Abstract
A heterologous expression system was used to evaluate activation of BlaR1, a sensor/signal transducer protein of Staphylococcus aureus with a central role in resistance to β-lactam antibiotics. In the absence of other S. aureus proteins that might respond to antibiotics and participate in signal transduction events, we documented that BlaR1 fragmentation is autolytic, that it occurs in the absence of antibiotics, and that BlaR1 directly degrades BlaI, the gene repressor of the system. Furthermore, we disclosed that this proteolytic activity is metal ion-dependent and that it is not modulated directly by acylation of the sensor domain by β-lactam antibiotics.
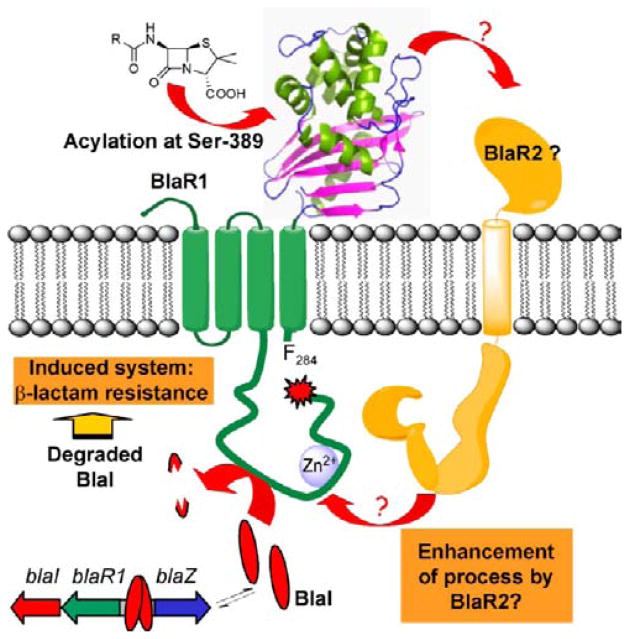
Over the past ~60 years, Staphylococcus aureus has become progressively more resistant to antibiotics, a transformation that underscores why this organism has become a clinical scourge.1–4 Among the mechanisms of resistance that have emerged in this organism, that of inducible resistance to β-lactam antibiotics is as elaborate as it is effective.5,6 Resistance to β-lactam antibiotics in strains that have come to be known as methicillin-resistant S. aureus (MRSA) is virtually complete. A key player in resistance to β-lactam antibiotics in S. aureus is the membrane-bound BlaR1 protein (Figure 1).5,7,8 This protein decorates the surface of the plasma membrane with a sensor domain that binds covalently to β-lactam antibiotics at Ser-389.9–12 This information is transduced to the cytoplasmic end of the protein, most likely through conformational changes.9,13,14 This event is proposed to lead to activation of the cytoplasmic domain of BlaR1, a putative metalloprotease,5 which either directly or through intervention of other effectors degrades the gene repressor BlaI. This indirect involvement is invoked for an as-yet-unidentified hypothetical regulator, the product of the so-called R2 locus, designated subsequently as BlaR2, unlinked to the penicillinase plasmid.5,15 The gene repressor BlaI binds to the bla operator, thus repressing transcription of the genes of the operon.16–19 Proteolysis of BlaI derepresses transcription of the genes of the bla operon for BlaI, BlaR1, and the PC1 β-lactamase, which it controls (Figure 1).5,8,19–21 Expression of β-lactamase gives the phenotype of antibiotic resistance, but that for BlaR1 maintains recruitment of resources for the manifestation of resistance. Expression of BlaI sets the organism en route to transcriptional repression, once the antibiotic challenge is no longer there. Notwithstanding the long-standing interest in BlaR1, its low expression level in S. aureus has been problematic in its study. This study of heterologous expression of BlaR1 in Escherichia coli was undertaken to address this difficulty and to dissect its biochemical events. Most importantly, we documented the steps inherent to the activity of BlaR1 itself, and the steps that might require the hypothetical mediator protein BlaR2 from S. aureus, if it were to exist. The expectation would be that BlaR2 would not be present in the unrelated E. coli, as this organism has not undergone evolution of a resistance mechanism to β-lactams akin to that of MRSA.
Figure 1.
Schematic of the proposed mechanisms of activation and turnover of BlaR1 and BlaI in response to β-lactam antibiotics in S. aureus.
EXPERIMENTAL PROCEDURES
Expression of Recombinant BlaR1 in E. coli
The previously described construct pET-24a(+)_blaRHis6x was used to evaluate the expression of BlaR1 in E. coli.8 Briefly, the blaR1 gene was amplified from plasmid pI258 of S. aureus NRS128 using the polymerase chain reaction (PCR) and cloned in vector pET-24a(+) (Novagen) between NdeI and XhoI sites. This construct, designated pET-24a(+)_blaRHis6x, allowed isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible expression of BlaR1 in E. coli from the T7 promoter, with eight additional amino acids at the C-terminus (LEHHHHHH) (the first two amino acids introduced by the XhoI cloning site and the His6x tag). Recombinant BlaR1 (designated BlaR1.H6x) was expressed in E. coli BL21 Star(DE3) harboring plasmid pET-24a(+)_blaRHis6x. The cells were grown at 37°C until the OD625 reached 0.9, at which point the cultures were rapidly cooled to 28 °C, protein expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and cells were grown for 5 h at 28 °C. Membrane proteins were prepared by ultracentrifugation from a 500 mL culture, as previously described.8
Cloning, Expression, and Purification of T7.BlaI
The blaI gene from plasmid pI258 of S. aureus NRS128 was previously cloned in the lab to generate plasmid pET_blaI.17 To fuse an N-terminal T7.tag to BlaI (designated T7.BlaI), which would allow for detection of the protein with antibodies specific for the T7.tag, we amplified the blaI gene from plasmid pET_blaI using Pfu Ultra II DNA polymerase (Stratagene) and primers blaI_fw_BamHI (5′-cgcggatccatggccaataagcaagttg) and blaI_rv_HindIII (5′-atgcaagcttttactttttactaatatcatttaaaatg) (with BamHI and HindIII sites in bold, respectively). The PCR product and pET-24a(+) vector were digested with BamHI and HindIII endonucleases, purified from 1% agarose gel, and ligated. The ligation mixture was transformed into competent E. coli TOP10 cells. Colonies containing the construct were selected on LB agar containing 50 μg/mL kanamycin. Plasmids from different colonies were isolated and sequenced. The construct was designated pET-24a(+)_blaI. E. coli BL21 Star(DE3) competent cells were transformed with plasmid pET-24a(+)_blaI, and transformant cells were selected on LB/kanamycin/agar plates. The induction of the expression of T7.BlaI and the protein purification conditions were the same as those we described previously.17 The previously described protocol for purification of BlaI allowed purification of the 16.3 kDa T7.BlaI protein to homogeneity with a protein yield of 5 mg/L of culture.
Incubation of Recombinant BlaR1.H6x with β-Lactam Antibiotics
Three different protocols were compared for the evaluation of the effect of β-lactam antibiotics on BlaR1 activity. The first protocol consisted of the incubation of the cells with the corresponding antibiotic after BlaR1 expression had taken place (after induction of BlaR1 expression with IPTG, as described above). For this, the cell pellet from a 500 mL culture was suspended in 20 mL of buffer A [100 mM sodium phosphate (pH 7.5) and 50 mM NaHCO3] supplemented with the corresponding β-lactam antibiotic [455 μM oxacillin, 1 μM Bocillin FL, or 20 μM 2-(2′-carboxyphenyl)benzoyl-6-aminopenicillanic acid (CBAP), each at its final concentration]. As a control, the cell pellet from a 500 mL culture was suspended in 20 mL of buffer A. The cells were incubated for 15 min at 37 °C, and afterward, the contents of the cells were liberated and the membrane protein fractions prepared as previously described.8 After ultracentrifugation, each pellet (membranes with membrane proteins) was suspended in 0.5 mL of buffer A with or without the corresponding β-lactam antibiotic (same concentration used before). The total protein concentration was determined using the BCA Protein Assay Kit (Pierce). The same protocol was used to prepare E. coli membranes that did not contain BlaR1, from a culture of E. coli BL21 Star(DE3) cells transformed with pET-24a(+) (negative control). The second protocol consisted of incubation with the β-lactam antibiotic after preparation of the membrane proteins fraction. After induction of protein expression with IPTG for 5 h at 28°C, the membrane proteins fraction was prepared in buffer A (no β-lactam antibiotic), as described before. For the evaluation of the effect of β-lactam antibiotics, the corresponding antibiotic was added to aliquots of the membranes containing 40 μg of total protein to reach a final concentration of 10 μM Bocillin FL, 10 μM oxacillin, or 10 μM CBAP, and the mixture was incubated for 10 min at 37 °C. The third protocol involved incubation of the cells with oxacillin at sub-MIC concentrations (76 and 152 μM) during the induction of recombinant BlaR1 expression with IPTG. The minimal inhibitory concentrations (MICs) of β-lactam compounds were determined by the broth microdilution method (Table S1 of the Supporting Information), as recommended by the Clinical and Laboratory Standards Institute.22 The MICs were determined in LB broth (same broth used for cell growth and BlaR1 protein induction) using a bacterial inoculum of 5 × 105 cfu/mL. All plates were incubated at 37 °C for 20 h, before the results were interpreted. Subsequent to BlaR1 induction and β-lactam exposure, the cells were collected by centrifugation and membrane proteins were prepared as previously described. The cell pellets and membranes were suspended in buffer A with the appropriate concentration of oxacillin (that is to say, that the proteins were consistently kept in contact with the antibiotic).
Detection of Bocillin FL-Acylated BlaR1.H6x Protein
After incubation of the bacteria expressing BlaR1 with Bocillin FL and preparation of the membrane protein fraction (or after incubation of the previously prepared membrane protein fractions with Bocillin FL), the proteins of the different membrane fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% gels), and the proteins covalently bound to Bocillin FL were detected by scanning the gel in a Storm Scanner in the fluorescence mode (excitation wavelength of 450 nm and emission wavelength of 540 nm). As a negative control, we used membrane protein preparations from E. coli BL21 Star(DE3) transformed with plasmid pET-24a(+). As a positive control, we expressed the sensor domain of BlaR1, BlaRS, as a soluble and active protein in the cytoplasm of E. coli.9 BlaRS in whole cell extracts (soluble fraction) or purified BlaRS was labeled via incubation with 1–10 μM Bocillin FL in buffer A for 10 min at 37 °C prior to loading of the sample for protein separation by SDS–PAGE.
T7.BlaI and BlaI Proteolysis Assays
To evaluate the proteolysis of T7.BlaI or BlaI by BlaR1 expressed and inserted into E. coli membranes, 50 ng of purified T7.BlaI or BlaI (25 ng/μL) was incubated for 1 or 2 h at 37 °C and 256 rpm, with 40 μg of E. coli membrane protein preparations with BlaR1.H6x prepared in the absence or presence of β-lactam antibiotics. As a control for possible T7.BlaI or BlaI proteolysis by E. coli proteases, 50 ng of purified T7.BlaI or BlaI protein (25 ng/μL) was incubated with 40 μg of E. coli membrane preparations without BlaR1 [from cells transformed with vector pET-24a(+)] prepared in the presence of β-lactam antibiotics. The total volume in the reaction was kept constant at 23 μL in all cases, and where necessary, the volume was completed with buffer A. A second control consisted of incubation of 50 ng of purified T7.BlaI or BlaI (25 ng/μL) in buffer A (same final volume). To evaluate the effect of the metal chelating agent ethylenediaminetetraacetic acid (EDTA) on proteolysis of T7.BlaI by BlaR1, the membranes (with or without BlaR1) were incubated with different concentrations of EDTA for 18 h at 15°C and 220 rpm prior to the incubation with T7.BlaI. Stock solutions of 1, 10, and 100 mM EDTA in buffer A were prepared from a 0.5 M EDTA solution (pH 8.0). After treatment of the membranes with EDTA, 50 ng of purified T7.BlaI (25 ng/μL) was added to 40 μg of E. coli membrane protein preparations with BlaR1.H6x and incubated for 1 or 2 h at 37 °C and 256 rpm. T7.BlaI or BlaI was detected by Western blotting (Supporting Information). In addition, aliquots of the membrane proteins fraction with BlaR1 were incubated with 1× Complete EDTA-free Protease Inhibitor Cocktail (Roche) or with 3 mM Pefabloc SC (Roche) for 18 h at 15°C and 220 rpm prior to the incubation with T7.Blal.
Site-Directed Mutagenesis of the blaR1 Gene and Cloning for Expression in E. coli
To produce the BlaR1 mutants H201A/H205A and S389A, we utilized the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Plasmid pET-24a(+)_blaRHis6x was used as the DNA template. For the mutant H201A/H205A, two non-overlapping mutagenic primers were used: BlaR_M2_fw (5′-atgctgctgctaaaaatagagatactttacatttaattatc), which introduced the H205A mutation, and BlaR_M2_rv (5′-attcagccaatattatatattt-tagtcttttgtcaattac), which introduced the H201A mutation into the reverse complementary strand. The same protocol was applied to obtain the S389A single mutant of BlaR1, using two overlapping mutagenic primers: BlaR_M4_fw (5′-ggtattctcc-taatgcaacttataaaatttatttagc) and BlaR_M4_rv (5′-gctaaataaattt-tataagttgcattaggagaatacc). After mutagenesis, the nucleotide sequence for the gene producing each mutant enzyme was verified, and the genes were recloned between the NdeI and XhoI sites of the pET-24a(+) expression vector, to give plasmids pET-24a(+)_blaRH6x_H201A/H205A and pET-24a(+)_blaRH6x_S389A, respectively. To produce the E202A single mutant of BlaR1, we conducted two PCRs to amplify two halves of the blaR1 gene from DNA template pET-24a(+)_blaRHis6x. To design the primers, we analyzed the region surrounding the GAA codon that encodes E202 in BlaR1 for the introduction of a unique endonuclease restriction site. Substitution of the GAA codon (Glu) for GCG (Ala) introduced the desired mutation and generated an SphI recognition site, which was appropriate for the mutagenesis strategy because there is no such site in the wild-type blaR1 sequence. In the first PCR, we amplified a fragment encompassing nucleotides 1-611 of blaR1 using a forward oligonucleotide that introduces an NdeI site [primer blaR_NdeI_fw (5′-gatatacatatggctaagttattaataatgagc)] and a reverse primer that introduces an SphI site and the E202A mutation [primer blaR_E202ASphI_rv (5′-gcatacgcatgcaatatta-tatattttagtc)]. The purified PCR product was digested with NdeI and SphI endonucleases, and the resulting fragment was purified from a 1% agarose gel. In the second PCR, we amplified a fragment encompassing nucleotides 594-1755 of blaR1 using a forward oligonucleotide that introduces an SphI site, and the E202A mutation [primer blaR_E202ASphI_fw (5′-aatattgcatgcgtatgctcatgc)] and a reverse primer with a XhoI site [primer XhoI_blaR_rv (5′-gtgctcgagttggccatttaaaac)]. The purified PCR product was digested with SphI and XhoI endonucleases, and the resulting fragment was purified from a 1% agarose gel. The two PCR fragments, digested with endonucleases as described above, were cloned between the NdeI and XhoI sites of the pET-24a(+) expression vector, to give plasmid pET-24a(+)_blaRH6x_E202A. The correct cloning and sequence of all the genes were verified before protein expression. Three sequencing reactions were required to cover the complete blaR1 gene, and to sequence the constructs for the E202A and H201A/H205A mutants, we used primer BlaR_SA_seq2 (5′-ttaaaagtgtaattgacaaaagac), apart from the T7 promoter and T7 terminator primers. For S389A blaR1, the third sequencing reaction was conducted using primer BlaR_SA_seq1, as for wild-type blaR1.8 The mutant proteins were expressed in E. coli BL21 Star(DE3), following the same protocol described for the wild-type enzyme. All mutant proteins were expressed as C-terminal His6x tags (as was the case for wild-type BlaR1-H6x).
RESULTS AND DISCUSSION
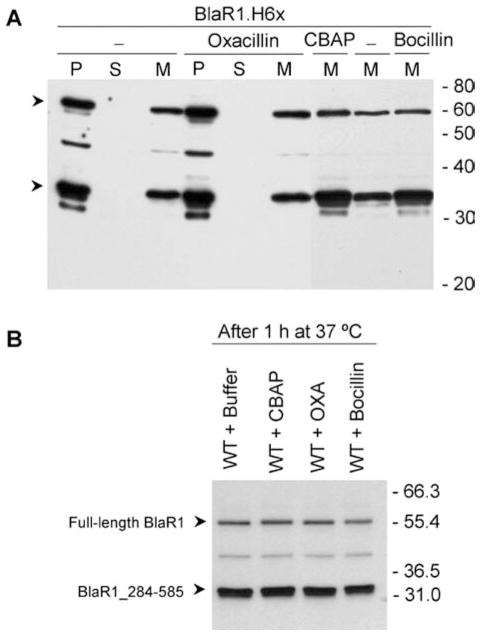
We expressed BlaR1 in E. coli BL21 Star(DE3) cells using construct pET-24a(+)_BlaRHis6x, which allows IPTG-inducible expression of BlaR1 with a C-terminal His6x tag under the control of a T7 promoter. Three protocols were followed to evaluate the effect of β-lactam antibiotics on BlaR1 activation. In the first protocol, bacteria harboring recombinant BlaR1 were exposed to different β-lactam antibiotics after induction of BlaR1 expression. The second procedure consisted of expression of BlaR1, isolation of the membrane fraction, and subsequent incubation with different β-lactam antibiotics. The level of expression of BlaR1 in E. coli was low but nonetheless significantly higher than in the case in S. aureus.8 BlaR1 was detected by Western blot analysis using antibodies specific to the C-terminal His6x tag. As shown in Figure 2A, recombinant BlaR1 localized to the membrane fraction (60 kDa full-length protein), and the fragmentation of BlaR1, to give the previously characterized 36.6 kDa fragment,8 took place in the absence of antibiotic. Incubation with CBAP, oxacillin, or Bocillin FL did not increase the degree of BlaR1 fragmentation: the pattern did not change nor did the relative quantities of the protein bands (Figure 2). In S. aureus, the level of BlaR1 in the membrane in the absence of the challenge by β-lactam antibiotics is very low (the basal level), and it increases upon exposure to the antibiotic, because of the regulatory events of the system.5,8 To better reproduce the situation in S. aureus, we evaluated a third protocol. The incubation with β-lactam antibiotics was performed simultaneously with the induction of BlaR1 expression with IPTG. For this purpose, BlaR1 expression was conducted in the presence of sub-MIC concentrations of oxacillin. This antibiotic was selected on the basis of MIC values determined for ampicillin, oxacillin, imipenem, and ceftazidime (Table S1 of the Supporting Information) and on the basis of the previously determined dissociation constants (KS) for the antibiotics with the sensor domain of BlaR1.9 To ensure that a significant fraction of BlaR1 in E. coli would be acylated by the β-lactam antibiotic, the concentration of antibiotic required for the sub-MIC assay should be higher than the KS value for the antibiotic with the sensor domain of BlaR1. Of the four antibiotics that we examined, oxacillin was the one with MIC values sufficiently high for this condition to be met (Table S1 of the Supporting Information). Furthermore, we have recently shown that oxacillin is one of the best inducers of the bla system in S. aureus,8 and hence, its selection is appropriate for the evaluation of induction of BlaR1 in this heterologous system. As disclosed for the other two protocols described above, there was no increase in the degree of BlaR1 fragmentation upon exposure to sub-MIC concentrations of oxacillin (76-152 μM) during induction of BlaR1 expression by IPTG (Figure S1 of the Supporting Information).
Figure 2.
Recombinant BlaR1 expressed in E. coli is a membrane-bound protein and is proteolyzed, an event that is not modulated by β-lactam antibiotics. (A) Detection of BlaR1.H6x in different cell fractions, in the absence of β-lactam antibiotic or after incubation with 455 μM oxacillin, 1 μM Bocillin FL, or 20.6 μM CBAP. Abbreviations: P, cell debris pellet after centrifugation of the lysed cell extract (approximately 40 μg of protein); S, soluble fraction after ultracentrifugation (40 μg of protein); M, membrane fraction after ultracentrifugation (6 μg of protein). (B) Comparison of the BlaR1.H6x fragments observed after incubation of membrane proteins with buffer or with 10 μM CBAP, oxacillin, or Bocillin FL. In this case, the antibiotics were added after preparation of the membranes and incubated for 1 h at 37 °C. In each case, a total of 6 μg of membrane proteins was resolved by SDS-PAGE. In panels A and B, recombinant BlaR1 was detected by Western blotting, using antibodies specific to the C-terminal His6x tag. The arrowheads for each panel indicate the 60 kDa band corresponding to full-length BlaR1 and the 36.6 kDa band corresponding to the C-terminal fragment BlaR1_284-585. Numbers on the right indicate the positions of migration of the molecular mass markers (kilodaltons). BlaR1 fragmentation gives a major fragment of 33–35 kDa, which was previously shown to correspond to a membrane-anchored protein fragment with a theoretical mass of 36.6 kDa, with the same N-terminal sequence as the fragment found in S. aureus (F284NGKKSLLKR...).
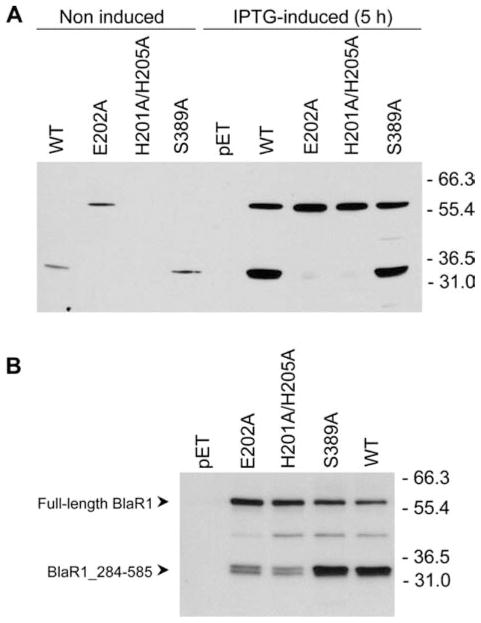
The cytoplasmic domain of BlaR1 has the ...H201EXXH... sequence motif within it, which is conserved in metalloproteases as the requisite zinc-binding site.23 Residues His-201 and His-205 are proposed to be involved in zinc ion coordination, with Glu-202 serving as the general base that promotes a water molecule for the hydrolytic function.5 With mutagenesis at these sites, we attempted to document whether the observed BlaR1 fragmentation is autolytic or catalyzed by a different protease. Mutant proteins E202A and H201A/H205A showed significant reductions in the level of accumulation of the 36.6 kDa BlaR1 fragment (Figure 3). This experiment within the heterologous expression system revealed that the process is autolytic, either in an intramolecular or in an intermolecular sense. To further investigate whether acylation of the surface sensor domain by β-lactam antibiotics directly resulted in activation of the cytoplasmic domain, we prepared the S389A BlaR1 mutant protein. In terms of its fragmentation behavior, as evaluated by Western blot analysis of the membrane proteins fraction, the S389A BlaR1 mutant protein behaved very much like the wild-type protein, notwithstanding the fact that it cannot be acylated by the antibiotics (Figure 3). Significantly, this experiment indicated that BlaR1 autoproteolysis is not modulated by the presence or absence of the antibiotic. Hence, elimination of the site of β-lactam acylation in the sensor domain did not result in the loss of the autoproteolytic activity of BlaR1.
Figure 3.
Fragmentation within the cytoplasmic domain of BlaR1.H6x in E. coli is an autoproteolytic event. (A) The wild-type (WT) and mutant (E202A, H201A/H205A, and S389A) BlaR1.H6x proteins were detected in whole cell extracts (40 μg of protein), after induction of protein expression by IPTG for 5 h at 28 °C. Noninduced samples correspond to whole cell extracts from aliquots taken before supplementation of the cultures with IPTG. The negative control consisted of cells transformed with the empty expression vector [pET-24a(+)]. (B) Detection of full-length BlaR1.H6x and the BlaR1_284-585_H6x fragment in membranes, for the wild-type (WT) protein and the E202A, H201A/H205A, and S389A mutants. In each case, 6 μg of total membrane protein was separated by SDS-PAGE. Recombinant BlaR1 was detected by Western blotting, using antibodies specific to the C-terminal His6x tag. Numbers on the right indicate the positions of migration of the molecular mass markers (kilodaltons).
To document that the recombinant proteins expressed in E. coli had a functional sensor domain, we evaluated acylation of the different BlaR1 variants by the fluorescent penicillin Bocillin FL, which would give rise to protein bands that would become fluorescent upon binding to the antibiotic. After exposure of the membrane protein preparation harboring the wild-type BlaR1 protein to Bocillin FL, both the full-length protein and the major fragmentation product (36.6 kDa) were detected as fluorescent bands, labeled with Bocillin FL (Figure 4). The S389A mutant protein would not be expected to become fluorescent, as the site of acylation by the antibiotic (S389) has been mutated to alanine, which was exactly the case. The two mutant variants in the ...H201EXXH... sequence motif (E202A and H201A/H205A) showed largely the full-length version of the protein, as the lack of the metal-binding site or of the amino acid that promotes the hydrolytic water molecule eliminated the opportunity for autolytic fragmentation. As a control experiment, the purified recombinant surface domain, BlaRS, was shown to bind to the same antibiotic with the attendant detection of fluorescence. One notices additional bands in the membrane preparations harboring the wild-type and mutant versions of BlaR1, which are also present in membrane protein preparations from cells harboring the empty expression vector (lane pET); hence, they are not BlaR1-related. These additional bands are due to penicillin-binding proteins of E. coli, which also bind to the fluorescent penicillin Bocillin FL.24,25 Similar results were obtained when whole cells were exposed to Bocillin FL before cell lysis and the preparation of the membrane protein fraction (Figure 4).
Figure 4.
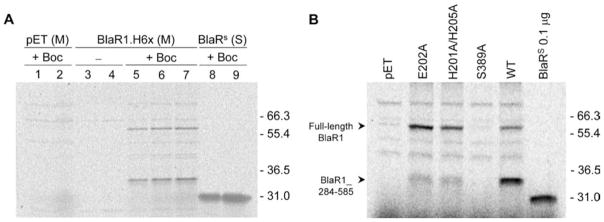
Detection of BlaR1 covalently linked to the fluorescent penicillin Bocillin FL in membrane protein preparations. (A) Acylation of wild-type BlaR1.H6x by Bocillin FL in membrane protein preparations from bacteria incubated for 15 min at 37 °C with buffer or buffer supplemented with 1 μM Bocillin FL before cell disruption: negative control (binding of Bocillin FL to E. coli PBP proteins), membrane protein preparations from cultures of E. coli BL21 Star(DE3) cells transformed with pET-24a(+) (pET) and incubated with Bocillin FL; lanes 1 and 2, 40 and 80 μg, respectively, of membrane proteins from E. coli cells transformed with pET-24a(+) and incubated with Bocillin FL; lanes 3 and 4, 40 and 80 μg, respectively, of membrane proteins from E. coli cells transformed with pET-24a(+)_BlaRH6x (BlaR1.H6x), without antibiotic; lanes 5–7, 40, 60, and 80 μg, respectively, of membrane proteins from E. coli cells transformed with pET-24a(+)_BlaRH6x (BlaR1.H6x) and incubated with Bocillin FL; lanes 8 and 9, 40 and 80 μg, respectively, of total soluble proteins from E. coli BL21 Star(DE3) cells expressing BlaRS, incubated with Bocillin FL for 15 min at 37 °C. The two fluorescently labeled bands detected in the BlaR1.H6x extracts incubated with Bocillin FL correspond in size to the 60 and 36.6 kDa bands detected by Western blotting using the antibodies specific to the C-terminal His6x tag. (B) Acylation of wild-type (WT) and mutant (E202A, H201A/H205A, and S389A) BlaR1.H6x by Bocillin FL in membrane protein preparations. After preparation of the membrane protein fraction, 40 μg of total proteins was incubated with 10 μM Bocillin FL (in a total volume of 15 μL) for 10 min at 37 °C. As a control, 0.1 μg of purified BlaRS was incubated with 10 μM Bocillin FL (in a total volume of 15 μL) for 10 min at 37 °C. In panels A and B, the proteins were separated by SDS-PAGE and the Bocillin FL-labeled bands were detected using a Storm scanner in the fluorescence mode. Numbers on the right indicate the positions of migration of the molecular mass markers (kilodaltons).
The expression of BlaR1 in the heterologous E. coli system reported here, in the absence of other proteins from S. aureus, also allowed us to document for the first time in vitro that BlaR1 is able to degrade purified BlaI directly, with no involvement of another intermediary protein (Figure 5A). The proteolytic activity was abrogated upon incubation of BlaR1 with EDTA, which indicates that this activity is metal ion-dependent (Figures 5B,C and 6). Furthermore, consistent with the lack of inducibility of the BlaR1 autolytic activity upon exposure to antibiotics, we did not see a difference between the rate of BlaI degradation in the presence and absence of antibiotic (Figure 6). No proteolytic activity was observed when BlaI was incubated with E. coli membrane preparations that lacked BlaR1. Importantly, the purified T7-BlaI was also degraded in the presence of membranes from bacteria that expressed S389A mutant BlaR1, and this activity was also inhibited by treatment with EDTA (Figure 5A,B). The two mutant variants at the ...H201EXXH... sequence motif (E202A and H201A/H205A) resulted in a total lack of discernible processing of BlaI, and no difference was observed in the presence or absence of EDTA with these mutant proteins (Figure 5A–C). The same results were obtained when nontagged wild-type BlaI was used as the substrate (Figure 5D).
Figure 5.
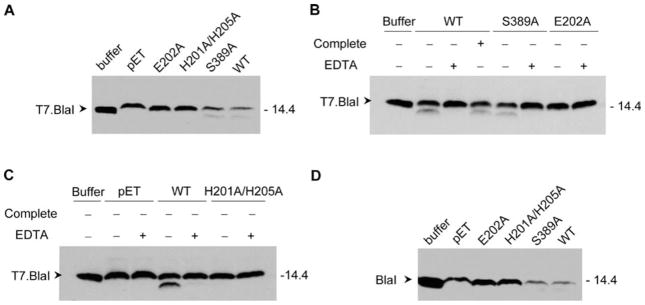
Proteolysis of T7.BlaI and BlaI by recombinant wild-type and mutant (E202A, H201A/H205A, and S389A) BlaR1.H6x and the effect of protease inhibitors. (A) A 40 μg total protein aliquot of membrane preparations with recombinant wild-type (WT) BlaR1 and its mutants was incubated with 50 ng of purified T7.BlaI for 1 h at 37 °C and 256 rpm (in a final volume of 23 μL). As a control, T7.BlaI was incubated with buffer A. (B) Membrane preparations containing recombinant wild-type (WT) BlaR1.H6x and the S389A and E202A mutants were preincubated in buffer A or buffer A supplemented with 13 mM EDTA for 18 h at 15 °C and 220 rpm, prior to incubation with T7.BlaI. Wild-type BlaR1.H6x was also preincubated in 1× Complete EDTA-free Protease Inhibitor Cocktail (Roche), which inhibits all proteases with the exception of metalloproteases. A 50 ng aliquot of purified T7.BlaI was then added to each membrane protein preparation and incubated for 1 h at 37 °C and 256 rpm. (C) Membrane preparations containing recombinant wild-type BlaR1.H6x or its H201A/H205A mutant or without BlaR1 [prepared from E. coli BL21 Star(DE3) cells transformed with pET-24a(+); lanes labeled as pET] were preincubated in buffer A or buffer A supplemented with 13 mM EDTA for 18 h at 15 °C and 220 rpm, prior to incubation with T7.BlaI. In panels A–C, the proteins in the samples were separated by SDS-PAGE in 15% gels, and T7.BlaI was detected by Western blotting, using antibodies specific to the T7 tag. (D) A 40 μg total protein aliquot of membrane preparations with recombinant wild-type (WT) and mutant BlaR1.H6x was incubated with 50 ng of purified BlaI for 1 h at 37 °C and 256 rpm (in a final volume of 23 μL). As a control, BlaI was incubated with buffer A. The proteins in the samples were separated by SDS-PAGE in 15% gels, and BlaI was detected by Western blotting, using antibodies specific for purified BlaI (anti-BlaI antibodies). Numbers on the right indicate the positions of migration of the molecular mass markers (kilodaltons).
Figure 6.
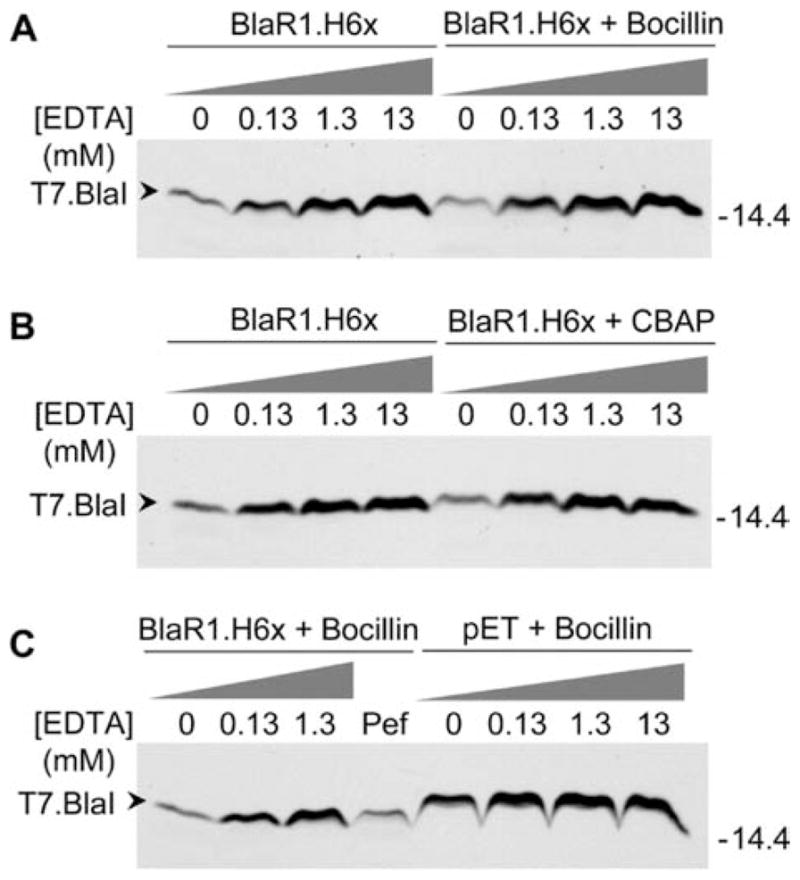
Proteolysis of T7.BlaI catalyzed by recombinant BlaR1.H6x in the absence and presence of β-lactam antibiotics and inhibition of proteolysis by preincubation of BlaR1.H6x with EDTA. (A) Membrane preparations with recombinant BlaR1.H6x in the absence of antibiotic or in the presence of 1 μM Bocillin FL were preincubated in buffer A or buffer A supplemented with increasing concentrations of EDTA (0.13, 1.3, and 13 mM) for 18 h at 15 °C and 220 rpm, prior to incubation with T7.BlaI. (B) Membrane preparations with recombinant BlaR1.H6x in the absence of antibiotic or in the presence of 20.6 μM CBAP were preincubated in buffer A or buffer A supplemented with increasing concentrations of EDTA (0.13, 1.3, and 13 mM) for 18 h at 15 °C and 220 rpm, prior to incubation with T7.BlaI. (C) Membrane preparations with recombinant BlaR1.H6x in the presence of 1 μM Bocillin FL and E. coli membranes without BlaR1 [prepared from E. coli BL21 Star(DE3) cells transformed with pET-24a(+)] in the presence of 1 μM Bocillin FL (pET + Bocillin) were preincubated in buffer A or buffer A supplemented with increasing concentrations of EDTA (0.13, 1.3, and 13 mM) for 18 h at 15 °C and 220 rpm, prior to incubation with T7.BlaI. The lane labeled Pef corresponds to membranes preincubated with the protease inhibitor Pefabloc SC (3 mM), which belongs to the family of sulfonyl fluorides that irreversibly block serine proteases. In panels A–C, after the corresponding treatment of the membranes with BlaR1, 50 ng of T7.BlaI was incubated with the corresponding membrane preparation (40 μg of total protein) for 2 h at 37 °C and 256 rpm. The proteins in the samples were separated via SDS-PAGE in 15% gels, and T7.BlaI was detected by Western blotting, using antibodies specific for the T7 tag. Numbers on the right indicate the positions of migration of the molecular mass markers (kilodaltons).
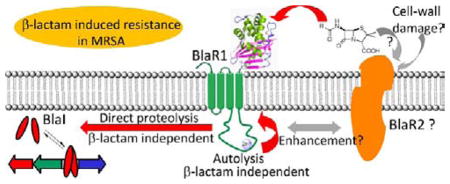
In this study, we document that the membrane protein BlaR1 from S. aureus is expressed in an active form in E. coli. In addition, the data show that the same mutations that prevent activation of the system in response to antibiotics in S. aureus resulted in abrogation of the ability of BlaR1 both to autoproteolyze and to degrade BlaI. Even though the bla operon is constituted by three genes, experiments conducted in S. aureus do not allow us to rule out the possibility that other proteins could be involved in the activation cascade or that other systems could be in cross-talk with the bla system. The experiments reported here with wild-type BlaR1 and the mutant variants expressed in E. coli membranes, in the absence of other S. aureus proteins, allowed us to conclude that BlaR1 has an autoproteolytic activity, which is not enhanced by mere acylation of the sensor domain by antibiotics, as is understood to be the case presently. Last, but not least, we were able to show that BlaR1 can directly act on BlaI and degrade it (metal ion-dependent activity). Hence, no other S. aureus protein is involved in the process, because such a protein would be unlikely to be present in the membrane of the unrelated bacterium E. coli. We do not rule out the possibility of the involvement of such a mediator in other steps in signal transduction at this point. The recombinant BlaR1 in E. coli merely did not show modulation of its metalloprotease activity in response to the antibiotic. It is possible that this hypothetical mediator, if present in S. aureus, would enhance the rates of these processes in response to antibiotics (Figure 1).
Supplementary Material
Acknowledgments
Funding
L.I.L. was a Pew Latin American Fellow in the Biomedical Sciences, supported by The Pew Charitable Trusts. The opinions expressed are those of the authors and do not necessarily reflect the views of The Pew Charitable Trusts.
ABBREVIATIONS
- MRSA
methicillin-resistant S. aureus
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- CBAP
2-(2′-carboxyphenyl)benzoyl-6-aminopenicillanic acid
- MIC
minimal inhibitory concentration
- EDTA
ethylenediaminetetraacetic acid
- BlaRS
C-terminal sensor domain of BlaR1
Footnotes
Author Contributions
L.I.L. executed all experiments. L.I.L. and S.M. wrote the manuscript.
The authors declare no competing financial interest.
BlaRS and BlaI recombinant proteins and specific antibodies, Western blot procedures, Figure S1, and Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 2.Loffler CA, Macdougall C. Update on prevalence and treatment of methicillin-resistant Staphylococcus aureus infections. Expert Rev Anti-Infect Ther. 2007;5:961–981. doi: 10.1586/14787210.5.6.961. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Chambers HF. Pathogenesis of staphylococcal infection: A manner of expression. J Infect Dis. 2009;199:291–293. doi: 10.1086/595983. [DOI] [PubMed] [Google Scholar]
- 5.Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF. A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 6.Llarrull LI, Fisher JF, Mobashery S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new β-lactams that meet the challenge. Antimicrob Agents Chemother. 2009;53:4051–4063. doi: 10.1128/AAC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackbarth CJ, Chambers HF. blaI and blaR1 regulate β-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1144–1149. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Llarrull LI, Toth M, Champion MM, Mobashery S. Activation of BlaR1 protein of methicillin-resistant Staphylococcus aureus, its proteolytic processing, and recovery from induction of resistance. J Biol Chem. 2011;286:38148–38158. doi: 10.1074/jbc.M111.288985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golemi-Kotra D, Cha JY, Meroueh SO, Vakulenko SB, Mobashery S. Resistance to β-lactam antibiotics and its mediation by the sensor domain of the transmembrane BlaR signaling pathway in Staphylococcus aureus. J Biol Chem. 2003;278:18419–18425. doi: 10.1074/jbc.M300611200. [DOI] [PubMed] [Google Scholar]
- 10.Birck C, Cha JY, Cross J, Schulze-Briese C, Meroueh SO, Schlegel HB, Mobashery S, Samama JP. X-ray crystal structure of the acylated β-lactam sensor domain of BlaR1 from Staphylococcus aureus and the mechanism of receptor activation for signal transduction. J Am Chem Soc. 2004;126:13945–13947. doi: 10.1021/ja044742u. [DOI] [PubMed] [Google Scholar]
- 11.Wilke MS, Hills TL, Zhang HZ, Chambers HF, Strynadka NC. Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 β-lactam sensor of Staphylococcus aureus. J Biol Chem. 2004;279:47278–47287. doi: 10.1074/jbc.M407054200. [DOI] [PubMed] [Google Scholar]
- 12.Borbulevych O, Kumarasiri M, Wilson B, Llarrull LI, Lee M, Hesek D, Shi Q, Peng J, Baker BM, Mobashery S. Lysine Nζ-decarboxylation switch and activation of the β-lactam sensor domain of BlaR1 protein of methicillin-resistant Staphylococcus aureus. J Biol Chem. 2011;286:31466–31472. doi: 10.1074/jbc.M111.252189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thumanu K, Cha J, Fisher JF, Perrins R, Mobashery S, Wharton C. Discrete steps in sensing of β-lactam antibiotics by the BlaR1 protein of the methicillin-resistant Staphylococcus aureus bacterium. Proc Natl Acad Sci USA. 2006;103:10630–10635. doi: 10.1073/pnas.0601971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanique S, Colombo ML, Goormaghtigh E, Soumillion P, Frere JM, Joris B. Evidence of an intramolecular interaction between the two domains of the BlaR1 penicillin receptor during the signal transduction. J Biol Chem. 2004;279:14264–14272. doi: 10.1074/jbc.M313488200. [DOI] [PubMed] [Google Scholar]
- 15.Cohen S, Sweeney HM. Constitutive penicillinase formation in Staphylococcus aureus owing to a mutation unlinked to the penicillinase plasmid. J Bacteriol. 1968;95:1368–1374. doi: 10.1128/jb.95.4.1368-1374.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke SR, Dyke KG. Studies of the operator region of the Staphylococcus aureus β-lactamase operon. J Antimicrob Chemother. 2001;47:377–389. doi: 10.1093/jac/47.4.377. [DOI] [PubMed] [Google Scholar]
- 17.Llarrull LI, Prorok M, Mobashery S. Binding of the gene repressor BlaI to the bla operon in methicillin-resistant Staphylococcus aureus. Biochemistry. 2010;49:7975–7977. doi: 10.1021/bi101177a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safo MK, Zhao Q, Ko TP, Musayev FN, Robinson H, Scarsdale N, Wang AH, Archer GL. Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: Insights into transcriptional regulation of the bla and mec operons. J Bacteriol. 2005;187:1833–1844. doi: 10.1128/JB.187.5.1833-1844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory PD, Lewis RA, Curnock SP, Dyke KG. Studies of the repressor (BlaI) of β-lactamase synthesis in Staphylococcus aureus. Mol Microbiol. 1997;24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 20.Clarke SR, Dyke KG. The signal transducer (BlaRI) and the repressor (BlaI) of the Staphylococcus aureus β-lactamase operon are inducible. Microbiology. 2001;147:803–810. doi: 10.1099/00221287-147-4-803. [DOI] [PubMed] [Google Scholar]
- 21.Lewis RA, Curnock SP, Dyke KG. Proteolytic cleavage of the repressor (BlaI) of β-lactamase synthesis in Staphylococcus aureus. FEMS Microbiol Lett. 1999;178:271–275. doi: 10.1111/j.1574-6968.1999.tb08687.x. [DOI] [PubMed] [Google Scholar]
- 22.Wikler MA, Cockerill FR, III, Bush K, Dudley MN, Eliopoulos GM, Hardy DJ, Hecht DW, Hindler JF, Patel JB, Powell M, Turnidge JD, Weinstein MP, Zimmer BL, Ferraro MJ, Swenson JM. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard. 7. Clinical and Laboratory Standards Institute; Wayne, PA: 2006. [Google Scholar]
- 23.Jiang W, Bond JS. Families of metalloendopeptidases and their relationships. FEBS Lett. 1992;312:110–114. doi: 10.1016/0014-5793(92)80916-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother. 1999;43:1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies TA, Page MG, Shang W, Andrew T, Kania M, Bush K. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob Agents Chemother. 2007;51:2621–2624. doi: 10.1128/AAC.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.