroreport. 2000;11:R35–44. doi: 10.1097/00001756-200011270-00013. [DOI] [PubMed] [Google Scholar]
- 34.Fritzsch B, Beisel KW. Evolution and development of the vertebrate ear. Brain Res Bull. 2001;55:711–721. doi: 10.1016/s0361-9230(01)00558-5. [DOI] [PubMed] [Google Scholar]
- 35.Caldwell JC, Eberl DF. Towards a molecular understanding of Drosophila hearing. J Neurobiol. 2002;53:172–189. doi: 10.1002/neu.10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghysen A, Dambly-Chaudiere C. A genetic programme for neuronal connectivity. Trends Genet. 2000;16:221–226. doi: 10.1016/s0168-9525(99)01969-1. [DOI] [PubMed] [Google Scholar]
- 37.Pauley S, Matei V, Beisel KW, Fritzsch B. Wiring the ear to the brain: the molecular basis of neurosensory development, differentiation, and survival. In: Kelley MK, Wu DK, Popper AN, Fay RR, editors. Development of the inner ear. New Yrok: Springer Verlag; 2005. pp. 85–121. [Google Scholar]
- 38.Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 41.Karis A, Pata I, van Doorninck JH, Grosveld F, de Zeeuw CI, et al. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J Comp Neurol. 2001;429:615–630. doi: 10.1002/1096-9861(20010122)429:4<615::aid-cne8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 42.Torres M, Gomez-Pardo E, Gruss P. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development. 1996;122:3381–3391. doi: 10.1242/dev.122.11.3381. [DOI] [PubMed] [Google Scholar]
- 43.Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M. Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 1998;125:3063–3074. doi: 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- 44.Raft S, Nowotschin S, Liao J, Morrow BE. Suppression of neural fate and control of inner ear morphogenesis by Tbx1. Development. 2004;131:1801–1812. doi: 10.1242/dev.01067. [DOI] [PubMed] [Google Scholar]
- 45.Hatini V, Ye X, Balas G, Lai E. Dynamics of placodal lineage development revealed by targeted transgene expression. Dev Dyn. 1999;215:332–343. doi: 10.1002/(SICI)1097-0177(199908)215:4<332::AID-AJA5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 46.Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004;231:640–646. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- 48.Zou D, Silvius D, Fritzsch B, Xu PX. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reim G, Brand M. Spiel-ohne-grenzen/pou2 mediates regional competence to respond to Fgf8 during zebrafish early neural development. Development. 2002;129:917–933. doi: 10.1242/dev.129.4.917. [DOI] [PubMed] [Google Scholar]
- 50.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 51.Blokzijl A, ten Dijke P, Ibanez CF. Physical and functional interaction between GATA-3 and Smad3 allows TGF-beta regulation of GATA target genes. Curr Biol. 2002;12:35–45. doi: 10.1016/s0960-9822(01)00623-6. [DOI] [PubMed] [Google Scholar]
- 52.Ohuchi H, Yasue A, Ono K, Sasaoka S, Tomonari S, et al. Identification of cis-element regulating expression of the mouse Fgf10 gene during inner ear development. Dev Dyn. 2005;233:177–187. doi: 10.1002/dvdy.20319. [DOI] [PubMed] [Google Scholar]
- 53.Nicolussi A, D’Inzeo S, Santulli M, Colletta G, Coppa A. TGF-beta control of rat thyroid follicular cells differentiation. Mol Cell Endocrinol. 2003;207:1–11. doi: 10.1016/s0303-7207(03)00238-7. [DOI] [PubMed] [Google Scholar]
- 54.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 55.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, et al. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 57.Zhao Y, Wang Y, Wang Z, Liu H, Shen Y, et al. Sonic hedgehog promotes mouse inner ear progenitor cell proliferation and hair cell generation in vitro. Neuroreport. 2006;17:121–124. doi: 10.1097/01.wnr.0000198439.44636.49. [DOI] [PubMed] [Google Scholar]
- 58.Mowbray C, Hammerschmidt M, Whitfield TT. Expression of BMP signalling pathway members in the developing zebrafish inner ear and lateral line. Mech Dev. 2001;108:179–184. doi: 10.1016/s0925-4773(01)00479-8. [DOI] [PubMed] [Google Scholar]
- 59.Farinas I, Jones KR, Tessarollo L, Vigers AJ, Huang E, et al. Spatial shaping of cochlear innervation by temporally regulated neurotrophin expression. J Neurosci. 2001;21:6170–6180. doi: 10.1523/JNEUROSCI.21-16-06170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fritzsch B, Beisel KW, Jones K, Farinas I, Maklad A, et al. Development and evolution of inner ear sensory epithelia and their innervation. J Neurobiol. 2002;53:143–156. doi: 10.1002/neu.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 62.Radde-Gallwitz K, Pan L, Gan L, Lin X, Segil N, Chen P. Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J Comp Neurol. 2004;477:412–421. doi: 10.1002/cne.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, et al. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu M, Pereira FA, Price SD, Chu MJ, Shope C, et al. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000;14:2839–2854. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fritzsch B, Beisel KW. Molecular conservation and novelties in vertebrate ear development. Curr Top Dev Biol. 2003;57:1–44. doi: 10.1016/s0070-2153(03)57001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 68.Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, et al. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satoh T, Fekete DM. Clonal analysis of the relationships between mechanosensory cells and the neurons that innervate them in the chicken ear. Development. 2005;132:1687–1697. doi: 10.1242/dev.01730. [DOI] [PubMed] [Google Scholar]
- 70.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 71.Calof AL, Bonnin A, Crocker C, Kawauchi S, Murray RC, et al. Progenitor cells of the olfactory receptor neuron lineage. Microsc Res Tech. 2002;58:176–188. doi: 10.1002/jemt.10147. [DOI] [PubMed] [Google Scholar]
- 72.Beites CL, Kawauchi S, Crocker CE, Calof AL. Identification and molecular regulation of neural stem cells in the olfactory epithelium. Exp Cell Res. 2005;306:309–316. doi: 10.1016/j.yexcr.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 73.Kawauchi S, Beites CL, Crocker CE, Wu HH, Bonnin A, et al. Molecular signals regulating proliferation of stem and progenitor cells in mouse olfactory epithelium. Dev Neurosci. 2004;26:166–180. doi: 10.1159/000082135. [DOI] [PubMed] [Google Scholar]
- 74.Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, et al. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- 75.Wu HH, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, et al. Autoregulation of Neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/s0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]
- 76.Lee YS, Liu F, Segil N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development. 2006;133:2817–2826. doi: 10.1242/dev.02453. [DOI] [PubMed] [Google Scholar]
- 77.Jones JM, Montcouquiol M, Dabdoub A, Woods C, Kelley MW. Inhibitors of differentiation and DNA binding (Ids) regulate Math1 and hair cell formation during the development of the organ of Corti. J Neurosci. 2006;26:550–558. doi: 10.1523/JNEUROSCI.3859-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 79.Pozniak CD, Pleasure SJ. A tale of two signals: Wnt and Hedgehog in dentate neurogenesis. Sci STKE. 2006:pe5. doi: 10.1126/stke.3192006pe5. [DOI] [PubMed] [Google Scholar]
- 80.Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, et al. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- 82.Ladher RK, Wright TJ, Moon AM, Mansour SL, Schoenwolf GC. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–613. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martynoga B, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 84.Yao J, Liu Y, Lo R, Tretjakoff I, Peterson A, Stifani S. Disrupted development of the cerebral hemispheres in transgenic mice expressing the mammalian Groucho homologue transducin-like-enhancer of split 1 in postmitotic neurons. Mech Dev. 2000;93:105–115. doi: 10.1016/s0925-4773(00)00278-1. [DOI] [PubMed] [Google Scholar]
- 85.Lanford PJ, Kelley MW. Notch signaling and cell fate determination in the vertebrate inner ear. In: Kelley MW, KWD, Popper AN, Fay RR, editors. Development of the Inner Ear. Volume SHAR 26, Springer Handbook of Auditory Research. New York, NY: Springer; 2005. pp. 122–157. [Google Scholar]
- 86.Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, et al. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- 87.Kiernan AE, Xu J, Gridley T. The Notch Ligand JAG1 Is Required for Sensory Progenitor Development in the Mammalian Inner Ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- 89.Daudet N, Lewis J. Two contrasting roles for Notch activity in chick inner ear development: specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development. 2005;132:541–551. doi: 10.1242/dev.01589. [DOI] [PubMed] [Google Scholar]
- 90.Ohtsuka T, Imayoshi I, Shimojo H, Nishi E, Kageyama R, McConnell SK. Visualization of embryonic neural stem cells using Hes promoters in transgenic mice. Mol Cell Neurosci. 2006;31:109–122. doi: 10.1016/j.mcn.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 91.Zine A, Aubert A, Qiu J, Therianos S, Guillemot F, et al. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J Neurosci. 2001;21:4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lanford PJ, Shailam R, Norton CR, Gridley T, Kelley MW. Expression of Math1 and HES5 in the cochleae of wildtype and Jag2 mutant mice. J Assoc Res Otolaryngol. 2000;1:161–171. doi: 10.1007/s101620010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Westendorf JJ. Transcriptional co-repressors of Runx2. J Cell Biochem. 2006;98:54–64. doi: 10.1002/jcb.20805. [DOI] [PubMed] [Google Scholar]
- 95.Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marcal N, Patel H, Dong Z, Belanger-Jasmin S, Hoffman B, et al. Antagonistic effects of Grg6 and Groucho/TLE on the transcription repression activity of brain factor 1/FoxG1 and cortical neuron differentiation. Mol Cell Biol. 2005;25:10916–10929. doi: 10.1128/MCB.25.24.10916-10929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mitsui K, Shirakata M, Paterson BM. Phosphorylation inhibits the DNA-binding activity of MyoD homodimers but not MyoD-E12 heterodimers. J Biol Chem. 1993;268:24415–24420. [PubMed] [Google Scholar]
- 98.Ninkovic J, Tallafuss A, Leucht C, Topczewski J, Tannhauser B, et al. Inhibition of neurogenesis at the zebrafish midbrain-hindbrain boundary by the combined and dose-dependent activity of a new hairy/E(spl) gene pair. Development. 2005;132:75–88. doi: 10.1242/dev.01525. [DOI] [PubMed] [Google Scholar]
- 99.Amoutzias GD, Weiner J, Bornberg-Bauer E. Phylogenetic profiling of protein interaction networks in eukaryotic transcription factors reveals focal proteins being ancestral to hubs. Gene. 2005;347:247–253. doi: 10.1016/j.gene.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 100.Amoutzias GD, Robertson DL, Oliver SG, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004;5:274–279. doi: 10.1038/sj.embor.7400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qian D, Radde-Gallwitz K, Kelly M, Tyrberg B, Kim J, et al. Basic helix-loop-helix gene Hes6 delineates the sensory hair cell lineage in the inner ear. Dev Dyn. 2006;235:1689–1700. doi: 10.1002/dvdy.20736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koyano-Nakagawa N, Kim J, Anderson D, Kintner C. Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development. 2000;127:4203–4216. doi: 10.1242/dev.127.19.4203. [DOI] [PubMed] [Google Scholar]
- 103.Kang SA, Seol JH, Kim J. The conserved WRPW motif of Hes6 mediates proteasomal degradation. Biochem Biophys Res Commun. 2005:332–36. doi: 10.1016/j.bbrc.2005.04.089. [DOI] [PubMed] [Google Scholar]
- 104.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 105.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 106.Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126:1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- 107.Zhang N, Martin GV, Kelley MW, Gridley T. A mutation in the Lunatic fringe gene suppresses the effects of a Jagged2 mutation on inner hair cell development in the cochlea. Curr Biol. 2000;10:659–662. doi: 10.1016/s0960-9822(00)00522-4. [DOI] [PubMed] [Google Scholar]
- 108.Yagi M, Kanzaki S, Kawamoto K, Shin B, Shah PP, et al. Spiral ganglion neurons are protected from degeneration by GDNF gene therapy. J Assoc Res Otolaryngol. 2000;1:315–325. doi: 10.1007/s101620010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Matter-Sadzinski L, Puzianowska-Kuznicka M, Hernandez J, Ballivet M, Matter JM. A bHLH transcriptional network regulating the specification of retinal ganglion cells. Development. 2005;132:3907–3921. doi: 10.1242/dev.01960. [DOI] [PubMed] [Google Scholar]
- 110.Szeder V, Grim M, Halata Z, Sieber-Blum M. Neural crest origin of mammalian Merkel cells. Dev Biol. 2003;253:258–263. doi: 10.1016/s0012-1606(02)00015-5. [DOI] [PubMed] [Google Scholar]
- 111.Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yucel R, et al. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- 112.Xiang M, Maklad A, Pirvola U, Fritzsch B. Brn3c null mutant mice show long-term, incomplete retention of some afferent inner ear innervation. BMC Neurosci. 2003;4:2. doi: 10.1186/1471-2202-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haeberle H, Fujiwara M, Chuang J, Medina MM, Panditrao MV, et al. Molecular profiling reveals synaptic release machinery in Merkel cells. Proc Natl Acad Sci USA. 2004;101:14503–14508. doi: 10.1073/pnas.0406308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leonard JH, Cook AL, Van Gele M, Boyle GM, Inglis KJ, et al. Proneural and proneuroendocrine transcription factor expression in cutaneous mechanoreceptor (Merkel) cells and Merkel cell carcinoma. Int J Cancer. 2002;101:103–110. doi: 10.1002/ijc.10554. [DOI] [PubMed] [Google Scholar]
- 115.Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, et al. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- 116.Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- 117.Conrad R, Barrier M, Ford LP. Role of miRNA and miRNA processing factors in development and disease. Birth Defects Res C Embryo Today. 2006;78:107–117. doi: 10.1002/bdrc.20068. [DOI] [PubMed] [Google Scholar]
- 118.Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 119.Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fekete DM, Wu DK. Revisiting cell fate specification in the inner ear. Curr Opin Neurobiol. 2002;12:35–42. doi: 10.1016/s0959-4388(02)00287-8. [DOI] [PubMed] [Google Scholar]