Abstract
miRNAs are small non-coding RNAs that have emerged as crucial post-transcriptional regulators of gene expression. They are key players in various critical cellular processes such as proliferation, cell cycle progression, apoptosis and differentiation. Self-renewal capacity and differentiation potential are hallmarks of stem cells. The switch between self-renewal and differentiation requires rapid widespread changes in gene expression. Since miRNAs can repress the translation of many mRNA targets, they are good candidates to regulate cell fates. In the past few years, miRNAs have appeared as important new actors in stem cell development by regulating differentiation and maintenance of stem cells. In this chapter we will focus on the role of miRNAs in various stem cell populations. After an introduction on microRNA biogenesis, we will review the recent knowledge on miRNA expression and function in pluripotent cells and during the acquisition of stem cell fate. We will then brie fly examine the role of miRNAs in adult and cancer stem cells.
Keywords: miRNA, Embryonic stem cells, Reprogramming, Adult stem cells, Cancer stem cells
18.1 Introduction
Mature microRNAs (miRNAs) are endogenous single-stranded non-protein coding RNAs of 20–23 nucleotides in length that regulate translation through interaction with mRNA transcripts [1]. They are members of the family of small non coding RNAs that also comprise endogenous small Mature microRNAs (miRNAs) are endogenous interfering RNAs (endo-siRNAs) and PIWI-single-stranded non-protein coding RNAs of 20–23 interacting RNAs (pi-RNAs) [2, 3]. In this chapter we will focus on the role of miRNAs in the regulation of stem cell populations.
The first miRNA was identified in 1993 in C. elegans [4]. Ambros and colleagues showed that lin-4 can control developmental timing by negatively regulating the level of LIN-14 protein via an antisense RNA-RNA interaction. This mode of gene regulation was thought to be restricted to nematodes, however since the term “microRNA” was used for the first time in 2001, thousands of miRNAs have been found in many other organisms from plants to mammalians.
In the past decade, great progress has been made in studying miRNA expression and function. miRNAs have emerged as crucial regulators of gene expression at a post-transcriptional level and have been shown to be involved in many different processes such as proliferation, cell cycle progression, programmed cell death (apoptosis) and differentiation [5]. They are highly evolutionary conserved, and deregulation of miRNA expression is associated with several diseases including cancer and neurodegeneration [6, 7]. Further, miRNAs have been shown to be important new players in regulation of stem cell development by playing a critical role in differentiation and maintenance of stem cells [8]. Stem cells are defined by their capacity to self-renew and their potential to differentiate into many other different cell types. This requires a massive and rapid transformation in cell phenotype and major changes in proteome network in a very short time. miRNAs are able to repress the translation of many mRNA targets, thus inducing widespread changes in gene expression [9]. Around 1,600 (miRbase) miRNAs have been identified so far in the human genome, thus making the miRNAs one of the most abundant class of gene-regulatory molecules in animals [10]. They are predicted to regulate most of the genome.
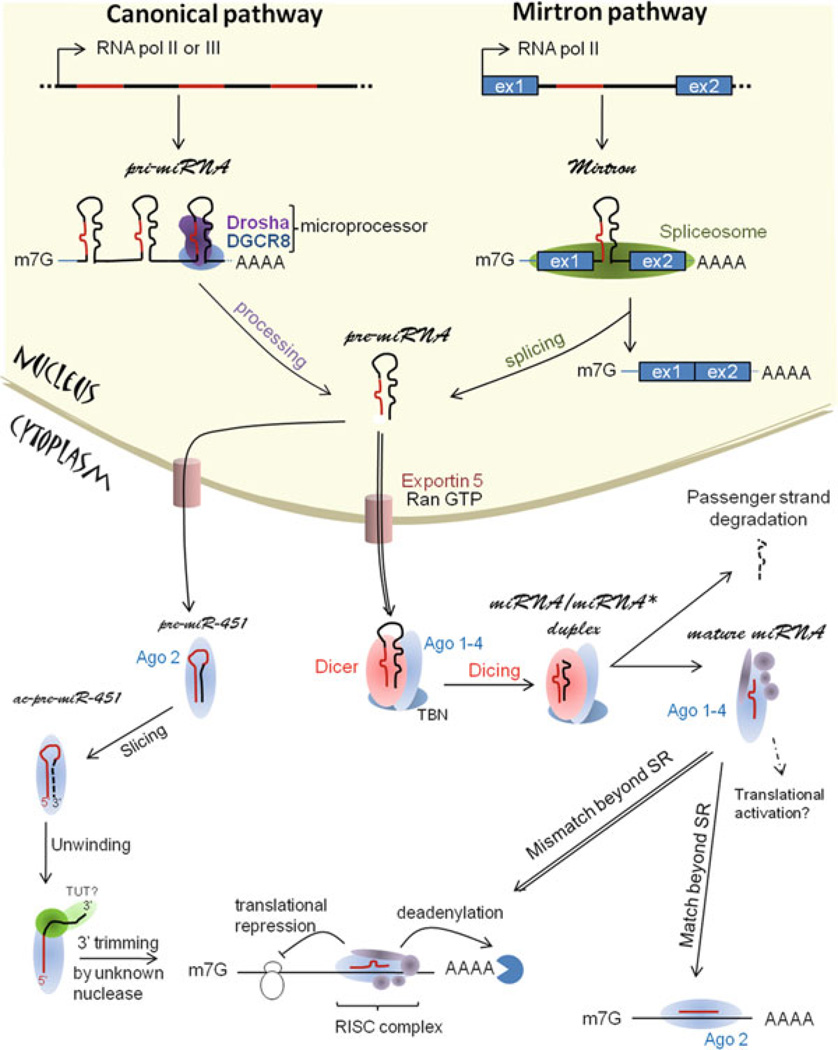
miRNAs are scattered throughout the genome. They can be found as isolated transcript units or clustered and co-transcribed as polycistronic primary transcripts [11]. Mature miRNAs are generated by multiple processing steps of sequential endonucleolytic cleavages (Fig. 18.1). The miRNAs can either be encoded within protein coding genes in both introns and exons or transcribed from independent genes in intergenic regions. Even if miRNA transcription by RNA polymerase III has been described [12], these genes are mostly transcribed by RNA polymerase II in the nucleus to form large primary transcripts (pri-miRNAs) [13]. Pri-miRNAs are thousands nucleotide long capped and polyadenlylated hairpin-shaped transcripts. In the most commonly used canonical pathway, pri-miRNAs are recognized and cleaved in the nucleus by the “microprocessor” complex composed of the RNAse III enzyme Drosha and its double strand RNA binding domain partner DiGeorge syndrome critical region gene 8 (DGCR8) [14, 15]. Drosha has also been found in a complex with other proteins, including RNA-binding proteins, RNA helicases and the Ewing’s sarcoma family of proteins [16]. The resulting 60–70-nucleotide hairpins from the Drosha processing step are called pre-miRNAs. Several alternative miRNA biogenesis pathways have been proposed recently. For example, some miRNAs (“mirtrons”) are embedded in short mRNA introns and pre-miRNAs are produced from splicing and debranching, therefore bypassing Drosha cleavage [17]. In both the canonical and mirtron pathways, pre-miRNAs are transported into the cytoplasm by exportin 5 and Ran-GTP, where they are further cleaved by another RNAse III enzyme, Dicer [14, 18]. The core ribonuclease Dicer interacts with other proteins, including TAR RNA binding protein (TRPB) and the PKR-activating protein (PACT) [19]. Cleavage by Dicer of the terminal loop end of pre-miRNA produces a short double-stranded RNA duplex measuring 20–23 bp in length. Following processing, the strand of the duplex with a less thermodynamically stable 5′ end (the guide strand, miRNA) is preferentially embedded with one of four human Argonaute proteins (Ago) to form the miRNA-induced silencing complex (RISC) [20] The other strand (the passenger strand, miRNA*) is released and degraded [21]. However, in some cases the passenger strand can also be incorporated into a RISC to function as miRNA. Of note, in mammals all but one miRNA, miR-451, seem to be processed by Dicer. The stem of predicted pre-miR-451 structure is only 17 bp long, which is too short to serve as a substrate for Dicer. Instead it requires a cleavage by Ago2 independently of Dicer and the 3′ end is generated by exonucleolytic trimming [22, 23].
Fig. 18.1. microRNA biogenesis and function.
miRNA genes are transcribed by RNA polymerase II or III and processed in two steps. The first step involved either the microprocessor containing Drosha and DGCR8 (canonical pathway) or the splicing machinery (mirtron pathway). The second cleavage is performed by Dicer for most mammalian miRNAs, but miR-451. Mature miRNAs assemble with the RISC complex and regulate gene expression by inhibiting translation, inducing mRNA degradation or, less commonly, upregulating translation. SR = seed region/seed sequence. See text for more details
The mature miRNA associated to the RISC binds to the 3′ UTR, or in few cases the coding region, of the target mRNA transcript based on complementarity between the miRNA and the mRNA target [24, 25]. Nucleotides 2–8 (counted from the 5′ end) of the mature miRNA, called the “seed sequence”, is critical for target recognition and hybridizes nearly perfectly with the target mRNA [5, 26]. The mechanism of regulation of the target depend on the degree of complementarity between the miRNA and the target mRNA. When the complementarity is perfect, the miRNA induces degradation of the target mRNA through Ago2 endonuclease activity. While frequent in plants, to our knowledge there is only one example in mammalian cells so far of a miRNA inducing the cleavage of a single target [27]. Partial pairing results in repression of target mRNA translation at the initiation step or at the elongation step, and/ or sequestration of target mRNAs into cytoplasmic processing bodies (P-Bodies) where the mRNA will be degraded through deadenylation pathways [28]. However those mechanisms are not entirely understood. It has also been proposed that in some cases miRNAs could activate the translation of its targets [29], but additional experiments are required to have a better understanding of the extend of this phenomenon.
The degree of downregulation of a target is quantitatively modest (generally less than 50 %) but miRNAs can induce subtle changes and fine tune gene expression [1]. A define spatiotemporal pattern of miRNAs is very important and regulatory mechanisms by which cells control miRNA production and function have recently come into light [21]. The transcription of miRNAs is regulated in a similar manner to that of protein coding genes, for example by DNA methylation and histone modification of the miRNA promoter or by DNA-bindings factors such as p53 or cMyc. miRNA biogenesis itself is subject to tight regulation at multiple steps [30]. miRNA processing could be controlled by regulation of the levels and activity of Drosha, Dicer and their partners [30]. One of the targets of let-7, lin-28, can physically interact with pre-let-7 in the cytoplasm of embryonic stem cells, promotes its polyuridylation, directly inhibits Dicer processing and induces degradation of pre-let-7 [31, 32]. The importance of this double negative feedback loop on stem cell function will be discussed further later. miRNAs are highly stable molecules with a half-life of hours or even days [21], however to allow developmental transition miRNAs decay need to be tightly regulated and proteins involved in miRNA turnover are under investigation [21]. miRNA function can also be regulated at the level of Ago proteins and other components of the RISC complex. Many pri-miRNAs accumulate without being fully processed until the right developmental or environmental stimuli arise [33]. Interestingly, miRNAs from the same polycistronic transcript can be expressed differently [34]. Further, many mRNAs are expressed in a developmental-stage and tissue-specific manner. Many types of cells seem to have unique miRNA patterns. One miRNA can target hundreds of mRNAs simultaneously and several miRNAs can target a single mRNA. miRNAs are major contributors of cell-type profiles of protein expression and thus play a critical role in stem cell identity and function. In this review, we will discuss the recent advances on the functions of miRNAs in pluripotent stem cells, adult stem cells and cancer stem cells.
18.2 microRNA Function in Pluripotent Stem Cells
18.2.1 Role of microRNAs in Embryonic Stem Cells (ESCs)
ESCs are derived from the inner cell mass of the blastocyst stage of the embryo and have been isolated in 1981 in mouse (mESC) [35] and in 1998 in human (hESC) [36]. They represent a powerful tool for developmental studies, in vitro diseases modeling and for potential cellular therapeutic in regenerative medicine. This is mainly due to the two important properties that defined them: pluripotency and self-renewal. ESCs are indeed pluripotent, they are able to differentiate into the three germ layers and give rise to cell types found in all tissues and organs of the body. They also possess an unlimited self-renewal capacity with continuous cell division in vitro. Undifferentiated ESCs display a particular abbreviated cell cycle profile critical for fast growth during early embryonic development [37]. The decision of an ESC to self-renew or differentiate is regulated by a complex set of factors, including transcription factors, chromatin modifications, signaling pathways and non coding RNAs [38]. The unique molecular program of ESCs needs to be conserved in order to maintain the undifferentiated pluripotent stage, whereas ESCs undergo major epigenetic and gene expression changes when cells are engaged in a differentiation process resulting in a massive transformation of cell phenotype. By their capacities to regulate simultaneously hundreds of targets, miRNAs represent good candidates for such rapid and large transformation.
A complex network of extrinsic and intrinsic factors in conjunction with chromatic remodeling is necessary to keep the ESC fate. A core network of transcription factors and RNA binding proteins has been defined in the past few years [39]. Among those, Oct4, Sox2 and Nanog, play a central role in the maintenance and acquisition of “stemness”, the ensemble of properties that define the stem cell fate. Various positive autoregulatory loops exist between those three transcription factors since each of them is able to bind to its own promoter and to the promoters of the two other members. Chromatin immunoprecipitation (ChiP) assays have revealed that they also control the transcription of many other players of the regulatory network of pluripotency, including Lin28, cMyc, Klf4, Tcf3 or Stat3 [39, 40]. Recently, a combination of a subset of those factors has been shown to reprogram somatic cells into pluripotent cells (induced pluripotent stem cells, iPSCs) possessing very similar properties to ESCs [41–43]. Interestingly, some of those stem cell core regulators are able to activate the promoters of several miRNAs in ESCs, including miR-290–295, miR-302/367 and miR-92 clusters [44].
In the past few years, miRNAs have appeared as central players in ESC self-renewal and differentiation. In this section, we will review our current knowledge of the role of miRNAs in ESC functions.
ESCs Display a Defined miRNA Signature
miRNA expression has been examined in mESCs and hESCs using cloning, qPCR, microarray and deep sequencing technologies by comparing undifferentiated ESCs to their differentiated counterparts [45–48]. Those experiments revealed that the global miRNA expression is low in ESCs compared to differentiated cells. Some of the ESC-enriched miRNAs are limited to ESCs, while other are more widely expressed but decrease significantly during differentiation. Thus, ESCs seem to be characterized by a unique miRNAs signature. We will use the term “ESC-enriched miRNA” in this review when referring to the miRNAs whose levels decrease as ESCs differentiate.
Depending on the method used to analyze miRNA expression, the ESC lines and the differentiation protocols, small variations were observed. However all those studies share miRNAs described as ESC-enriched. The miR-290 family and miR-302 clusters account for the majority of all miRNAs expressed in undifferentiated mESCs. hESC-enriched miRNAs can be categorized in four major groups: miRNAs from the miR-302 cluster, miRNAs from the miR-17 family, miRNAs from the miR-371–373 cluster and miRNAs from C19MC (chromosome 19 miRNA cluster) [48]. Two additional families have been found enriched in hESCs in some studies: miR-130 and miR-200 [48]. The promoters of most of those miRNAs can be activated by Oct4, Sox2 and Nanog [44].
miRNAs whose expression increases during differentiation are also of importance since their low expression might keep ESCs in an undifferentiated state and will be discussed further later. The most studied among this group of miRNAs activated during differentiation are let-7, miR-145 (in human) and miR-134 (in mouse).
Majority of the hESC-enriched miRNA clusters are transcribed as polycistronic transcripts, suggesting that they share common upstream regulators and the same pattern of expression. Moreover, several of those ESC-enriched miRNAs have the same or a similar seed sequence, so can target a common group of mRNAs [48].
Global Disruption of Mature miRNAs
The fact that many miRNAs have the same seed sequence and that a single miRNA can target multiple mRNAs make difficult to study their function individually. Removal of all miRNAs can be achieved by deleting the genes encoding the enzymes involved in the processing of miRNAs, Drosha and Dicer, or their partners, such as DGCR8. Individual miRNAs can then be reintroduced as mimics to assess their functions. Homozygous Dicer1 knockout (KO) mice die early in the development [49] while conditional Dicer1 mutant mESCs are viable in culture but are defective in differentiation [50]. Dicer loss also leads to severe growth defects of mESCs and slightly prolongs G1 and G0 phases of their cell cycle [51]. In addition to its role in miRNA processing, Dicer has been shown to be involved in the biogenesis of endo-siRNAs and other small RNAs [52]. Therefore, the studies of DGCR8 KO in mice might better depict the functions of most miRNAs in mESCs. Similar to Dicer mutant, DGRC8 deficient mice are not viable, and DGCR8 KO mESCs exhibit a proliferation defect and fail to differentiate [53] Knockdown (KD) of Dicer or Drosha also dramatically attenuates cell division in hESCs and results in the formation of stem cells with high levels of stem cell factors, correlating with delayed differentiation [54]. Altogether those studies show that miRNAs are critical for ESC self-renewal and differentiation.
microRNAs Regulate ESC Proliferation
A tight regulation of stem cell division is primordial to sustain the self-renewal capacity of ESCs. Observed proliferation defects of Dicer, Drosha and DGCR8 mutant ESCs suggest that miRNAs are involved in the regulation of their cell cycle. ESCs exhibit a very specific expedited cell cycle due to a short transition from G1 to S phase [55]. Cell cycle checkpoints control progression through the phases of the cell cycle and are regulated by the sequential activation and inactivation of cyclin-dependent kinases (CDKs) by cyclins. However, contrary to somatic cells, ESCs express very low level of cell cycle inhibitors (p21cip1, p27Kip1 and p16INK4a) and exhibit an atypical cell cycle, in which the major point of regulation does not take place in the Restriction checkpoint [54, 56, 57].
Many of the defects in Dicer-deficient mouse ESCs can be reversed by transfection with members of the miR-290 cluster [58]. In accordance with this study, a screen performed to identify miRNAs that can rescue the proliferation defect observed in DGCR8 KO cells uncovered members of the miR-290 and miR-302 clusters as important mESC cell cycle regulators [53]. Those miRNAs were called ESCC (for ESC cell Cycle promoting) miRNAs. They share a common seed sequence, suggesting that ESCC miRNAs regulate a common set of genes. A search for their targets has revealed that they function by suppressing several key regulators of the Restriction checkpoint, thus enabling rapid proliferation of ESCs. Indeed those targets are inhibitors of the CyclinE/CDK2 pathway, known to regulate the G1/S transition and include p21, the Retinoblastoma like 2 protein (Rbl2) and Last2. ESCC miRNAs post-transcriptionally downregulate those inhibitors and increase CyclinE/CDK2 activity [53].
Interestingly, the promoters of miR-290 and miR-302 clusters are directly regulated by pluripotency factors and in turn ESCC miRNAs maintain the expression of the pluripotency factors by inhibiting their epigenetic silencing. For example miR-290 cluster in mice has been shown to target Rbl2 and decrease the expression of de novo DNA methyltransferases [53] Similarly, the proposed human ortholog for the mouse miR-290 family, miR-372, might regulate human Rbl2 [54].
Using a similar approach, miRNAs are also shown to be critical for human ESC self-renewal and proliferation [54]. Knocking-down Dicer or Drosha by lentivirus-delivered shRNA dramatically affected cell division in hESCs. Dicer and Drosha KD induced G1/S and G2/M transition delays compared to cells infected with lentivirus controls. Re-introducing ESC-enriched miRNAs as mature miRNA mimics into Dicer KD hESC showed that both miR-372 and miR-195 could partially rescue the cell cycle defect. Moreover, miR-195 overexpression in wild-type H1 hESCs was sufficient to increase cell proliferation. miR-195 alone was able to rescue the G2 defect in the Dicer-KD line by directly targeting WEE1 kinase, a negative regulator of the CyclinB/CDK complex in the G2/M transition. Introduction of miR-372 mimics dramatically reduced the levels of the G1/S transition inhibitor p21 in Dicer KD and overexpression of p21 affected hESC proliferation, suggesting that miR-372 regulates hESC cell cycle by modulating p21 expression [54] Another hESC-enriched miRNA, miR-92b, has also been shown to target p21 [54, 59]
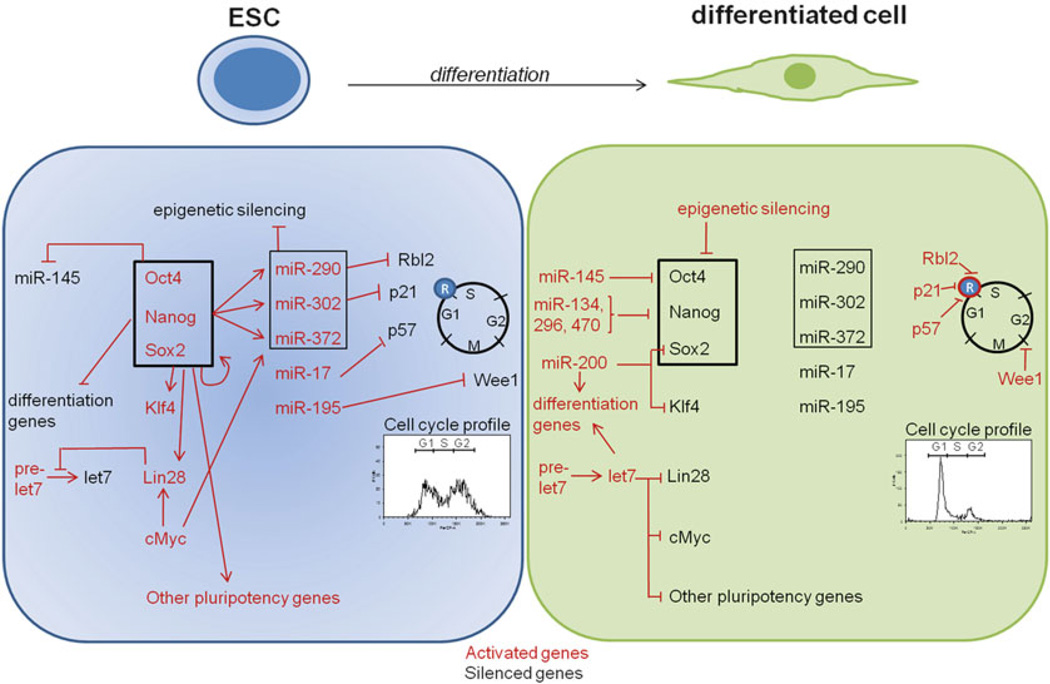
Overall, these data suggest that miRNAs can cooperate in maintaining the proliferative capacity of ESCs and appear as major players in the control of embryonic stem cell division (Fig. 18.2).
Fig. 18.2. Role of miRNAs in ESC self-renewal, proliferation and differentiation.
ESCs express a unique signature of miRNAs whose transcription is regulated by a core pluripotency factors (Oct4, Sox2, Nanog). ESC-enriched miRNAs control the specific ESC cell cycle by targeting regulatory proteins involved in G1/S and G2/M transitions. ESC-enriched miRNAs maintain self-renewal capacities of ESCs as well as their pluripotency potential. Differentiated cells express miRNAs such as miR-145 and let-7 that target pluripotency factors and activate differentiation genes. Moreover, cell cycle inhibitors are expressed and cells exhibit a cell cycle dependent of the restriction point (R)
microRNAs Regulate ESC Differentiation
In addition to the proliferation defect, Dicer KO and DGCR8 KO mESCs fail to downregulate pluripotency factors upon differentiation [50, 53, 58, 60, 61]. Similarly, in hESCs the levels of Nanog, Oct4 and Sox2 are upregulated in Dicer- and Drosha-knockdown while most early differentiation markers fail to be expressed when cultured under differentiation-inducing conditions [54]. Re-introduction of ESCC miRNAs into Dicer and DGCR8 mutant mESC did not rescue the differentiation defect, suggesting that other miRNAs are involved in the maintenance of pluripotency and the induction of ESC differentiation [53, 54]. Several miRNAs have been reported to target the ESC transcriptional network and therefore be involved in silencing the self-renewal capacities of hESCs and mESCs during the early stages of their differentiation [62].
miR-145 is significantly upregulated upon differentiation of hESCs [48]. An increase of miR-145 represses the expression of pluripotency genes and facilitates differentiation, while the loss of miR-145 impairs differentiation and induces the expression of Oct4, Sox2, and Klf4 [63]. miR-145 controls ESC differentiation by directly targeting the stem cell factors, thereby silencing the self-renewal program. Interestingly, miR-145 promoter is repressed by OCT4 in hESCs, creating a double negative feedback loop [63].
In mESCs several miRNAs have been shown to promote differentiation by targeting genes encoding transcription factors involved in the maintenance of stem cell identity. miR-200c, miR-203 and miR-183 cooperate to repress Sox2 and Klf4 [64]. Upon retinoic-acid-induced differentiation of mESC, miR-134, miR-296 and miR-470 are up-regulated and target coding regions of Nanog, Oct4, and Sox2 [65].
When ESCs are engaged in a differentiation process, they need both to silence their self-renewal program and activate specific differentiated programs. It has recently been shown that let-7 is an important pro-differentiation factor that tightly controls the level of stem cell factors [66]. let-7 was one of the first miRNAs discovered for its role in the developmental timing of C. elegans [67]. pri-let-7 is transcribed in ESCs and pre-let-7 is found in their cytoplasm, however mature let-7 is not detected in undifferentiated ESCs while highly expressed in somatic cells. A study by Melton et al. revealed that let-7 can repress the mESC pluripotency program upon differentiation [66]. Re-introduction of mature let-7 family members into DGCR8 KO mESCs can rescue the differentiation defect by directly targeting transcripts of the self-renewal factors nMyc, Lin28 and Sal4. However let-7 family members had no effect when co-transfected with members of the ESCC miRNAs family, and let-7 did not induce differentiation in wild type mESCs. A model has been proposed in which let-7 and ESCC miRNA families oppose each other’s functions on ESC self-renewal: let-7 miRNAs repress pluripotency genes that are indirectly activated by ESCC miRNAs through an unknown target. Interestingly, as discussed earlier let-7 processing is negatively regulated by lin28 [31, 32] and lin28 expression is under the control of stem cell transcription factors cMyc, Oct4, Sox2 and Nanog [62]. Those results highlight how miRNAs are intricately integrated into the molecular network of pluripotency and are involved in switches crucial for cell fate decisions (Fig. 18.2).
While some miRNAs like miR-145, let-7 family or miR-200 family seem to reduce the pluripo-tency of ESCs, other miRNAs are involved in direct differentiation of ESCs toward a specialized lineages or terminally differentiated cell types. For example miR-133 and miR-1 are essential for the differentiation of ESCs into cardiomyocytes [68] and miR-9 promotes the differentiation into neuronal progenitors [69].
18.2.2 Role of microRNAs in Cellular Reprogramming
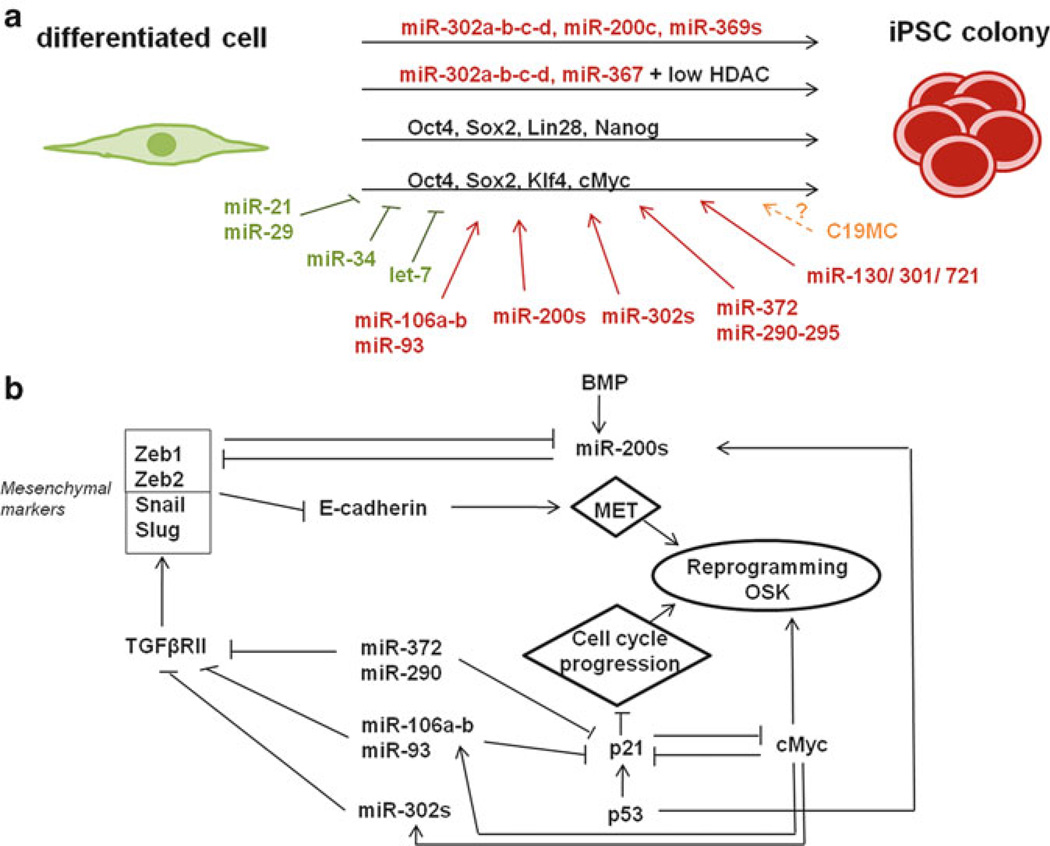
A huge breakthrough in the stem cell research field was achieved when Yamanaka group showed that it is possible to reprogram mouse embryonic fibroblasts into pluripotent cells, later called iPSCs, by ectopic expression of only four factors, Oct4, Sox2, Klf4 and cMyc (OSKM, Yamanaka factors) [43]. Omission of the oncogene cMyc from that cocktail still results in formation of iPSC colonies, though with a lower efficiency. This result has been repeated by several groups in human to reprogram various cell types from different tissues [42, 70, 71]. Besides the Yamanaka factors, another set of four factors can induce the generation of iPSCs, Oct4, Sox2, Lin28 and Nanog (OSLN, Thomson factors) [41]. Despite great efforts, the molecular mechanisms underlying the events of reprogramming remain mostly unknown. A growing numbers of studies are reporting an important role of miRNAs in reprogramming (Fig. 18.3a). This is not very surprising since, as mentioned earlier, miRNAs are critical for the balance between self-renewal and proliferation of ESCs.
Fig. 18.3. Functions of miRNAs in cellular reprogramming.
(a) Overview of the effects of miRNAs on iPSC formation. miRNAs beneficial for iPSC induction are represented in red while miRNAs shown to repress iPSC formation are in green. In orange are the microRNAs whose function has not been tested yet during reprogramming of somatic cells into pluripotent stem cells. (b) Mechanisms of action of miRNAs during the reprogramming process. OSK: Oct4, Sox2, Nanog. MET mesenchymal to epithelial transition
Live cell monitoring of iPSC generation from human fibroblasts using miRNAs reporter vectors shows that miR-302s, the most abundant hESC miRNAs, are expressed during the early stage of the OSKM-induced reprogramming [72]. miRNA pro filing and qPCR analysis revealed that other ESC-enriched miRNAs are induced early during iPSC formation, including the miR-17 family [73]. This was expected since Oct4 and Sox2, two of the transcription factors used to induce reprogramming, can activate the promoters of miR-302 and miR-106 clusters [44, 62]. Fully reprogrammed iPSCs have a similar miRNA pro file than ESCs. However imperfectly reprogrammed mouse cells have been shown to inappropriately silence the Dlk1-Dio3 locus, containing about 50 miRNAs [74]. Despite expression of genes associated with pluripotency, cells with a silenced Dlk1-Dio3 locus contribute poorly to chimaeras and seem to have limited capacities to differentiate into certain type of tissue-specific cells. Moreover, recent studies suggest that iPSCs might retain a memory of the cell of origin they come from [75]. It would be interesting to determine if this memory could be linked to miRNA expression.
Disruption of miRNA maturation or function by knock-down of Drosha, Dicer or Ago2 using lentiviral vectors dramatically reduces the number of iPSC colonies induced by OSKM or OSK in mouse embryonic fibroblasts (MEF), suggesting that some miRNAs are essential for the reprogramming process [73].
Introduction of ESC-Enriched microRNAs Enhances Reprogramming
One of the first evidence of the involvement of miRNAs in the formation of iPSCs comes from a study by Judson et al. They demonstrated that several members of the miR-290 cluster can increase the efficiency of OSK-induced reprogramming of MEF to a similar ef fi ciency as OSKM. Interestingly, introduction of a miR-294 mimic did not enhance OSKM-induced reprogramming, suggesting that miR-294 acts as a downstream target of c-Myc, and that miR-290s can substitute for cMyc contribution in cellular reprogramming. Indeed cMyc can bind to the promoter region of the mir-290–295 cluster [76], and bioinformatic analysis suggest that miR-294 may regulate a subset of c-Myc target genes [77]. ESCC miRNAs can promote cell cycle progression in ESCs by targeting inhibitors of the G1/S transition like p21 [53, 54]. Moreover, it has been shown that cMyc can repress p21 expression by downregulating its transcription [78] or at the post-transcriptional level through members of the miR-17 family [79]. Several groups have also reported that p53 and its downstream effectors antagonize iPSC induction, and knock-down of p21 in mouse fibroblasts increases reprogramming ef fi ciency [80–82]. Therefore, inhibition of p21 by miRNA and subsequent activation of proliferation could partly explain why ESCC miRNAs enhance reprogramming efficiency (Fig. 18.3b). However, unlike with cMyc, a homogeneous population of fully reprogrammed colonies was observed with miR-294, suggesting that ESCC miRNAs also have functions independent of cMyc’s [76].
Later, Li et al. proposed that miR-93 and miR-106b are key regulators of reprogramming activity. They found that they can enhance OSK and OSKM iPSC-induction in mouse by directly targeting p21 and TGF βR2 [73]. Ectopic expression in MEF by a retroviral vector of the miR-106a cluster (containing among others miR-20b) also increases reprogramming efficiency in OSK and OSKM iPSC-induction but with a greater effect with the three factors induction [83]. This result can be explained by the fact that cMyc can activate miR-106a cluster [38]. Members of the miR-302 cluster enhance reprogramming in both mouse and human, as well as miR-372 in human [76, 83, 84] and miR-130/301/721 in mouse [85].
In order to uncover the mechanisms behind this effect, a time course microarray analysis of the three factors plus or minus the miR-106a or miR-302 clusters have been performed in mouse [82]. This analysis showed that pathways changing at early time point during reprogramming with the addition of miR-106a cluster fall into three main groups: cell cycle, epigenetic modification and mesenchymal to epithelial transition (MET). Proteins belonging to those three groups were also found important miR-302 and miR-372 targets during OSK-induced reprogramming of human fibroblasts [84]. Direct targets include TGRβR2 and RHOC. Both are involved in MET. However miR-302a, b, c or d and 372 alone (without OSK reprogramming factors) were not able to induce the expression of epithelial markers [84]. The importance of MET in the reprogramming process has been highlighted recently and will be discussed in more details in the next section. Members of the miR-200 family have also been shown to facilitate the MET and improve reprogramming in mouse [86–88] (Fig. 18.3b).
Of note, all those studies were done with the Yamanaka factors. It will be interesting to see whether ESC-enriched miRNAs also enhance reprogramming induced by Thomson factors.
Other hESC-enriched miRNAs have been identified, but their potential role in reprogramming has not been investigated yet. In particular, some miRNAs of the C19MC cluster, containing miR-515 and miR-520 families, have different seed sequences than miRNAs enhancing iPSC formation, like miR-302, miR-372, miR-200 and miR-106. It will be critical in the future to test the function of these other hESC-enriched miRNAs in iPSC induction.
Inhibition of Tissue-Specific microRNAs Promotes Formation of iPSCs
Several studies came to the same conclusion that ESC-enriched miRNAs can enhance human and mouse reprogramming by targeting proteins involved in cell cycle, epigenetic modification and MET. miRNAs having a negative effect on those pathways have been shown to inhibit iPSC formation. During the dedifferentiation of somatic cells, important changes need to occur in their molecular signature : they have to acquire ESC-like signature but also have to down-regulate the tissue specific signature. miR-21 and miR-29a are the most abundant miRNAs in mouse fibroblasts and are downregulated during reprogramming by more than 50 %. It has recently been shown that inhibition of miR-21 and miR-29a using miR antagomirs enhances reprogramming efficiency through p53 downregulation [89]. miR-34 is also a target of p53 early during iPSC formation and constitutes a barrier for somatic cells reprogramming since genetic ablation of miR-34 in mice significantly promotes iPSC generation [90]. Moreover, cMyc can repress let7 family members indirectly through upregulation of lin28. Opposite effects of let-7 and ESCC miRNAs prompted researchers to test whether inhibition of let-7 has an effect on reprogramming. Indeed, antisens inhibitors of let7 modestly enhance reprogramming efficiency of MEF induced by OSK or OSKM [66].
Introduction of microRNAs Can Induce Reprogramming Without Other Factors
It was reported previously that introduction of a polycistronic cassette expressing miR-302a–b-c-d was sufficient to generate cells highly resembling to hESC from cancer cell lines and human hair follicule cells [91, 92]. Those cells, named miR-iPSC re-expressed hESC stem cell factors, their global gene expression was very close to hESC’s and they were able to differentiate into various lineages. However iPSC isolation and characterization were not well described and incomplete.
More recently, two independent groups have convincingly shown that human and mouse iPSCs can be derived from fibroblasts without the requirement of exogenous transcription factors by adding microRNAs [93, 94]. Anokye-Danso et al. demonstrated that lentiviral expression of the miR-302/367 cluster is able to reprogram MEF and human foreskin and dermal fibroblasts in a very rapid and efficient way [93]. miR-302/367-iPSC display similar self-renewal and pluripotency characteristic to OSKM-iPSC. miR-367, which has a different seed sequence than miR-302s, is required for reprogramming. Moreover low level of the histone deacetylase HDAC2 is also required, confirming the importance of chromatin modeling in iPSC reprogramming. Until now, the generation of iPSC from somatic cells was a very slow and inefficient process. In Anokye-Danso et al. study, not only miR-302/367 cluster improves the temporal kinetics of iPSC colony apparition, but also increases the efficiency by two orders of magnitude compared to existing protocols, when using similar viral titers. With a percentage approaching 10 % of human fibroblasts generating iPSCs, this method could be used in large-scale iPSC formation. The authors propose that such high efficiency could be explained by the nature of miRNAs themselves since a single miRNA can target hundreds of mRNAs simultaneously, hence coordinating several pathways and allowing a major phenotype change of the identity of the cell. miRNA derived-iPSCs have been called mi-iPSCs.
Shortly after this work was published, the Miyoshi et al. reprogrammed human and mouse multipotent adipose stromal cells as well as human dermal fibroblasts into pluripotent stem cells using seven miRNAs: 200c, 302a, 302b, 302c, 302d, 369-3p and 369-5p [94]. miRNAs were introduced by four transfections of mature double-stranded miRNAs within the first 8 days of reprogramming. The efficiency of generating mouse mi-iPSCs was similar to that seen in the original report of Yamanaka using OSKM induction in MEF. However the efficiency was considerably lower in human mi-iPSCs generated from human fibroblasts. More repeated transfections during the course of reprogramming might increase the efficiency of iPSC formation. Nonetheless, this study brings proof of principle that iPSCs can be obtained with miRNAs without the need for genomic integration of foreign DNA and might hold significant potential for both biomedical research and regenerative medicine. The miRNAs used in the Miyoshi et al. study belong to three families of miRNAs. The use of members of the miR-302 family confirmed previous studies showing that miR-302s can enhance OSK-induced iPSC formation or generate iPSC without other stem cell factors. miR-302 family appears as the most important miRNA family involved in reprogramming from human cells. As mentioned before, the promoter of miR-302 cluster is directly activated by Oct4 [44] and miR-302 have been shown to facilitate the MET during dedifferentiation of fibroblasts [83, 84]. We can wonder if miR-302 could be the equivalent of Oct4 in reprogramming since in any combination of stem cell factors, Oct4 is necessary for iPSC generation. It will be interesting to determine whether a combination of miRNA without miR-302 could also induce iPSC formation and whether miR-372 could replace miR-302s since they share the same seed sequence. Contrarily to the Anokye-Danso et al. study, miR-367 was not required to induce the reprogramming, but could be replaced by miR-200c, a miRNA important for MET, and members of the miR-369 family. Target prediction softwares suggest that miR-369s could also be involved in MET. Moreover, interestingly miR-369-3 is one of the few miRNAs that can up-regulate the translation of its target mRNAs on cell cycle arrest [29]. miR-302s, 367, 200c and 369 have different seed sequences, so both Anokye-Danso et al. and Miyoshi et al. protocols are likely to induce reprogramming through targeting of different mRNAs and pathways. Further studies should tell which combination of mature miRNAs is the best one and when each miRNA is involved during the course of iPSC formation. This would allow to determinate the best cocktail and timing of miRNA introduction in order to reach the maximum efficiency. It will also be interesting to investigate whether miR-induced reprogramming follows the same steps as OSKM or OSLN-induced reprogramming.
miRNAs can be powerful tools for reprogramming and consequently for therapeutic applications since they avoid integration of factors into the genome and can be used for large scale production of iPSCs.
18.2.3 Role of microRNAs in Cell Fate Transitions
Mesenchymal to Epithelial Transition
The epithelial-to-mesenchymal transition (MET) is the set of coordinated changes in cell-cell and cell-matrix interactions leading to loss of mesenchymal features and acquisition of epithelial characteristics. MET has been shown to play a pivotal role during embryonic development and its reverse process, the epithelial to mesenchymal transition (EMT), is important for cancer progression and invasion [95]. The process of reprogramming of fibroblasts resembles MET since it consists of transformation from single layer of adherent cells into tightly packed clusters of round ESC-like cells. MET seems to be a hallmark of the initiation phase characterized by an increase of epithelial-associated genes and a decrease of mesenchymal factors [96] siRNA against epithelial markers, in particular E-cadherin, totally inhibit the formation of iPSCs [86]. Therefore MET appears as a crucial step of fibroblasts dedifferentiation. Signaling pathways involved in the regulation of MET affect the efficiency of reprogramming and several miRNAs can regulate reprogramming by targeting proteins involved in the MET (Fig. 18.3b). As mentioned, members of the miR-200 family synergize with OSKM or other miRNAs to promote MEF reprogramming via regulation of MET by downregulating mesenchymal markers such as Zeb1 and Zeb2 [86–88, 94, 97, 98] Moreover miR-106a, miR-106b, miR-17, miR-93, and miR-302 cluster function in reprogramming is dependent of the fact that they all target TGFβR2, resulting in an increase of E-cadherin expression during fibroblast reprogramming [73, 83, 84]
The epithelial-to-mesenchymal transition (MET) is the set of coordinated changes in cell-cell and cell-matrix interactions leading to loss of mesenchymal features and acquisition of epithelial characteristics. MET has been shown to play a pivotal role during embryonic development and its reverse process, the epithelial to mesenchymal transition (EMT), is important for cancer progression and invasion [95]. The process of reprogramming of fibroblasts resembles MET since it consists of transformation from single layer of adherent cells into tightly packed clusters of round ESC-like cells. MET seems to be a hallmark of the initiation phase characterized by an increase of epithelial-associated genes and a decrease of mesenchymal factors [96] siRNA against epithelial markers, in particular E-cadherin, totally inhibit the formation of iPSCs [86]. Therefore MET appears as a crucial step of fibroblasts dedifferentiation. Signaling pathways involved in the regulation of MET affect the efficiency of reprogramming and several miRNAs can regulate reprogramming by targeting proteins involved in the MET (Fig. 18.3b). As mentioned, members of the miR-200 family synergize with OSKM or other miRNAs to promote MEF reprogramming via regulation of MET by downregulating mesenchymal markers such as Zeb1 and Zeb2 [86–88, 94, 97, 98] Moreover miR-106a, miR-106b, miR-17, miR-93, and miR-302 cluster function in reprogramming is dependent of the fact that they all target TGFβR2, resulting in an increase of E-cadherin expression during fibroblast reprogramming [73, 83, 84]
Fibroblasts are mesenchymal cells, however iPSCs have also been generated from other cell types and not all cells have to go through MET during reprogramming. A question comes to mind: would the miRNAs shown to enhance or induce reprogramming through MET activation have the same effect on reprogramming of epithelial somatic cells such as keratinocytes.
Transition Between Different Pluripotent States
It has been shown recently that expression of specific miRNAs can define the developmental state of ESCs and iPSCs [48]. hESCs are likely to be the in vitro equivalent of mouse epiblast stem cells (EpiSCs), derived from the post-implantation epiblast stage, while mESCs are derived from the inner cell mass of pre-implanted embryos and represent an earlier stage of embryonic development [99]. Low concentrations of sodium butyrate, a HDAC inhibitor, can induce hESCs to go back to an earlier developmental stage [100]. This method constitutes a useful tool to study the expression of miRNAs in early steps of human development. miR-372 cluster is expressed at higher levels in butyrate-treated hESCs than in hESCs while miR-302 cluster expression was slightly lower [48]. It would be important to analyze miR-302 and miR-372 expression levels in newly derived hESCs that might represent an earlier state of development. miR-302 cluster was expressed at considerably higher levels in EpiSCs than in mESCs [48]. miRNAs can be good indicators of the state of pluripotency, in particular miR-302 could be used as a marker for the epiblast stage in mouse. Moreover, it will be interesting to assess whether overexpression of miR-372 in hESCs can make them regress to an earlier developmental stage and whether over-expression of miR-302 in mESCs can in the contrary differentiate them toward an EpiSClike stage.
18.3 microRNA Function in Adult Stem Cells
Adult stem cells are undifferentiated cells which primary role is to replenish dying cells and repair damaged tissue. They can self-renew to maintain the pool of undifferentiated cells and they are multipotent, they are able to differentiate into progeny of cell types of the tissue in which they reside. Adult stem cells induce the regeneration of a specific tissue throughout adult life. They have been isolated from various tissues, including skin, brain, muscle and hematopoietic system. Adult stem cells reside in a specialized microenvironment, called niche, which regulates the balance between self-renewal, differentiation and quiescence. miRNAs have been shown to play an important role in the maintenance and differentiation of adult stem cell populations and are therefore essential regulators of the homeostasis of somatic tissues. In this section we will review very briefly the important miRNAs discovered in some adult stem cell populations.
18.3.1 Germline Stem Cells (GSCs)
GSCs have the potential to self-renew to maintain the GSC pool or to differentiate to give rise to gametes that are responsible for passing on their genetic material to the next generation. Mechanisms of maintenance and differentiation of GSCs have been well studied in a model organism, Drosophila M. In this fruit fly model microRNAs have shown to play a critical role in GSC regulation (reviewed in [101] ). Dicer-1 mutant GSCs are defective in cell cycle control and present a delayed G1/S transition due to an increase expression of Dacapo, a p21/p27 homolog [102]. mir-7 and miR-278 can target Dacapo, and GSC mutant for those miRNAs are partially defective in GSC division [103]. Moreover, Dicer-1 mutant GSCs are rapidly lost from the niche if the clones are generated during adult development, suggesting that miRNAs also control GSC maintenance and self-renewal [104–107]. Interestingly while loss of Dicer-1 in adult flies induces GSC loss, it does not induce a maintenance defect if the miRNAs processing enzyme is lost already during development [107]. This suggests that a dynamic compensation process takes place during development and emphasizes the capacity of maturing animal to protect the key cells for the continuity of the individual and the species, germ line stem cells. Some specific miRNAs, including miR-184, bantam and miR-7, have shown to regulate GSCs and their differentiation [107–109]. The results obtained in Drosophila ovarian GSCs demonstrate that miRNAs play a key role in GCS control, regulating maintenance, self-renewing division and differentiation.
Since Dicer KO reduces all microRNAs, the Dicer KO phenotype may be more complex than the phenotype caused by a loss of any individual microRNA. Nevertheless, Dicer KO experiments have been invaluable in clarifying the requirements of miRNAs and endo-siRNAs in different cell types. For example in mouse, Dicer KO causes dramatic gametogenesis defects suggesting that miRNAs or endo-siRNAs play key roles in these processes [110, 111]. Mouse spermatogenesis is a continuous, highly active process in adult males, indicating that self-renewing adult GSCs exist in testes lending fertile ground for detailed miRNA analysis in adult mammalian GSCs. Interestingly, recent highthroughput sequencing analysis has revealed that miR-21, along with miR-34c, −182, −183, and −146a, are enriched in male GSCs. Furthermore, reduction of miR-21 can induce male GSC apop-tosis [112].
The precursors for GSCs, primordial germ-cells (PGCs) are established during Epiblast stage in mouse development [113]. It therefore will be important to define whether key miRNAs in EpiSCs and hESCs control PGS differentiation. Recent paper addresses that pivotal question [114], a cluster of ESC-enriched miRNAs, miR-290–295 is shown to be critical for PGC migration. miR-290–295−/− PGCs show a significant reduction in their early developmental migration. However, the male germ line due to its GSC prolonged self-renewal capacity is able to recover, while females are sterile due to ovarian failure.
18.3.2 microRNAs and Hematopoietic Stem Cells (HSCs)
HSCs continuously replenish all cells in our blood throughout our lifetime and thereby are an excellent example of self-renewing adult stem cell population. The hematopoiesis is a complex process in which a common immature precursor differentiates into increasingly specialized populations of blood cells. This process is tightly regulated by a combination of transcription factors, epigenetic modifications, miRNAs and extrinsic signals from the niche. miRNAs have appeared critical in almost every stage of hematopoiesis (reviewed in [115] ). miR-181, miR-150 and miR-155 control lymphocyte development [116, 117], while miR-223 is involved in the regulation of both myeloid and erythroid differentiation [116, 118–120]. miR-130a and miR-10a are important for megakaryocytic maturation. miR-221 and miR-222 are downregulated during eryth-roid differentiation and they both target KIT receptor, an important regulator of the proliferation of hematopoietic cells [115].
miRNAs can regulate hematopoietic differentiation but can also regulate the HSC reservoir. For example, miR-125a controls HSC population size by inhibiting their apoptosis via translational repression of the pro-apoptotic protein Bak1 [28]. Similarly, miR-125b is highly expressed in HSCs and regulates their survival. In addition, miR-125b promotes lymphoid fate decision [121] and block G-CSF-induced granulocytic differentiation [122].
18.3.3 microRNAs and Neural Stem Cells (NSCs)
NSCs are localized in few specific zones within the brain. They can self-renew or produce progenitors that can engage in neurogenesis and give rise to neuronal and glial lineages. Recent evidences highlight the function of miRNAs in the regulation of NSC self-renewal and neurogenesis. miR-124, the most abundant miRNA in adult brain, is expressed at low level in NSCs and is upregulated in adult neurons. mir-124 has been shown to induce neural differentiation by directly targeting Sox9 and RE1-silencing transcription factor (REST) [32, 123]. miR-9, another brain specific miRNA, regulates NSC differentiation by suppressing NSC factors such as REST or the orphan nuclear receptor TLX [69]. Similarly, let-7b overexpression inhibits NSC proliferation and accelerates neural differentiation by targeting TLX and cyclinD1 involved in the regulation of cell cycle progression [69].
18.3.4 microRNAs and Muscle Stem Cells
One of the main players of adult muscle regeneration is the skeletal adult stem cell found in mature muscle and called satellite cell. Satellite cells have the capacity to differentiate and fuse with each others to form muscle fibers. miRNAs have been shown to be important regulators of muscle cell fate decision. For example miR-1, miR-206 and miR-486 downregulate Pax7, a protein required to maintain the muscle stem cell population. Overexpression of those miRNAs induces differentiation of satellite cells and myoblasts [124, 125], while miR-221 and miR-222 play a role in the progression from myoblasts to myocytes [126]. In the contrary, miR-125b negatively regulates myoblast differentiation [127].
18.3.5 microRNAs and Skin Stem Cells
Skin stem cells are localized in the basal layer of the epidermis or at the base of hair follicles. miR-203 the most abundant miRNA in mammalian skin, has been identified as an inhibitor of “stemness” in epidermal stem cells [128]. miR-203 promotes epidermal differentiation by repressing p63, resulting in restrictive proliferative potential and induction of cell-cycle exit [128, 129]. miR-125b is preferentially expressed in skin stem cells and is implicated in the balance between self-renewal and early lineage commitment [130]. The authors propose that miR-125b regulates the number of divisions that a progenitor undergoes prior to committing to a lineage.
As mentioned, miR-125b also regulates HSCs and muscle stem cells, suggesting that some miRNAs might be common regulators of various adult stem cells.
18.4 microRNA Function in Cancer Stem Cells
18.4.1 Cancer Stem Cells (CSCs)
A tumor contains a very heterogeneous population of cells at various stages of differentiation [131]. Some cancer cells have been shown to share similarities with stem cells, specially the capacity to self-renew with uncontrolled division and the ability to produce differentiated progenies. Those cells were named cancer stem cells or tumor-initiating cells because of their potential to regenerate the entire heterogeneous tumor [132]. Such cells are believed to be highly aggressive and resistant to chemotherapy and have been presented as a possible explanation to relapse. The first CSCs were identified in leukemia [133]. Since then, CSC populations have been isolated in various different tissues, including breast, prostate, brain, colon and head and neck cancer [134–138]. However, the exact nature of CSC is still not known [139]. Recent data suggest that poorly differentiated aggressive human tumors and CSCs possess hESC-like gene expression signature [140–142]. Furthermore hESC markers such as Oct4, Sox2, Nanog or Lin28 are overex-pressed in CSCs and can promote transformation [143, 144].
It is not quite understood where CSCs come from. Evidences show that they could arise from either transformation of adult stem cells, progenitor cells or reprogramming of cancer cells [145]. Their number within the tumor seems to vary greatly between cancer cell types [146]. The hypothesis that generation of CSCs could be a dynamic process regulated by the microenvironment of the cells has emerged recently [147]. For example recent evidence suggests that hypoxia can dedifferentiate cancer to a more potent and aggressive stem cell stage [144]. Hypoxia can induce the expression of CSC and hESC markers in cancer cells, correlating with tumor aggressiveness and apparition of stem cell-like populations [144]. Cancer cells cultured under hypoxia also express a higher level of ESC-enriched miRNAs compared to cancer cells cultured under normoxia [144]
18.4.2 miRNAs in CSCs
As discussed in the previous section, miRNAs have been associated with ESC self-renewal and differentiation. They can also be prognosis factors in various cancers [148], and act as either tumor suppressors or oncogenes (oncomiR) [149]. Since miRNAs are critical for both cancer and stem cell properties, their role in CSC self-renewal, proliferation and differentiation is under intense study and they hold great hope for therapy. Recent pro filing data performed in newly isolated CSCs map miRNAs that are up or down regulated in CSCs compared to non-stem cancer cells or normal stem cell population arising from the same tissue (Table 18.1, [171]). CSCs seem to display a distinct signature of miRNAs that varies depending on the tissue of origin [171]. Some variations were also observed among CSCs from the same tissue, however this could be explained by the different methods used to isolate CSCs. Functional studies show that miRNAs are major components of acquisition and maintenance of “stemness” of CSCs [171]. It is interesting that several of the miRNAs found to control CSC properties are miRNAs involved in the regulation of ESC self-renewal and differentiation as well as in the reprogramming of fibroblasts into iPSCs. For example, miR-371–373 cluster is upregulated in undifferentiated aggressive hepatocellular cancer cells [164]. In breast cancer cells, C19MC cluster and miR-371–373 cluster are linked to high aggressiveness and promote tumor invasion and metastasis by targeting CD44 [157]. Those clusters are also activated by chromosomal rearrangement in a subgroup of thyroid adenoma [158]
Table 18.1.
List of miRNAs regulating tumor aggressiveness, invasion and CSC properties
| Upregulated in aggressive tumors and CSC | Downregulated in aggressive tumors and CSC | ||||
|---|---|---|---|---|---|
| miRNA | Cancer typed | Ref | miRNA | Cancer type | Ref |
| miR-130b | Liver | [150] | miR-125b | Glioma | [151] |
| miR-17-92 | Leukemia | [152] | miR-200s | Lung, ovary, head and neck, liver, pancreas, breast | [153–156] |
| C19MC | Thyroid, breast | [157, 158] | let-7 | Lung, breast, liver, head and neck | [153, 159–163] |
| miR-371-373 | Liver, thyroid, breast | [157, 158, 164] | miR-34 | Pancreas, stomach, glioma, prostate | [165–169] |
| miR-145 | Ewing sarcoma | [170] | |||
miR-17–92 polycistron is more abundant in leukemic stem cells than in non-stem leukemic cells or than in their normal counterpart precursors [152]. miR-17–92 cluster regulates mixed lineage leukemia (MLL) stem cells by targeting p21, resulting in more proliferative cells [152]. In addition, miR-17–92 cluster increases self-renewal and leukemic stem cell potential [152]. miR-130b is upregulated in CD133+ liver cancer stem cells compared to CD133- cells and promotes liver CSC growth and self-renewal via targeting of TP53INP1 [150]. Overexpression of miR-130b increases tumorigenicity in vivo as well as resistance to chemotherapeutic agents, while inhibition of miR-130b has inverse effects.
While ESC-enriched miRNAs seem to be important in CSC functions, miRNAs shown to be repressing pluripotency are inhibiting CSC properties. Members of the let-7 family have a role of tumor suppressor by targeting K-Ras and cMyc, and their expression is repressed in lung, breast, liver and head and neck CSCs [153, 159–162]. Overexpression of let-7 decreases the stemness signature of various CSCs and increases their chemosensitivity. In particular, it has been shown that let-7 can regulate breast CSCs properties. Indeed let-7 overexpression reduces proliferation, mammosphere formation, and the proportion of undifferentiated cells in vitro, as well as tumor formation and metastasis in vivo [163].
Unexpectedly, members of the miR-200 family, which are enriched in ESCs and play a role in iPSC induction, are downregulated in CSCs isolated from lung, ovarian, head and neck, liver, pancreatic and breast cancer compared to their non-stem cancer counterparts [153, 154]. As discussed earlier, members of the miR-200 family are major activators of the MET by targeting mesenchymal markers, which results in expression of epithelial markers. Therefore expression of miR-200 represses EMT. The process of EMT plays an important role in the progression of cancer by promoting invasion and metastasis [95] A recent hypothesis proposes that cells undergoing EMT have very similar properties than CSCs and several reviews discuss the relationship between those two populations of cells [172–174]. In pancreatic cancer cells, Notch1 has been shown to inhibit miR-200b and miR-200c and induces EMT consistent with CSC phenotype as monitored by pancreatosphere and expression of CSC markers [155]. Members of miR-200 family also inhibit migration and metastasis of ovarian CSCs and head and neck squamous cell CSCs via repression of Zeb1 and Zeb2 [154]. In breast cancer, miR-200c targets proteins involved in invasiveness, resistance to apoptosis and induction of breast CSC characteristics [156].
miRNAs shown to be regulators of various adult stem cell functions also play important roles in CSCs regulation. For instance, miR-125b suppresses glioma SC proliferation by targeting CDK6 an CDC25A, thus inducing cell cycle arrest in G1 [151], and miR-21 induces “stemness” in colon cancer cells [175].
Another miRNA playing an important function in CSCs is miR-34. As mentioned, miR-34 reduces reprogramming efficiency [90]. Its expression is downregulated in many cancers and its target mRNAs code for proteins involved in the inhibition of apoptosis, cell cycle progression and migration such as E2F3, Notch, CyclinD or Bcl2 [165]. Those mechanisms are involved in CSC self-renewal and survival. In particular Bcl2 has been involved in chemo- and radio-resistance of CSCs by preventing apoptosis. When miR-34 is overexpressed pancreatic and gastric cancer cells are more sensitive to chemotherapeutic drugs, while tumor growth and tumosphere formation are inhibited [166, 167]. miR-34 also has a function of tumor suppressor in brain tumor and glioma SC and induces glioma SC differentiation [168]. Very recently miR-34 has been shown to directly target CD44, one of the markers of prostate CSCs. Introduction of miR-34a by liposome-based delivery in prostate CSCs decreases the clonogenicity in vitro and the tumor growth in vivo [169]. In this work, the authors achieved an efficient systemic delivery of miRNAs and opened new avenues for cancer therapy by targeting CSC maintenance.
miRNAs play important roles in CSC proliferation, differentiation and tumor formation. Therapy targeting CSC could potentially eliminate cancer at its source and avoid relapses to occur. The potential advantage of using miRNAs is that they can simultaneously silence several molecules that regulate CSCs. However such strategy presents challenges. Only CSCs should be destroyed, while normal stem cells should be left intact. Moreover it would be necessary to kill all CSCs since a single CSC could potentially re-grow an entire tumor.
18.5 Conclusion
In less than 10 years, miRNAs have experienced a radical shift in people’s mind, once considered as junk RNA not useful for the individual, they now appear as critical regulators of most cellular events. By their ability to target hundreds of mRNAs they can induce a rapid switch in cell fate and fine tune genome expression. They are now accepted as major post-transcriptional regulators. In this chapter we discussed the central role of miRNAs in controlling proliferation, survival, self-renewal and differentiation of various stem cells. We particularly focused on miRNA function in pluripotent cells and acquisition of stem cell fate, for instance during the reprogramming process. We then briefly reviewed the role of miRNAs in adult stem cells and tumor initiating cells, “cancer stem cells”, that share properties with stem cells.
miRNAs regulate target genes involved in key cellular processes regulating stem cell biology. Those miRNAs and their targets are under intense investigation. Among hundreds of predicted targets, few of them have been identified experimentally. In the next few years, more investigations should shed light on miRNA targets and interactions with stem cell markers, signaling pathways, and epigenetic regulatory mechanisms. This should allow us to build a more comprehensive understanding of stem cell identity and behavior. One mRNA can be targeted by several miRNAs, and because of the unique signature of miRNAs in particular cells at a given time and conditions, the combination of miRNA expression can lead to a specific outcome.
miRNAs are key players in the control of the abbreviated cell cycle of ESC proliferation and are therefore implicated in proliferation of stem cells and their self-renewal capacities. They also regulate the differentiation and de-differentiation of cells. Indeed, it has been shown that specific miRNAs can enhance or repress iPSC formation. Several groups have also demonstrated that miRNAs alone can reprogram somatic cells into pluripotent stem cells, underlying their fundamental role in cell fate decision.
Expression pro files revealed miRNAs enriched in pluripotent stem cells, adult stem cells and CSCs. Interestingly some miRNAs regulating ESCs are found important in regulating the CSC phenotype. Indeed ESC-enriched miRNAs such as miR-17–92 cluster, miR-371–373 cluster, C19CM or miR-130 induce “stemness” and aggressiveness in cancer cells, while let-7 and miR-200 inhibit CSC properties. Despite its central role in ESC maintenance and reprogramming, miR-302 has not been shown directly implicated in CSC formation, proliferation or maintenance. However miR-302 is upregulated after exposure of cancer cells to hypoxia and correlates with increase of stem cell properties [144]. Moreover miR-302 is activated by stem cells factors regulating the “stemness” of CSCs. More data are needed to understand the role of miR-302 family in CSC functions.
miRNAs present a great potential in regenerative medicine and cancer therapy. miRNAs can be used to efficiently generate induced pluripotent stem cells without DNA integration from patient cells in order to model diseases and obtain a reservoir of cells. They can also be used to differentiate pluripotent cells into cells of particular lineages for potential cell therapies. Moreover, targeting specific miRNAs in cancer treatment could constitute a new approach to eradicate CSCs and avoid relapses. Full understanding of miRNA functions in stem cell fate and differentiation will be essential to take advantage of miRNA therapeutic promises.
References
- 1.Baek D, Villen J, Shin C, Camargo FD, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genom Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 7.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 8.Yi R, Fuchs E. MicroRNAs and their roles in mammalian stem cells. J Cell Sci. 2011;124(Pt 11):1775–1783. doi: 10.1242/jcs.069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 10.Berezikov E, Guryev V, van de Belt J, Wienholds E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Kim M, Han J, Yeom KH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Ahn C, Han J, Choi H, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 15.Denli AM, Tops BB, Plasterk RH, Ketting RF, et al. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 16.Gregory RI, Yan KP, Amuthan G, Chendrimada T, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 17.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38(3):323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz DS, Hutvagner G, Du T, Xu Z, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 21.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 22.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465(7298):584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cifuentes D, Xue H, Taylor DW, Patnode H, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328(5986):1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 25.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69(8):3245–3248. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 26.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 27.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 28.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 30.Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem. 2010;148(4):381–392. doi: 10.1093/jb/mvq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heo I, Joo C, Cho J, Ha M, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32(2):276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Visvanathan J, Lee S, Lee B, Lee JW, et al. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21(7):744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20(16):2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchida A, Ohno S, Wu W, Borjigin N, et al. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011;102(12):2264–2271. doi: 10.1111/j.1349-7006.2011.02081.x. [DOI] [PubMed] [Google Scholar]
- 35.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 36.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 37.Becker KA, Ghule PN, Therrien JA, Lian JB, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209(3):883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 38.Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol. 2011;13(5):490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 39.Boyer LA, Lee TI, Cole MF, Johnstone SE, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loh YH, Wu Q, Chew JL, Vega VB, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 41.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi K, Tanabe K, Ohnuki M, Narita M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Marson A, Levine SS, Cole MF, Frampton GM, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5(2):351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 46.Bar M, Wyman SK, Fritz BR, Qi J, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26(10):2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stadler B, Ivanovska I, Mehta K, Song S, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19(7):935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernstein E, Kim SY, Carmell MA, Murchison EP, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 50.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murchison EP, Partridge JF, Tam OH, Cheloufi S, et al. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babiarz JE, Ruby JG, Wang Y, Bartel DP, et al. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev. 2008;22(20):2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Baskerville S, Shenoy A, Babiarz JE, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40(12):1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi J, Yu JY, Shcherbata HR, Mathieu J, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8(22):3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fluckiger AC, Marcy G, Marchand M, Negre D, et al. Cell cycle features of primate embryonic stem cells. Stem Cells. 2006;24(3):547–556. doi: 10.1634/stemcells.2005-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Becker KA, Ghule PN, Lian JB, Stein JL, et al. Cyclin D2 and the CDK substrate p220(NPAT) are required for self-renewal of human embryonic stem cells. J Cell Physiol. 2010;222(2):456–464. doi: 10.1002/jcp.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faast R, White J, Cartwright P, Crocker L, et al. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a) Oncogene. 2004;23(2):491–502. doi: 10.1038/sj.onc.1207133. [DOI] [PubMed] [Google Scholar]
- 58.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15(3):259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 59.Sengupta S, Nie J, Wagner RJ, Yang C, et al. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells. 2009;27(7):1524–1528. doi: 10.1002/stem.84. [DOI] [PubMed] [Google Scholar]
- 60.Benetti R, Gonzalo S, Jaco I, Munoz P, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15(3):268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Medvid R, Melton C, Jaenisch R, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tiscornia G, Izpisua Belmonte JC. MicroRNAs in embryonic stem cell function and fate. Genes Dev. 2010;24(24):2732–2741. doi: 10.1101/gad.1982910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, et al. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 64.Wellner U, Schubert J, Burk UC, Schmalhofer O, et al. The EMT-activator ZEB1 promotes tum-origenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 65.Tay Y, Zhang J, Thomson AM, Lim B, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 66.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463(7281):621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 68.Takaya T, Ono K, Kawamura T, Takanabe R, et al. MicroRNA-1 and MicroRNA-133 in spontaneous myocardial differentiation of mouse embryonic stem cells. Circ J Off J Jpn Circ Soc. 2009;73(8):1492–1497. doi: 10.1253/circj.cj-08-1032. [DOI] [PubMed] [Google Scholar]
- 69.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16(4):365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park IH, Lerou PH, Zhao R, Huo H, et al. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3(7):1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 71.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13(5):497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kamata M, Liang M, Liu S, Nagaoka Y, et al. Live cell monitoring of hiPSC generation and differentiation using differential expression of endogenous microRNAs. PLoS One. 2010;5(7):e11834. doi: 10.1371/journal.pone.0011834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30(5):823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465(7295):175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim K, Doi A, Wen B, Ng K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27(5):459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanina SA, Mifsud W, Down TA, Hayashi K, et al. Genome-wide identification of targets and function of individual MicroRNAs in mouse embryonic stem cells. PLoS Genet. 2010;6(10):e1001163. doi: 10.1371/journal.pgen.1001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor in fluences the outcome of the p53 response to DNA damage. Nature. 2002;419(6908):729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 79.Wang Z, Liu M, Zhu H, Zhang W, et al. Suppression of p21 by c-Myc through members of miR-17 family at the post-transcriptional level. Int J Oncol. 2010;37(5):1315–1321. [PubMed] [Google Scholar]
- 80.Utikal J, Polo JM, Stadtfeld M, Maherali N, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460(7259):1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marion RM, Strati K, Li H, Murga M, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hong H, Takahashi K, Ichisaka T, Aoi T, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liao B, Bao X, Liu L, Feng S, et al. MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem. 2011;286(19):17359–17364. doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Subramanyam D, Lamouille S, Judson RL, Liu JY, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29(5):443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pfaff N, Fiedler J, Holzmann A, Schambach A, et al. miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep. 2011;12(11):1153–1159. doi: 10.1038/embor.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, et al. Functional genomics reveals a BMPdriven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7(1):64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 87.Li R, Liang J, Ni S, Zhou T, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 88.Gregory PA, Bert AG, Paterson EL, Barry SC, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 89.Yang CS, Li Z, Rana TM. MicroRNAs modulate iPS cell generation. RNA. 2011;17(8):1451–1460. doi: 10.1261/rna.2664111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi YJ, Lin CP, Ho JJ, He X, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13(11):1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin SL, Chang DC, Chang-Lin S, Lin CH, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14(10):2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin SL, Chang DC, Lin CH, Ying SY, et al. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39(3):1054–1065. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miyoshi N, Ishii H, Nagano H, Haraguchi N, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8(6):633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 95.Cannito S, Novo E, di Bonzo LV, Busletta C, et al. Epithelial-mesenchymal transition: from molecular mechanisms, redox regulation to implications in human health and disease. Antioxid Redox Signal. 2010;12(12):1383–1430. doi: 10.1089/ars.2009.2737. [DOI] [PubMed] [Google Scholar]
- 96.Polo JM, Hochedlinger K. When fibroblasts MET iPSCs. Cell Stem Cell. 2010;7(1):5–6. doi: 10.1016/j.stem.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 97.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 98.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283(22):14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 100.Ware CB, Wang L, Mecham BH, Shen L, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4(4):359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reynolds S, Ruohola-Baker H. In: StemBook (ed) The stem cell research community. Cambridge, MA: Harvard Stem Cell Institute; Sep 15, 2008. The role of microRNAs in germline differentiation. StemBook http://www.stembook.org 2008, PMID: 20614619. [PubMed] [Google Scholar]
- 102.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, et al. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435(7044):974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 103.Yu JY, Reynolds SH, Hatfield SD, Shcherbata HR, et al. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development. 2009;136(9):1497–1507. doi: 10.1242/dev.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang Y, Xu S, Xia L, Wang J, et al. The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009;5(4):e1000444. doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park JK, Liu X, Strauss TJ, McKearin DM, et al. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol CB. 2007;17(6):533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- 106.Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol CB. 2007;17(6):539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 107.Shcherbata HR, Ward EJ, Fischer KA, Yu JY, et al. Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell. 2007;1(6):698–709. doi: 10.1016/j.stem.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pek JW, Lim AK, Kai T. Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Dev Cell. 2009;17(3):417–424. doi: 10.1016/j.devcel.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 109.Iovino N, Pane A, Gaul U. miR-184 has multiple roles in Drosophila female germline development. Dev Cell. 2009;17(1):123–133. doi: 10.1016/j.devcel.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 110.Murchison EP, Stein P, Xuan Z, Pan H, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21(6):682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hayashi K, de Sousa C, Lopes SM, Kaneda M, Tang F, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3(3):e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Niu Z, Goodyear SM, Rao S, Wu X, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2011;108(31):12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hayashi K, Ohta H, Kurimoto K, Aramaki S, et al. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146(4):519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 114.Medeiros LA, Dennis LM, Gill ME, Houbaviy H, et al. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc Natl Acad Sci U S A. 2011;108(34):14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vasilatou D, Papageorgiou S, Pappa V, Papageorgiou E, et al. The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol. 2010;84(1):1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- 116.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 117.Xiao C, Calado DP, Galler G, Thai TH, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 118.Felli N, Pedini F, Romania P, Biffoni M, et al. MicroRNA 223-dependent expression of LMO2 regulates normal erythropoiesis. Haematologica. 2009;94(4):479–486. doi: 10.3324/haematol.2008.002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fazi F, Rosa A, Fatica A, Gelmetti V, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 120.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 121.Ooi AG, Sahoo D, Adorno M, Wang Y, et al. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci U S A. 2010;107(50):21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, et al. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood. 2011;117(16):4338–4348. doi: 10.1182/blood-2010-06-289058. [DOI] [PubMed] [Google Scholar]
- 123.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12(4):399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen JF, Tao Y, Li J, Deng Z, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190(5):867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dey BK, Gagan J, Dutta A. miR-206 and −486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31(1):203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cardinali B, Castellani L, Fasanaro P, Basso A, et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4(10):e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192(1):69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452(7184):225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lena AM, Shalom-Feuerstein R, di Val R, Cervo P, Aberdam D, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15(7):1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 130.Zhang L, Stokes N, Polak L, Fuchs E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell. 2011;8(3):294–308. doi: 10.1016/j.stem.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138(5):822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 132.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124(6):1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 133.Lapidot T, Sirard C, Vormoor J, Murdoch B, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 134.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Singh SK, Hawkins C, Clarke ID, Squire JA, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 136.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 137.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, et al. Identification and expansion of human coloncancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 138.Hermann PC, Huber SL, Herrler T, Aicher A, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 139.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 140.Ben-Porath I, Thomson MW, Carey VJ, Ge R, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wong DJ, Liu H, Ridky TW, Cassarino D, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2(4):333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chiou SH, Wang ML, Chou YT, Chen CJ, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70(24):10433–10444. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- 144.Mathieu J, Zhang Z, Zhou W, Wang AJ, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71(13):4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wu XZ. Origin of cancer stem cells: the role of self-renewal and differentiation. Ann Surg Oncol. 2008;15(2):407–414. doi: 10.1245/s10434-007-9695-y. [DOI] [PubMed] [Google Scholar]
- 146.Quintana E, Shackleton M, Sabel MS, Fullen DR, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Borovski T, De Sousa EMF, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer Res. 2011;71(3):634–639. doi: 10.1158/0008-5472.CAN-10-3220. [DOI] [PubMed] [Google Scholar]
- 148.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 149.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 150.Ma S, Tang KH, Chan YP, Lee TK, et al. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7(6):694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 151.Shi L, Zhang J, Pan T, Zhou J, et al. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res. 2010;1312:120–126. doi: 10.1016/j.brainres.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 152.Wong P, Iwasaki M, Somervaille TC, Ficara F, et al. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70(9):3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lo WL, Yu CC, Chiou GY, Chen YW, et al. MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J Pathol. 2011;223(4):482–495. doi: 10.1002/path.2826. [DOI] [PubMed] [Google Scholar]
- 154.Wu Q, Guo R, Lin M, Zhou B, et al. MicroRNA-200a inhibits CD133/1+ ovarian cancer stem cells migration and invasion by targeting E-cadherin repressor ZEB2. Gynecol Oncol. 2011;122(1):149–154. doi: 10.1016/j.ygyno.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 155.Bao B, Wang Z, Ali S, Kong D, et al. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett. 2011;307(1):26–36. doi: 10.1016/j.canlet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 156.Chang CJ, Chao CH, Xia W, Yang JY, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13(3):317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huang Q, Gumireddy K, Schrier M, le Sage C, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 158.Rippe V, Dittberner L, Lorenz VN, Drieschner N, et al. The two stem cell microRNA gene clusters C19MC and miR-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS One. 2010;5(3):e9485. doi: 10.1371/journal.pone.0009485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yu CC, Chen YW, Chiou GY, Tsai LL, et al. MicroRNA let-7a represses chemoresistance and tumourigenicity in head and neck cancer via stemlike properties ablation. Oral Oncol. 2011;47(3):202–210. doi: 10.1016/j.oraloncology.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 160.Yu F, Deng H, Yao H, Liu Q, et al. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29(29):4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 161.Kong D, Banerjee S, Ahmad A, Li Y, et al. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One. 2010;5(8):e12445. doi: 10.1371/journal.pone.0012445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yang X, Lin X, Zhong X, Kaur S, et al. Double-negative feedback loop between reprogramming factor LIN28 and microRNA let-7 regulates aldehyde dehydrogenase 1-positive cancer stem cells. Cancer Res. 2010;70(22):9463–9472. doi: 10.1158/0008-5472.CAN-10-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yu F, Yao H, Zhu P, Zhang X, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 164.Cairo S, Wang Y, de Reynies A, Duroure K, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A. 2010;107(47):20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 166.Ji Q, Hao X, Meng Y, Zhang M, et al. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ji Q, Hao X, Zhang M, Tang W, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4(8):e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Guessous F, Zhang Y, Kofman A, Catania A, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9(6):1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu C, Kelnar K, Liu B, Chen X, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Riggi N, Suva ML, De Vito C, Provero P, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24(9):916–932. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300(1):10–19. doi: 10.1016/j.canlet.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Floor S, van Staveren WC, Larsimont D, Dumont JE, et al. Cancer cells in epithelial-to-mesenchymal transition and tumor-propagating-cancer stem cells: distinct, overlapping or same populations. Oncogene. 2011;30:4609–4621. doi: 10.1038/onc.2011.184. [DOI] [PubMed] [Google Scholar]
- 173.Kong D, Li Y, Wang Z, Sarkar FH. Cancer stem cells and epithelial-to-mesenchymal transition (EMT)-phenotypic cells: are they cousins or twins? Cancers. 2011;3(1):716–729. doi: 10.3390/cancers30100716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Scheel C, Weinberg RA. Phenotypic plasticity and epithelial-mesenchymal transitions in cancer - and normal stem cells? Int J Cancer J Int du Cancer. 2011;129(10):2310–2314. doi: 10.1002/ijc.26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yu Y, Kanwar SS, Patel BB, Oh PS, et al. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFbetaR2) in colon cancer cells. Carcinogenesis. 2011;1(11):e32. doi: 10.1093/carcin/bgr246. [DOI] [PMC free article] [PubMed] [Google Scholar]