Abstract
NFBD1/MDC1 is involved in DNA damage checkpoint signaling and DNA repair. NFBD1 binds to the chromatin component γH2AX at sites of DNA damage, causing amplification of ataxia telangiectasia-mutated gene (ATM) pathway signaling and recruitment of DNA repair factors. Residues 508–995 of NFBD1 possess transactivation activity, suggesting a possible role of NFBD1 in transcription. Furthermore, NFBD1 influences p53-mediated transcription in response to adriamycin. We sought to determine the role of NFBD1 in ionizing radiation (IR)–responsive transcription and if NFBD1 influences transcription independently of p53.
Using microarray analysis, we identified genes altered upon NFBD1 knockdown. Surprisingly, most NFBD1 regulated genes are regulated in both the absence and presence of IR, thus pointing toward a novel function for NFBD1 outside of the DNA damage response. Furthermore, NFBD1 knockdown regulated genes mostly independent of p53 knockdown. These genes are involved in pathways including focal adhesion signaling, carbohydrate metabolism, and insulin signaling.
We found that CAV1 and CAV2 mRNA and protein levels are reduced by both NFBD1 knockdown and knockout independently of IR and p53. NFBD1-depleted cells exhibit some similar phenotypes to Cav1-depleted cells. Furthermore, like Cav1-depletion, NFBD1 shRNA increases Erk phosphorylation. Thus, Cav1 could act as a mediator of the DNA-damage independent effects of NFBD1 in mitogenic signaling.
Introduction
NFBD1 was discovered by a computational screen for genes encoding proteins with BRCA1 carboxy-terminal (BRCT) domains (1). NFBD1 is also known as MDC1 (mediator of DNA damage checkpoint 1). NFBD1 is a DNA damage response factor that modulates the ataxia telangiectasia-mutated gene (ATM)–Chk2 signaling pathway (2–6). Previous work indicates that NFBD1 amplifies ATM signals by binding directly to γH2AX, thus recruiting active ATM and other DNA damage response factors to sites of DNA damage. H2AX is a chromatin component that is phosphorylated by ATM and other factors to form γH2AX (7). The interaction between NFBD1 and γH2AX is important for homologous recombination in addition to checkpoint signaling (8).
Another mechanism by which NFBD1 influences checkpoint signaling is by binding to the MRE11/Rad50/NBS1 (MRN) complex, thus allowing for sustained MRN complex association with damaged chromatin (3, 5, 9–14). The MRN complex is essential for activation of ATM following DNA damage (15–22). Furthermore, Mre11 possesses nuclease activity that is important for DNA repair and genomic stability (23).
Although the best characterized function for NFBD1 is in DNA damage signaling, there is also evidence that NFBD1 possesses transactivation activity. Residues 508–995 activate transcription of a chloramphenicol acetyl transferase reporter in yeast and mammalian cells (24). That region of NFBD1 has not been reported to contain additional motifs or functions. Another study establishes a relationship between NFBD1 and p53. Treatment of A549 lung carcinoma cells with adriamycin (ADR) and other DNA damaging agents reduces NFBD1 protein and mRNA at late time points (6–12 hours). The decrease in NFBD1 levels occurs simultaneously with increased p53 phosphorylation on S15 and increased abundance of p53 targets. Forcing greater NFBD1 expression decreases p53 S15 phosphorylation and reduces expression of p53 targets at late time points after ADR treatment. Luciferase reporter assays using NFBD1 overexpression and knockdown show that NFBD1 expression levels are inversely correlated with transcription from the promoters of p21, MDM2, and BAX, which are p53-targets (25).
Although it is known that NFBD1 influences p53-mediated transcription in response to ADR, much remains unknown about the role of NFBD1 in regulation of gene expression. For example, it is unclear if NFBD1 regulates targets other than p21, MDM2, and BAX. Furthermore, the role of NFBD1 in ionizing radiation (IR)–induced transcription has not been examined. To elucidate those areas, we carried out microarray experiments to analyze global transcription with single and combined NFBD1 and p53 knockdowns in U2OS cells without and with irradiation. Surprisingly, the expression levels of most NFBD1-regulated genes identified in this study are changed by NFBD1 depletion in both the presence and absence of IR and p53. Two of the genes regulated by NFBD1 are Caveolin 1 (CAV1) and Caveolin 2 (CAV2). NFBD1-depleted cells show similar phenotypes to Cav1-depleted cells, including increased Erk phosphorylation, rounded cells with smaller focal adhesions, and impaired wound healing.
Materials and Methods
Cell culture
U2OS human osteosarcoma cells [American Type Culture Collection (ATCC)] were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and L-glutamate. NFBD1+/+ and NFBD1−/− mouse embryo fibroblasts (MEFs), a gift from Junjie Chen, MD Anderson Cancer Center (7), were grown in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, 1% sodium pyruvate, and 1% nonessential amino acids.
U2OS cells were transfected with a pcDNA3 vector (Invitrogen Corporation) containing the cDNA encoding human hemagglutinin (HA)–tagged Cav1 or the empty vector (26). U2OS cells stably expressing HA-Cav1 or the empty vector were obtained by selection with 500 μg/ml G418 for 5 days. Surviving cells were pooled and maintained in the presence of 200 μg/ml G418.
Human foreskin fibroblasts (HFF) were obtained from the Yale Skin Disease Research Center, and grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
shRNA and retroviral infection
pSIREN-RetroQ (BD Biosciences) shRNA-expressing retroviral vectors were described previously (27, 28). The shRNA sequences are as follows: NFBD1a: 5/-GCCAC-TAGGAGAAAGACAAATCGAAATTTGTCTTTCTCC-TAGTGGC-3/, NFBD1b: 5/-GCAGAAGCCAATCAG-CAAATTCAAGAGATTTGCTGATTGGCTTCTGC -3/(27), P53: 5′-GGAAGACTCCAGTGGTAATCTCGAAA-GATTACCACTGGAGTCTTCC-3′ (28), and CAV1: 5/-GATCCAGGGCAACATCTACAAGCTTTCAAGAGA-AGCTTGTAGATGTTGCCCTTTTTTTG-3/(adapted from (29)).
The negative control vector contains a scrambled sequence of a luciferase-directed shRNA (BD Biosciences).
shRNA-expressing retroviruses were produced by cotransfection of the retroviral plasmids, pVSV-G (Clontech) (encoding the G glycoprotein of the vesicular stomatitis virus), and pCL-ECO (30) (encoding Gag and Pol) into 293T cells using FuGene6 (Roche Diagnostics Corp.) per manufacturer’s protocol. Retrovirus was harvested in Opti-MEM (Invitrogen) daily for five days, pooled, and concentrated with Centricon plus-20 columns (Millipore Corporation). U2OS cells were infected at a multiplicity of infection of approximately 5.
RNA isolation and real-time PCR
RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer’s protocol following the optional DNAse treatment steps. cDNA was synthesized from the RNA using the iScript kit (Bio–Rad). Real-time PCR was carried out by mixing the cDNAs with TaqMan universal PCR master mix and premixed FAM-labeled TaqMan probes (Applied Biosystems). Samples were run on a Bio–Rad iCycler real-time PCR machine. Abundance of a given mRNA in relation to a GAPDH loading control was calculated using the 2-ΔΔCT method.
Illumina microarray analysis
Cells were infected with NFBD1 and/or p53 shRNA-expressing retroviruses. Two days later, cells were irradiated with a Shepherd Mark I137 Cs irradiator with a dose of 5 Gy or mock irradiated. Four hours later RNA was isolated and analyzed using Illumina Sentrix HumanWG6_v3 Expression bead chips (Illumina, San Diego, CA). The chips contain 47,296 probes derived from the following databases: Curated RefSeq (19,730), Genome Annotation RefSeq (6,368), Gnomon (9,576), and Unigene-163 (11,622).
Differential gene expression was determined using methods described in Ref. (31). Briefly, the expression data were background corrected, variance stabilizing transformed, and quantile normalized using the lumi software package for the R programming environment (available from: http://www.ncbi.nlm.nih.gov/pubmed/18467348). Differential gene expression between two treatment groups was analyzed using linear model methods from the R limma package (32). Moderated t tests were used to compare the mean intensities between different samples. Statistical significance was determined by calculating P values based on the change in intensity of each probe across three biological replicates. To control for false discovery rate, P values were adjusted for multiple testing using the method of Benjamini and Hochberg (33).
For Gene Ontology analysis, lists of differentially regulated genes were compared to lists of genes in known pathways or functional classes in the Gene Ontology database using the R GOstats package (available from: http://www.ncbi.nlm.nih.gov/pubmed/17098774). Pathways overrepresented in the lists of differentially regulated genes were identified by calculating the P-value of the number of genes in a pathway regulated by chance versus the actual number of regulated genes in a given pathway.
Cell lysis and immunoblotting
Cells lysates were prepared with 2× Laemmli sample buffer. Immunoblots on polyvinylidene fluoride (PVDF) membranes were blocked with 5% nonfat milk in PBST (Dulbecco’s phosphate buffered saline with 0.1% Tween-20). The following antibodies were incubated with membranes in 5% nonfat milk in PBST: NFBD1 (2), p53, HA, and GAPDH (Santa Cruz), Cav1 and Cav2 (BD Biosciences), β-tubulin, phospho-p42/44 MAPK (Erk1/2) T202/Y204, and p42/44, (Cell Signaling Technology). Membranes were then washed with PBST and incubated with horseradish peroxidase-conjugated secondary antibodies in 5% nonfat milk in PBST.
Immunofluorescence
Cells were plated on glass chamber slides coated with fibronectin (BD Biosciences) at a final concentration of 5 μg/cm2. The next day, cells were washed with phosphate buffered saline (PBS), fixed in 2% paraformaldehyde for 30 minutes, washed twice with PBS, washed once with 25 mmol/L NH4Cl in PBS, quenched with 25 mmol/L NH4Cl in PBS for 10 minutes, washed with 0.1% triton-X-100 in PBS, then permeabilized in 0.1% triton-X-100 in PBS for 10 minutes. Slides were then blocked in 5% goat serum in PBS for 45 minutes at room temperature and then incubated with primary antibodies toward vinculin (Sigma–Aldrich), NFBD1 (2), and/or Cav1 (BD Biosciences) diluted in 2% goat serum in PBS for 1 hour at room temperature. Slides were washed with PBS, incubated in alexa fluor 594 or alexa fluor 488-conjugated secondary antibodies (Invitrogen) diluted in 2% goat serum/PBS for 1 hour at room temperature, and then washed in PBS. Where indicated, texas red-conjugated phalloidin (Invitrogen) staining was carried out according to the manufacturer’s protocol. Slides were mounted with Prolong Gold antifade reagent (Invitrogen). Cells were visualized with a fluorescence microscope.
Scratch filling assay
Confluent cells were scratched with a pipette tip. Cells were then washed twice with DMEM containing 0.1% FBS, then incubated in DMEM containing 0.1% FBS. At the indicated time-points cells were visualized with a phase-contrast microscope and images were taken. Razor blade crossmarks on the undersides of plates were used so that the same part of each scratch could be analyzed at each time point.
Results
NFBD1 knockdown regulates gene expression in an IR-independent manner
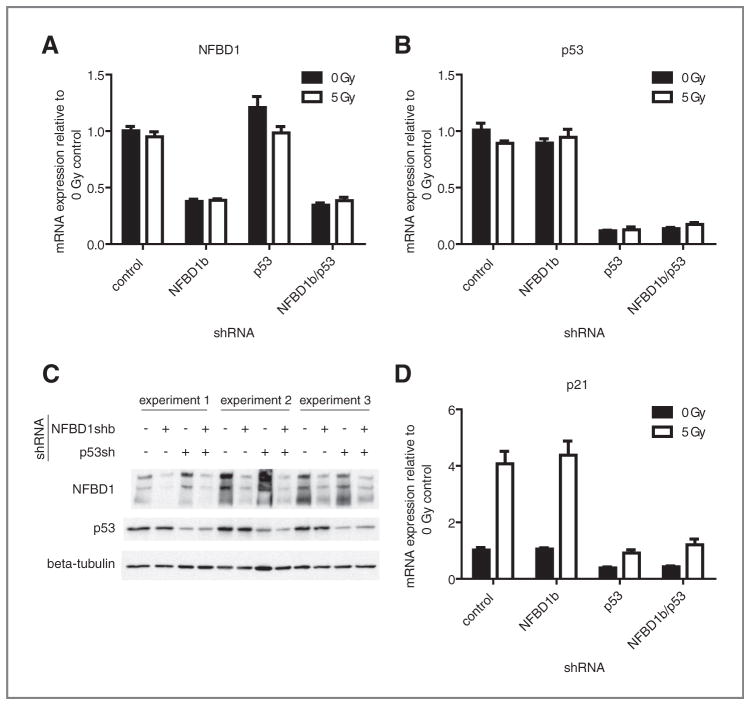
To determine the role of NFBD1 in global gene expression, shRNA infection was employed to deplete NFBD1. U2OS osteosarcoma cells, with wild type TP53, were used because NFBD1 interacts with p53. In order to separate p53-dependent and p53-independent effects of NFBD1 knockdown, p53 knockdowns were carried out singly and in combination with NFBD1 knockdown. Both NFBD1 and TP53 were effectively suppressed at the mRNA level (Fig. 1A, B) and protein level (Fig. 1C) by shRNA. For microarray analyses, cells infected with NFBD1 and/or p53 shRNA-expressing retroviruses were mock-irradiated or irradiated with 5 Gy. Four hours later, RNA was isolated for microarray analysis. Three independent infections, irradiations, and RNA isolations were carried out.
Figure 1.
Knockdown efficiencies of NFBD1b and p53 in U2OS cells. U2OS cells were infected with retroviruses expressing control, NFBD1b, or p53-targeted shRNAs. Two days later, cells were irradiated and RNA was isolated four hours after irradiation. One aliquot was used for the microarray experiment. cDNA was synthesized from the other aliquot of mRNA. Real-time PCR was carried out using primers toward A, NFBD1; B, p53; or D, p21. Error bars represent the standard error of the mean from six samples: three technical replicates from two biological replicates. C, western blots were carried out on cell lysates using the indicated antibodies.
Table 1 lists selected genes for which mRNA levels were altered by NFBD1 knockdown in irradiated cells. The selection was based on highest fold change and pathways that were of interest. The entire list will be available in the Gene Expression Omnibus (GEO). Interestingly, the vast majority of the genes regulated by NFBD1 knockdown in irradiated cells were also regulated by NFBD1 shRNA in nonirradiated cells (Table 1). Most of the genes that were regulated by NFBD1 shRNA only in the presence of IR showed a very small fold change. Therefore, these results suggest a noncanonical role of NFBD1 in regulation of gene expression in the absence of radiation. Genes regulated by NFBD1 knockdown fall into functional categories including focal adhesion, Jak–STAT pathway, insulin signaling, carbohydrate metabolism, and RNA splicing.
Table 1.
NFBD1 shRNA-regulated genes identified by microarray analysis
| Pathway | Symbol | Description | 0 Gy
|
5 Gy
|
||||
|---|---|---|---|---|---|---|---|---|
| log2FC NFBD1 | log2FC p53 | log2FC NFBD1/p53 | log2FC NFBD1 | log2FC p53 | log2FC NFBD1/p53 | |||
| focal adhesion | CAV2 | caveolin 2 | −0.973 | −0.899 | −0.839 | −0.852 | ||
| CAV2 | caveolin 2 | −0.716 | −0.743 | −0.662 | −0.664 | |||
| PPP1CC | Protein phosphatase 1, catalytic subunit, gamma isoform | −0.534 | −0.516 | −0.786 | −0.504 | |||
| CAV1 | Caveolin 1, caveolae protein, 22kDa | −0.664 | −0.656 | −0.604 | −0.498 | |||
| CAV1 | Caveolin 1, caveolae protein, 22kDa | −0.537 | −0.482 | −0.664 | ||||
| PTEN | Phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | −0.425 | −0.374 | −0.373 | −0.448 | |||
| PTEN | Phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | −0.293 | −0.324 | −0.431 | −0.389 | |||
| LAMA5 | Laminin, alpha 5 | −0.661 | −0.519 | |||||
| LAMB3 | Laminin, beta 3 | −0.319 | −0.239 | −0.346 | −0.392 | |||
| LAMB2 | Laminin, beta 2 (laminin S) | −0.267 | −0.347 | |||||
| RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 | −0.349 | −0.353 | |||||
| CCND2 | Cyclin D2 | 0.340 | 0.232 | |||||
| THBS3 | Thrombospondin 3 | −0.266 | ||||||
| TLN2 | Talin 2 | −0.243 | ||||||
| COL5A1 | Collagen, type V, alpha 1 | −0.492 | −0.536 | |||||
| GRB2 | Growth factor receptor-bound protein 2 | −0.254 | ||||||
| CAPN2 | Calpain 2, (m/II) large subunit | −0.191 | −0.180 | |||||
| PDGFRB | Platelet-derived growth factor receptor, beta polypeptide | −0.242 | −0.271 | |||||
| Jak-STAT | BCL2L1 | BCL2-like 1 | −0.541 | −0.232 | −0.560 | −0.523 | −0.343 | −0.771 |
| BCL2L1 | BCL2-like 1 | −0.443 | 0.179 | −0.351 | −0.523 | −0.263 | −0.394 | |
| IFNAR1 | Interferon (alpha, beta and omega) receptor 1 | −0.292 | −0.438 | |||||
| IL12A | Interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) | 0.180 | 0.294 | 0.186 | ||||
| STAM | Signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | −0.598 | −0.559 | −0.593 | −0.442 | |||
| LEPR | Leptin receptor | −0.360 | −0.307 | |||||
| IL21R | Interleukin 21 receptor | 0.115 | 0.171 | |||||
| IL7R | Interleukin 7 receptor | −0.277 | −0.230 | −0.364 | ||||
| IL11 | Interleukin 11 | 0.179 | 0.133 | 0.162 | ||||
| CCND2 | Cyclin D2 | 0.395 | 0.340 | 0.232 | ||||
| CCND2 | Cyclin D2 | 0.499 | ||||||
| IL13RA2 | Interleukin 13 receptor, alpha 2 | 0.245 | ||||||
| GRB2 | Growth factor receptor-bound protein 2 | −0.254 | ||||||
| GHR | Growth hormone receptor | −0.201 | ||||||
| SOCS2 | Suppressor of cytokine signaling 2 | 0.157 | ||||||
| Insulin | PPP1CC | Protein phosphatase 1, catalytic subunit, gamma isoform | −0.534 | −0.516 | −0.786 | −0.504 | ||
| NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | −0.448 | −0.430 | −0.459 | −0.316 | |||
| IRS1 | Insulin receptor substrate 1 | −0.382 | −0.353 | −0.347 | 0.288 | |||
| EXOC7 | Exocyst complex component 7 | −0.532 | ||||||
| RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 | −0.349 | −0.353 | |||||
| GRB2 | Growth factor receptor-bound protein 2 | −0.254 | ||||||
| MKNK2 | MAP kinase interacting serine/threonine kinase 2 | 0.160 | ||||||
| TSC1 | Tuberous sclerosis 1 | −0.221 | ||||||
| PRKAR2A | Protein kinase, cAMP-dependent, regulatory, type II, alpha | −0.267 | ||||||
| SOCS2 | Suppressor of cytokine signaling 2 | 0.57 | ||||||
| FLOT2 | Flotillin 2 | −0.276 | ||||||
| TGF-beta | INHBE | Inhibin, beta E | 0.670 | 0.471 | 0.461 | 0.572 | ||
| TGFBR3 | Transforming growth factor, beta receptor III | −0.505 | ||||||
| TIAF1 | TGFB1-induced anti-apoptotic factor 1 | −0.517 | ||||||
| SMAD6 | SMAD family member 6 | −0.280 | −0.386 | |||||
| SMAD3 | SMAD family member 3 | −0.474 | ||||||
| SKP1 | S-phase kinase-associated protein 1 | −0.420 | −0.321 | 0.249 | −0.365 | −0.290 | ||
| PPP2R2C | Protein phosphatase 2 (formerly 2A), regulatory subunit B, gamma isoform | −0.354 | ||||||
| THBS3 | Thrombospondin 3 | −0.266 | ||||||
| TGFBR1 | Transforming growth factor, beta receptor I (activin A receptor type II-like kinase, 53kDa) | −0.109 | ||||||
| carbohydrate metabolism | ADRA1B | Adrenergic, alpha-1B-, receptor | −0.447 | 0.240 | −0.396 | −0.227 | ||
| CS | Citrate synthase | −0.580 | −0.511 | −0.559 | −0.483 | |||
| IDH1 | Isocitrate dehydrogenase 1 (NADP+), soluble | −0.476 | −0.537 | −0.352 | −0.477 | |||
| LAMP2 | Lysosomal-associated membrane protein 2 | −1.257 | −1.225 | −1.014 | −1.065 | |||
| LAMP2 | Lysosomal-associated membrane protein 2 | −1.121 | −0.813 | −0.911 | −1.001 | |||
| PGM3 | Phosphoglucomutase 3 | −0.389 | −0.362 | −0.415 | −0.478 | |||
| PPP1CC | Protein phosphatase 1, catalytic subunit, gamma isoform | −0.786 | −0.504 | −0.534 | −0.516 | |||
| SLC2A3 | Solute carrier family 2 (facilitated glucose transporter), member 3 | −0.495 | −0.471 | −0.749 | −0.409 | −0.444 | −0.710 | |
| B4GALT4 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 | −0.292 | −0.377 | −0.402 | −0.439 | |||
| B4GALT4 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 | −0.730 | −0.569 | −0.646 | −0.536 | |||
| SUCLA2 | Succinate-CoA ligase, ADP-forming, beta subunit | −0.349 | −0.348 | −0.400 | −0.413 | |||
| SUCLA2 | Succinate-CoA ligase, ADP-forming, beta subunit | −0.466 | −0.246 | −0.392 | −0.362 | |||
| POFUT1 | Protein O-fucosyltransferase 1 | −1.046 | −1.012 | −0.906 | −0.864 | |||
| PGM2 | Phosphoglucomutase 2 | −0.413 | −0.378 | −0.385 | −0.405 | |||
| RNA splicing | NCBP1 | Nuclear cap binding protein subunit 1, 80kDa | −0.732 | −0.462 | −0.567 | −0.533 | ||
| PPP1R8 | Protein phosphatase 1, regulatory (inhibitor) subunit 8 | −0.301 | 0.249 | −0.261 | 0.188 | |||
| PTBP1 | Polypyrimidine tract binding protein 1 | −0.613 | −0.506 | −0.332 | −0.254 | |||
| PTBP1 | Polypyrimidine tract binding protein 1 | −0.599 | −0.353 | |||||
| RBM9 | RNA binding motif protein 9 | 0.208 | −0.222 | −0.201 | −0.102 | |||
| PTBP2 | Polypyrimidine tract binding protein 2 | 0.601 | 0.530 | 0.800 | 0.522 | |||
| PTBP2 | Polypyrimidine tract binding protein 2 | 0.534 | 0.448 | 0.635 | 0.591 | |||
| THOC3 | THO complex 3 | −0.631 | −0.568 | −0.724 | −0.722 | |||
| THOC3 | THO complex 3 | −0.607 | −0.417 | −0.712 | −0.687 | |||
NOTE: NFBD1 shRNA-regulated genes identified by Illumina microarray analysis are listed (second and third columns from the left). Genes with the highest fold change and representative pathways (leftmost column) are listed. The log2 fold change caused by NFBD1 or p53 shRNA is indicated in the presence and absence of IR (6 rightmost columns). If the gene did not appear in the list of top 1000 most significant (based on the P-value of the three biological replicates) genes regulated by a given shRNA or IR condition, the entry is left blank. “FC” = fold change.
The list of NFBD1-regulated genes was compared with lists of genes grouped by pathways and functions in the Gene Ontology database. A pathway is considered over-represented if more genes in that pathway are in the list of interest (Table 2, “Count”) than would occur by chance (“ExpCount”). Pathways overrepresented by NFBD1-regulated genes include carbohydrate metabolism, cellular respiration, and RNA splicing.
Table 2.
Gene Ontology pathways overrepresented in the list of NFBD1 shRNA-regulated genes
| P | ExpCount | Count | Term |
|---|---|---|---|
| 0 | 1 | 6 | Energy derivation by oxidation of organic compounds |
| 0.001 | 0 | 3 | Tricarboxylic acid cycle |
| 0.001 | 0 | 3 | Acetyl-CoA catabolic process |
| 0.001 | 3 | 11 | Carbohydrate metabolic process |
| 0.001 | 2 | 9 | Cellular carbohydrate metabolic process |
| 0.001 | 0 | 3 | Coenzyme catabolic process |
| 0.001 | 0 | 3 | Acetyl-CoA metabolic process |
| 0.001 | 0 | 2 | Visual learning |
| 0.002 | 0 | 3 | Cofactor catabolic process |
| 0.002 | 0 | 3 | Aerobic respiration |
| 0.003 | 0 | 2 | Visual behavior |
| 0.003 | 0 | 3 | Glycogen metabolic process |
| 0.004 | 1 | 4 | Response to light stimulus |
| 0.004 | 0 | 3 | Glucan metabolic process |
| 0.004 | 0 | 3 | Cellular respiration |
| 0.004 | 2 | 6 | Generation of precursor metabolites and energy |
| 0.005 | 0 | 3 | Energy reserve metabolic process |
| 0.006 | 0 | 2 | Vesicle organization and biogenesis |
| 0.008 | 0 | 1 | Positive regulation of T cell mediated cytotoxicity |
| 0.008 | 0 | 1 | Vasoconstriction of artery involved in baroreceptor response to lowering of systemic arterial blood pressure |
| 0.008 | 0 | 1 | Regulation of glycogen catabolic process |
| 0.008 | 0 | 1 | Succinyl-CoA pathway |
| 0.008 | 0 | 1 | Response to isoquinoline alkaloid |
| 0.008 | 0 | 1 | Positive regulation of mRNA 3/-end processing |
| 0.008 | 0 | 1 | Tyrosine phosphorylation of Stat4 protein |
| 0.008 | 0 | 1 | Regulation of tyrosine phosphorylation of Stat4 protein |
| 0.008 | 0 | 1 | Positive regulation of tyrosine phosphorylation of Stat4 protein |
| 0.008 | 0 | 1 | Response to morphine |
| 0.008 | 0 | 1 | Negative regulation of glycogen catabolic process |
| 0.008 | 0 | 1 | Positive regulation of glycogen catabolic process |
| 0.008 | 0 | 1 | Positive regulation of mRNA processing |
| 0.008 | 0 | 2 | Learning |
| 0.008 | 0 | 2 | Regulation of blood coagulation |
| 0.009 | 2 | 6 | RNA splicing |
| 0.009 | 1 | 4 | Response to radiation |
| 0.009 | 0 | 2 | Tricarboxylic acid cycle intermediate metabolic process |
NOTE: The list of NFBD1 shRNA-regulated genes in the absence of IR was compared with lists of genes grouped by pathways in the Gene Ontology database. The expected number of genes regulated by chance in each pathway was compared with the actual number of genes regulated by NFBD1 shRNA. Overrepresented pathways are listed.
Because the most established role of NFBD1 is modulation of the DNA damage response, it is surprising that NFBD1-regulated genes do not fall into categories related to the DNA damage response, and that NFBD1 seems to regulate gene expression in an IR-independent manner. IR-regulated genes in the microarray analysis fall into categories, including apoptosis, cell cycle, and DNA damage checkpoint (Supplementary Table S1). The microarray data and real-time PCR analysis show that p21 mRNA levels were increased by IR as expected (Table 1, Fig. 1D). Hence, IR-independence of the NFBD1 effect on gene expression was not due to ineffective irradiation.
NFBD1 knockdown regulates gene expression in a p53-independent manner
The majority of identified NFBD1-regulated genes were not regulated by p53 shRNA (Table 1). Exceptions include BCL2L1 (involved in apoptosis and the Jak–STAT pathway), SKP1 (a core component of SCF family ubiquitin ligases involved in TGF-β signaling), and SLC2A3 (involved in carbohydrate metabolism). Almost all of the genes that were regulated by the single NFBD1 knockdown were also regulated by the NFBD1/p53 double knockdown to a similar degree in the absence and presence of IR (Table 1). Therefore, NFBD1 depletion regulates global gene expression mostly in a p53-independent manner. Although NFBD1 knockdown increased TP21 transcription in the presence of ADR (25), NFBD1 knockdown did not affect p21 levels in the presence or absence of IR (Fig. 1D).
The p53-independence of NFBD-driven gene expression changes is not due to the lack of p53 signaling in U2OS cells. As expected, RNA levels of the p53 target TP21 were reduced by p53 knockdown in the presence and absence of IR (Fig. 1D). The integrity of p53 signaling in U2OS cells is also illustrated by Gene Ontology analysis of genes regulated by p53 knockdown. Pathways overrepresented in p53-regulated genes from this study include apoptosis, caspase activation, DNA damage response, and cell cycle arrest (Supplementary Table S2).
Confirmation of select microarray results by real-time PCR
A subset of NFBD1, p53, and IR-regulated genes were validated by real-time PCR analysis. cDNA was synthesized from two mRNA samples (biological replicates) from each condition used in the microarray study and quantitative real-time PCR was carried out (Table 3). With the exception of PTEN and LAMA5, the genes identified to be regulated by NFBD1 shRNA in the microarray study were shown to be regulated in the same direction in the real-time PCR experiments (Table 3A). Furthermore, p53- and IR-regulated genes were verified by real-time PCR analysis (Table 3B, C). Overall, 89% of the genes that were tested were regulated in the same direction in both microarray and real-time PCR experiments.
Table 3.
Confirmation of microarray results by real-time PCR
| (A) Select genes regulated by NFBD1 shRNA | |||||
|---|---|---|---|---|---|
| Gene | Pathway(s) | Fold change NFBD1sh 0Gy
|
Fold change NFBD1sh 5Gy
|
||
| Microarray | Real-time PCR | Microarray | Real-time PCR | ||
| MDC1(NFBD1) | shRNA control | −1.94 | −2.72 | −2.28 | −2.59 |
| Cav1 | focal adhesion | −1.52 | −1.63 | −1.55 | −1.61 |
| Cav2 | focal adhesion | −1.80 | −4.68 | −1.68 | −3.41 |
| PPP1CC | focal adhesion, insulin | −1.45 | −1.78 | −1.72 | −1.91 |
| PTEN | focal adhesion, p53 | −1.34 | −0.96 | −1.29 | 0.77 |
| LAMA5 | focal adhesion | 1.00 | −1.44 | −1.58 | −1.23 |
| CDH2 | cell adhesion molecules | −1.94 | −1.82 | −2.32 | −1.74 |
| STAM | Jak-STAT | −1.51 | −1.82 | −1.51 | −1.74 |
| BCL2L1 | Jak-STAT | −1.41 | −1.43 | −1.44 | −0.49 |
| IRS1 | Insulin | −1.30 | −1.44 | −1.27 | −1.58 |
| nRas | Insulin | −1.36 | −1.36 | −1.37 | −1.80 |
| SLC2A3 | carbohydrate metabolism | −1.41 | −1.46 | −1.33 | −1.58 |
| NCBP1 | RNA splicing | −1.66 | −1.57 | −1.48 | −1.89 |
| PTBP2 | RNA splicing | 1.48 | 2.45 | 1.64 | 2.33 |
| EIF1AX | translation | −1.63 | −2.04 | −1.51 | −2.38 |
| (B) Select genes regulated by p53 shRNA | |||||
|---|---|---|---|---|---|
| Gene | Pathway(s) | Fold change p53sh 0Gy
|
Fold change p53sh 5Gy
|
||
| Microarray | Real-time PCR | Microarray | Real-time PCR | ||
| p53 | shRNA control | −1.40 | −8.53 | −1.36 | −11.78 |
| p21 | IR control, p53 target | −1.68 | −2.78 | −2.55 | −0.18 |
| (C) Select genes regulated by IR | |||
|---|---|---|---|
| Gene | Pathway(s) | Fold change IR
|
|
| Microarray | Real-time PCR | ||
| p21 | IR control, p53 target | 2.43 | 4.16 |
| AURKA | IR-control, cell cycle | −2.03 | −2.57 |
NOTE: Real-time PCR, using primers toward the indicated genes, was carried out on the same mRNA samples that were used in two of the biological replicates in the microarray experiments. Two biological replicates were executed. The fold change in mRNA levels caused by (A) NFBD1 shRNA, (B) p53 shRNA, or (C) IR as determined by microarray and real-time PCR analysis is shown. 89% of the genes tested were regulated in the same direction according to both the microarray and real time PCR experiments.
Percent validation: 89%
Effect of NFBD1 depletion on CAV1 and CAV2 mRNA and protein levels
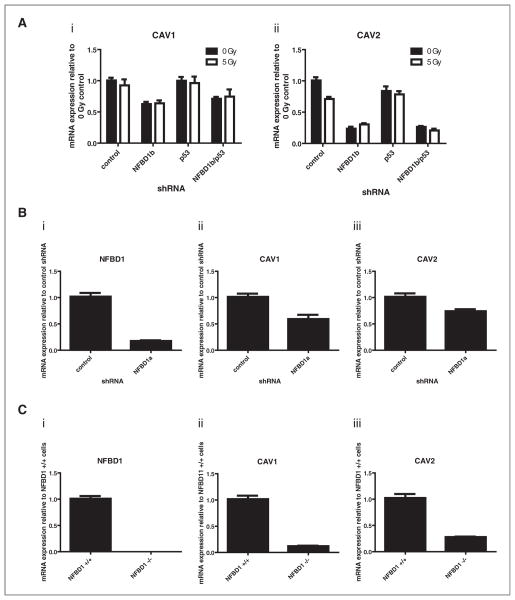
CAV1 and CAV2 mRNAs were both reduced with NFBD1 depletion independent of irradiation and/or p53 depletion. Real-time PCR analysis confirmed that result [Fig. 2A(i) and 2A(ii)]. To ensure that the aforementioned result was not a unique off-target effect of a specific shRNA sequence, U2OS cells were infected with a retrovirus expressing a different NFBD1-directed sequence (NFBD1a) (Fig. 2Bi). Real-time PCR analysis indicated that CAV1 and CAV2 mRNA levels are reduced by that shRNA as well [Fig. 2B(ii) and 2B(iii)]. Furthermore, NFBD1−/− mouse embryonic fibroblasts (MEFs) have greatly reduced CAV1 and CAV2 mRNA levels compared with NFBD1+/+ MEFs [Fig. 2C(i), 2C(ii), and 2C(iii)]. Thus, the observed relationship between NFBD1 and Cav1 occurs in cell types other than U2OS and is not an artifact of shRNA-expressing retroviral infection.
Figure 2.
Effect of NFBD1 depletion on CAV1 and CAV2 mRNA levels. A and B, U2OS cells were infected with retroviruses expressing control, NFBD1b, p53, or NFBD1a-targeted shRNAs. A, two days later, cells were irradiated and RNA was isolated four hours after irradiation. B, one day later, cells were treated with 0.6 μg/ml puromycin to select for infected cells. Four days later, RNA was isolated. A and B, cDNA was synthesized and real-time PCR was carried out using primers toward [A(i) and B(ii)] CAV1, [A(ii) and B(iii)] CAV2, or (Bi) NFBD. C, mRNA was isolated from NFBD1+/+ or NFBD1−/− MEFs. cDNA was synthesized and real-time PCR was carried out using primers toward (i) mNFBD1, (ii) mCAV1, or (iii) mCAV2. Error bars represent the standard error of the mean from A, six samples: three technical replicates from two biological replicates, B and C, nine samples: three technical replicates from three independent plates of cells for each condition.
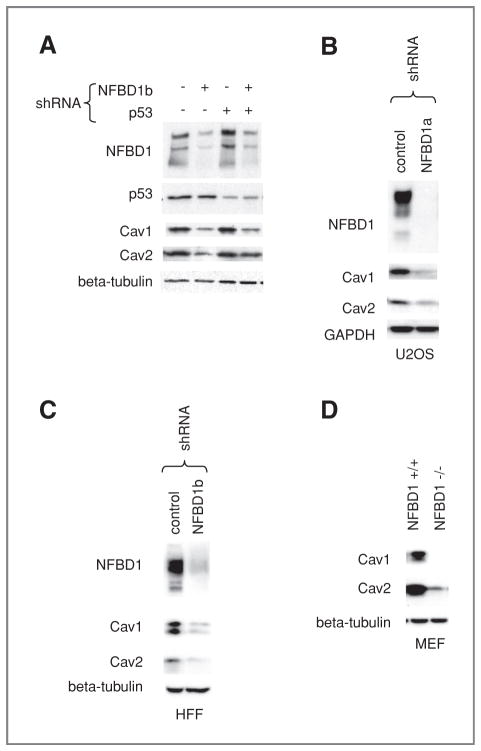
Next, we sought to determine if NFBD1 depletion reduces Cav1 and Cav2 protein levels in addition to mRNA levels. Lysates from irradiated and nonirradiated cells infected with shRNA-expressing retroviruses directed toward NFBD1 and/or p53 were immunoblotted for Cav1, Cav2, and the beta tubulin loading control (Fig. 3A). Consistent with the mRNA results, NFBD1 knockdown reduced Cav1 and Cav2 protein levels in an IR-independent, p53-independent manner. Cav1 and Cav2 protein levels were also reduced by a different NFBD1-directed shRNA-expressing retrovirus (Fig. 3B). A similar response was seen in HFF cells, where CAV1 and CAV2 mRNA and protein levels were reduced with NFBD1 knockdown (Supplementary Figure S1 and Fig. 3C). The reduction in Cav1 and Cav2 levels was also seen in NFBD1−/− MEFs as compared with NFBD1+/+ MEFs (Fig. 3D).
Figure 3.
Effect of NFBD1 depletion on Cav1 and Cav2 protein levels. A and B, U2OS cells or C, HFF cells were infected with the indicated shRNAs. A–C, two days after infection, cells were lysed. A, the NFBD1, p53, and beta–tubulin loading control blots are also shown in Fig. 1C (four left-most lanes). D, NFBD1+/+ and NFBD1−/− MEFs were lysed. A–D, western blots were carried out on lysates using the indicated antibodies.
Cav1 and Cav2 are structural components of caveolae, which are flask-shaped plasma membrane invaginations involved in endocytosis and regulation of signaling molecules (34). Antisense-mediated downregulation or knockout of Cav1 causes increased phosphorylation/activation of Erk1/2 and Mek1/2 (35, 36). Other signaling molecules negatively regulated by Cav1 include Akt, Gαs, c-Src, eNOS, H-Ras, and Neu (37–42). A region called the “scaffolding domain” (residues 82–101) is thought to negatively regulate several signaling molecules (39, 40). Specifically, a synthetic Cav1 peptide of residues 82–101 can inhibit the kinase activity of purified Erk2 and Mek1 (43).
CAV1−/− mice have increased mammary tumorigenesis and lung metastasis driven by a MMTV–PyMT (mouse mammary tumor virus-polyoma middle T antigen) trans-gene (44). Conversely, enhanced expression of CAV1 through gene transfer into MCF-7 human breast cancer cells reduces growth of these cells as mouse xenografts (45).
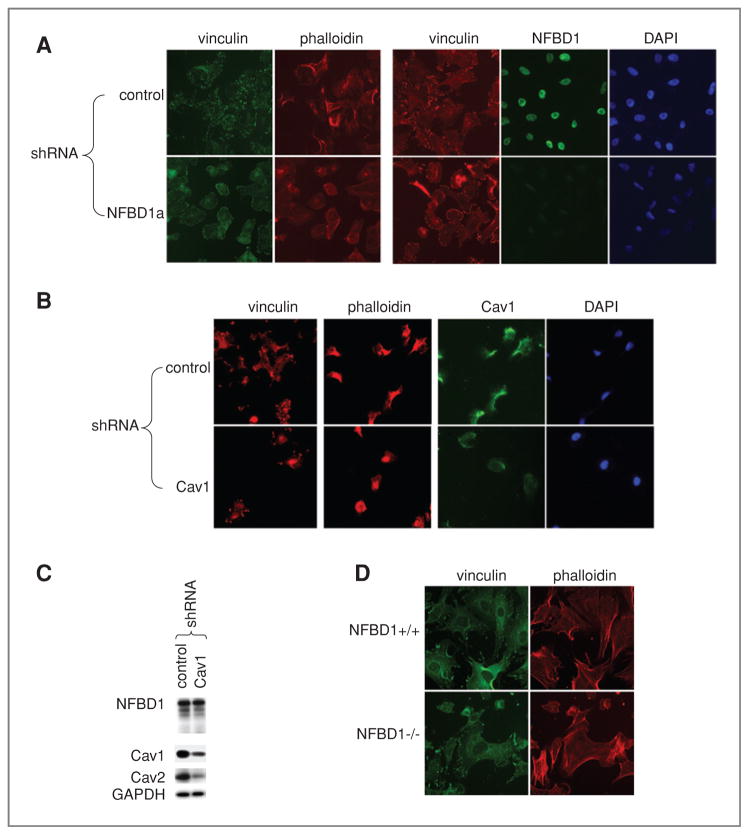
Effect of NFBD1 depletion on focal adhesions and actin cytoskeleton
Next, we sought to determine if NFBD1-depleted cells exhibit similar phenotypes to Cav1-depleted cells. CAV1−/− MEFs have a nonpolarized round shape, cortical actin rings (as opposed to bundled stress fibers) and smaller and more abundant focal adhesions (46). NFBD1 shRNA efficiently suppressed expression of NFBD1 in U2OS cells growing on fibronectin-coated glass coverslips (Fig. 4A). Like Cav1 depletion, NFBD1 depletion reduced the size of focal adhesions marked with immunofluorescence for vinculin (Fig. 4A, 4B). Furthermore, similar to Cav1 depletion, NFBD1 depletion caused cells to adopt a rounder morphology as visualized by phalloidin staining of the actin cytoskeleton (Fig. 4A, 4B). Because Cav1 depletion did not affect NFBD1 levels (Fig. 4C), the effect of Cav1 on cellular morphology is not due to reduced NFBD1 levels. Consistent with NFBD1 knockdown in U2OS cells, NFBD1−/− MEFs had rounder morphology and smaller focal adhesions (Fig. 4D).
Figure 4.
Effect of NFBD1 depletion on focal adhesions and actin cytoskeleton. A–C, U2OS cells were infected with control, Cav1, or NFBD1a shRNA-expressing retroviruses. A and B, two days after infection, cells were plated on glass chamber slides coated with fibronectin. D, NFBD1+/+ and NFBD1−/− MEFs were plated on fibronectin-coated glass chamber slides. A, B, D, immunofluorescence was carried out using vinculin, NFBD1, or Cav1 antibodies. Actin was stained with Texas Red–phalloidin. C, western blots were carried out on lysates using the indicated antibodies.
Effect of NFBD1 depletion on scratch filling
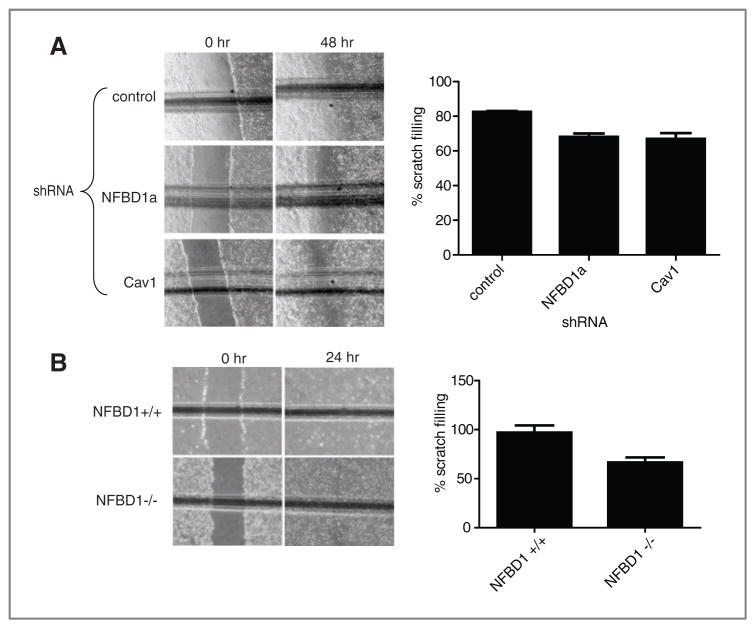
CAV1−/− MEFs have reduced migratory capability compared with CAV1+/+ MEFs as measured by scratch assays (46). Because NFBD1 depletion caused a reduction in Cav1, we sought to determine if NFBD1 depletion affects scratch filling. NFBD1 depletion in U2OS cells does in fact delay scratch filling as measured by scratch assays similarly to Cav1 depletion (Fig. 5A). Furthermore, NFBD1−/− MEFs exhibit slower scratch filling than NFBD1+/+ MEFs (Fig. 5B).
Figure 5.
Effect of NFBD1 depletion on scratch filling. A, U2OS cells were infected with control, Cav1, or NFBD1a shRNA-expressing retroviruses. Cells were allowed to grow three days after infection to reach confluency. B, NFBD1+/+ and NFBD1−/− MEFs were plated to reach confluency. A and B, a scratch was made with a pipette tip. Cells were immediately washed twice with DMEM with 0.1% FBS and then incubated in Dulbecco’s modified eagle medium (DMEM) with 0.1% FBS for the indicated time points. The horizontal line represents a scratch on the outside of the plate to ensure that the same part of the scratch was examined at both time points. The percentage difference between the width of the scratch at the 0-hour time point and the 24-hour or 48-hour time point is plotted. Error bars represent the standard deviation among six samples, three different scratches from each of A, three or B, two biological replicates.
Effect of NFBD1 depletion on Erk phosphorylation
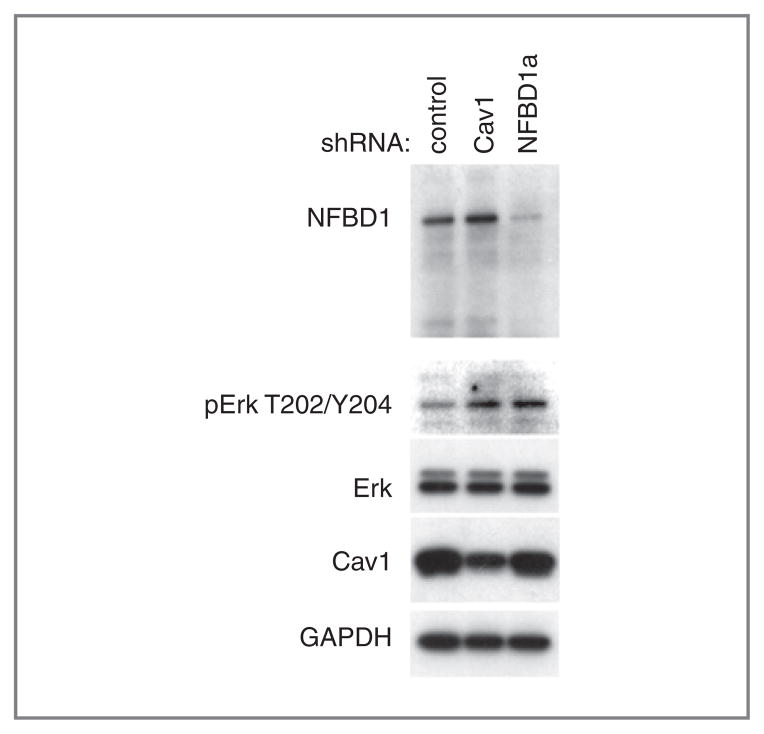
Because Cav1 depletion increases Erk phosphorylation (35–37), we examined Erk phosphorylation in control and NFBD1-depleted cells. U2OS cells infected with either an NFBD1 or Cav1-shRNA expressing retrovirus had higher Erk phosphorylation than cells infected with a control shRNA-expressing retrovirus (Fig. 6), consistent with the decrease in Cav1 levels (Fig. 3B). Total levels of Erk and GAPDH remain constant (Fig. 6).
Figure 6.
Effect of NFBD1 depletion on Erk phosphorylation. U2OS cells were infected with the indicated shRNA-expressing retroviruses. Three days after infection, cells were replated to a lower density (30%–50% confluency) then lysed one day later. Western blots were carried out on lysates using the indicated antibodies.
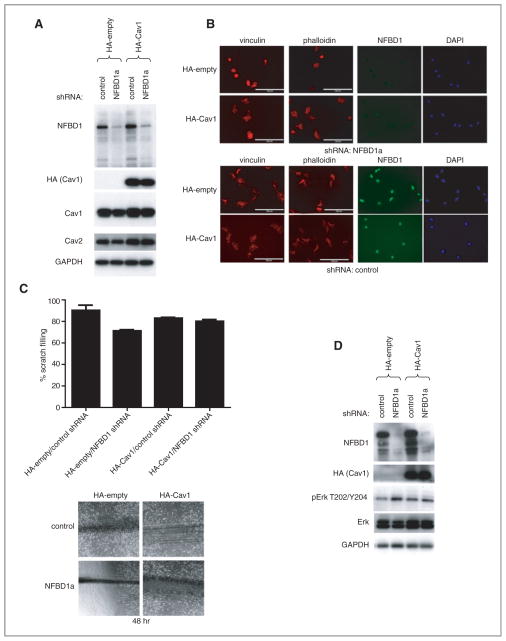
Effect of restoring Cav1 levels on NFBD1 depletion–induced phenotypes
Given the reduction in Cav1 levels upon depletion of NFBD1, and the similar phenotypes observed with NFBD1 depletion and Cav1 depletion, we sought to determine the effects of restoring Cav1 levels on NFBD1 depletion in U2OS cells. U2OS cells were stably transfected with either a control or an HA–Cav1 expression plasmid, and then infected with either control or NFBD1a shRNA-expressing retrovirus. Transfection with the HA–Cav1 expression plasmid restored expression of Cav1 and Cav2 to near wild-type levels in NFBD1-depleted cells (Fig. 7A). NFBD1 depletion in U2OS cells expressing the control plasmid resulted in cells with a rounder morphology and smaller less abundant focal adhesions than in cells expressing the HA–Cav1 plasmid (Fig. 7B). Furthermore, depletion of NFBD1 in cells expressing the control plasmid resulted in delayed scratch filling, whereas scratch filling was not delayed by NFBD1 depletion in cells expressing the HA–Cav1 plasmid (Fig. 7C). Finally, pERK levels were not increased upon NFBD1 depletion in cells expressing HA–Cav1 compared with cells expressing the control plasmid, whereas baseline levels of total ERK were increased in these cells, regardless of NFBD1 depletion (Fig. 7D).
Figure 7.
Effect of restoring Cav1 levels on NFBD1 depletion. U2OS cells were stably transfected with control or an HA-Cav1 plasmid. These cells were infected with either control or NFBD1a shRNA-expressing retroviruses. A and D, western blots were carried out on cell lysates using the indicated antibodies. B, cells were plated onto fibronectin coated chamber slides and immunofluorescent analysis was carried out using the indicated antibodies. Actin was stained with Texas Red-phalloidin. Scale bars are 100 μm. C, scratch assays were carried out as in Fig. 5 on the cells. (Top) Results are quantified, and error bars represent the standard deviation of three biological replicates. Representative images 48 hours after the scratches were made (bottom).
Discussion
NFBD1 regulates the mRNA levels of several genes independently of IR and p53
In this study, we sought to identify novel NFBD1-regulated genes. NFBD1-regulated genes identified by microarray analysis fell into the functional categories of focal adhesion, Jak–STAT, insulin signaling, carbohydrate metabolism, RNA splicing, and translation (Table 1, Table 2). Most of those genes were downregulated with NFBD1 shRNA treatment in an IR-independent and p53-independent manner (Table 1). Therefore, those results point toward a novel role for NFBD1 in altering levels of some mRNAs independently of DNA damage.
Previously, it was reported that residues 508–995 of NFBD1 possess transactivation activity (24). Therefore, NFBD1 might directly regulate expression of various genes at the transcriptional level. Alternatively, NFBD1 may operate indirectly by modulating expression or activity of other transcription factors. For example, the related protein BRCA1 physically interacts with transcription factors, including STAT1, Myc, and estrogen receptor (47). Identifying novel NFBD1-interacting proteins might reveal interactions between NFBD1 and transcription factors.
NFBD1 positively regulates Cav1 and Cav2 mRNA and protein levels
CAV1 and CAV2 are two genes for which mRNA and protein levels are reduced by NFBD1 depletion in an IR-and p53- independent manner (Figs. 2, and 3). The link between NFBD1 and Cav1 is of special interest because both proteins are expressed at reduced levels in some cancer tissues in comparison to normal tissues (48). For example, Cav1 expression is lower in lung cancer cell lines than in normal bronchial cell lines (49). CAV1 and CAV2 mRNA and protein levels are also reduced in follicular thyroid tumors as compared with normal thyroid tissue (50). A dominant negative CAV1 mutation (P132L) occurs in ~16 percent of human breast cancers (51). Furthermore, overexpression of oncogenes v-ABL, polyoma virus middle T antigen, H-RAS G12V, or NEUT in NIH 3T3 cells decreases Cav1 protein levels (41, 52). Our results indicate that, like oncogene overexpression, depletion of the transformation suppressor NFBD1 reduces Cav1 levels.
NFBD1 depletion evokes similar phenotypes to Cav1 depletion
To elucidate the biological significance of NFBD1 depletion, we sought to determine if NFBD1 depletion yields similar phenotypes to those induced by Cav1 depletion. NFBD1 depletion caused cells to adopt a more rounded morphology and have smaller focal adhesions, similar to CAV1−/− cells and cells expressing Cav1-directed shRNA (Fig. 4) (46). Consistent with the focal adhesion defect, NFBD1 depletion by either shRNA or knockout caused a scratch-filling defect (Fig. 5). U2OS cells depleted of Cav1 and CAV1−/− MEFs also exhibit a scratch filling defect [Fig. 5 and (46)]. Furthermore, we found that, like Cav1 depletion, NFBD1 depletion increases Erk phosphorylation [Fig. 6 and (35–37)].
Although ERKs are best-known for their roles in growth factor-regulated pathways, some studies do suggest crosstalk with DNA damage response systems. For example, ATM has been reported to influence Erk phosphorylation. Specifically, Erk phosphorylation is increased in ATM−/− compared with ATM+/+ astrocytes (53, 54). As NFBD1 is a component of the ATM pathway, our finding that NFBD1 increases Erk phosphorylation is consonant with that result. BRCA1, a DNA damage response factor with partial sequence homology to NFBD1 and some functional overlap, inhibits Erk activation induced by estradiol (27, 55). Furthermore, BRCA1 depletion, like NFBD1 depletion, reduces Cav1 levels (56).
Restoration of Cav1 levels rescues effects of NFBD1 depletion
Forced expression of Cav1 rescued the effects of NFBD1 depletion. Specifically, cells expressing the control plasmid were rounder, had smaller less abundant focal adhesions, were delayed for scratch filling, and showed an increase in pERK levels upon NFBD1 depletion compared with cells expressing the HA–Cav1 plasmid (Fig. 7). These results indicate that ectopic Cav1 expression can compensate for NFBD1 depletion. Thus, these data suggest that the effects of NFBD1 depletion are mediated by the reduction in Cav1 levels seen with NFBD1 knockdown, at least with regards to the phenotypes analyzed here.
Overall, these studies reinforce a general model in which the DNA damage response system, in addition to promoting DNA repair, negatively regulates general cellular transformation processes in response to genotoxins and also endogenous DNA damage. Documented roles for DNA damage response proteins in the absence of exogenous DNA damaging agents are not abundant. An example is ADP ribosylation factor (ARF) and p53-dependent cell cycle arrest and senescence induced by oncogenic Ras (57). Furthermore, overexpression of the oncogene Mos in human fibroblasts is known to cause activation of the ATM pathway, as measured by phosphorylation of the downstream factors Chk2, H2AX, and p53 (58). Now we find that, even without DNA damage, NFBD1 also collaterally reduces growth factor pathway signaling.
NFBD1 expression, as measured by immunohistochemistry, is lower in some breast and lung cancers compared with normal tissue (48). Determining if Cav1 and Cav2 levels correlate with NFBD1 levels in those cancers would be important, as it may provide further insight into the mechanism by which NFBD1 antagonizes carcinogenesis. It will be of interest to determine if Erk phosphorylation is increased in NFBD1-deficient tumors. If Erk is activated in tumors deficient in NFBD1, Erk, and Akt inhibitors might be effective in treating those cancers.
Supplementary Material
Acknowledgments
We thank Daniel DiMaio and Kristin Yates (Yale University) for the p53 shRNA retroviral plasmid, Junjie Chen (M.D. Anderson) and Zhenkun Lou (Mayo Clinic) for the NFBD1+/+ and NFBD1−/− MEFs, and the Stern lab for insightful discussions.
Grant Support
This work was supported by USPHS grant R01CA82257 and USAMRMC predoctoral fellowship W81XWH-06–1-0743 in Breast Cancer Research awarded to K. A. Wilson.
Footnotes
Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. Faseb J. 1997;11:68–76. [PubMed] [Google Scholar]
- 2.Xu X, Stern DF. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J Biol Chem. 2003;278:8795–803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- 3.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–6. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 4.Shang YL, Bodero AJ, Chen PL. NFBD1, a novel nuclear protein with signature motifs of FHA and BRCT, and an internal 41-amino acid repeat sequence, is an early participant in DNA damage response. J Biol Chem. 2003;278:6323–9. doi: 10.1074/jbc.M210749200. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–6. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 6.Lou Z, Minter-Dykhouse K, Wu X, Chen J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature. 2003;421:957–61. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- 7.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–57. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, et al. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–40. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–26. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci U S A. 2008;105:11200–5. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X, Stern DF. NFBD1/MDC1 regulates ionizing radiation-induced focus formation by DNA checkpoint signaling and repair factors. Faseb J. 2003;17:1842–8. doi: 10.1096/fj.03-0310com. [DOI] [PubMed] [Google Scholar]
- 14.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. Embo J. 2004;23:2674–83. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horejsi Z, Falck J, Bakkenist CJ, Kastan MB, Lukas J, Bartek J. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene. 2004;23:3122–7. doi: 10.1038/sj.onc.1207447. [DOI] [PubMed] [Google Scholar]
- 16.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–21. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. Embo J. 2003;22:6610–20. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerosaletti K, Concannon P. Independent Roles for Nibrin and Mre11-Rad50 in the Activation and Function of Atm. J Biol Chem. 2004;279:38813–9. doi: 10.1074/jbc.M404294200. [DOI] [PubMed] [Google Scholar]
- 19.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 20.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–6. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 21.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM Activation and Its Recruitment to Damaged DNA Require Binding to the C Terminus of Nbs1. Mol Cell Biol. 2005;25:5363–79. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shull ER, Lee Y, Nakane H, Stracker TH, Zhao J, Russell HR, et al. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–80. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozaki T, Nagase T, Ichimiya S, Seki N, Ohiri M, Nomura N, et al. NFBD1/KIAA0170 is a novel nuclear transcriptional transactivator with BRCT domain. DNA Cell Biol. 2000;19:475–85. doi: 10.1089/10445490050128403. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi M, Ozaki T, Yamamoto H, Hanamoto T, Kikuchi H, Furuya K, et al. NFBD1/MDC1 associates with p53 and regulates its function at the crossroad between cell survival and death in response to DNA damage. J Biol Chem. 2007;282:22993–3004. doi: 10.1074/jbc.M611412200. [DOI] [PubMed] [Google Scholar]
- 26.Dávalos A, Fernáandez-Hernando C, Sowa G, Derakhshan B, Lin MI, Lee JY, et al. Quantitative proteomics of caveolin-1-regulated proteins: characterization of polymerase i and transcript release factor/CAVIN-1 IN endothelial cells. Mol Cell Proteomics. 9:2109–24. doi: 10.1074/mcp.M110.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson KA, Stern DF. NFBD1/MDC1, 53BP1 and BRCA1 have both redundant and unique roles in the ATM pathway. Cell Cycle. 2008;7:3584–94. doi: 10.4161/cc.7.22.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yates KE, Korbel GA, Shtutman M, Roninson IB, DiMaio D. Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging Cell. 2008;7:609–21. doi: 10.1111/j.1474-9726.2008.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X, Liu Y, Ma Q, Wang X, Jin H, Mehrpour M, et al. Caveolin-1 negatively regulates TRAIL-induced apoptosis in human hepatocarcinoma cells. Biochem Biophys Res Commun. 2009;378:21–6. doi: 10.1016/j.bbrc.2008.10.123. [DOI] [PubMed] [Google Scholar]
- 30.Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng EC, Luo Q, Bruscia EM, Renda MJ, Troy JA, Massaro SA, et al. Role for MKL1 in megakaryocytic maturation. Blood. 2009;113:2826–34. doi: 10.1182/blood-2008-09-180596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 34.Hnasko R, Lisanti MP. The biology of caveolae: lessons from caveolin knockout mice and implications for human disease. Mol Interv. 2003;3:445–64. doi: 10.1124/mi.3.8.445. [DOI] [PubMed] [Google Scholar]
- 35.Galbiati F, Volonte D, Engelman JA, Watanabe G, Burk R, Pestell RG, et al. Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. Embo J. 1998;17:6633–48. doi: 10.1093/emboj/17.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen AW, Park DS, Woodman SE, Williams TM, Chandra M, Shirani J, et al. Caveolin-1 null mice develop cardiac hypertrophy with hyperactivation of p42/44 MAP kinase in cardiac fibroblasts. Am J Physiol Cell Physiol. 2003;284:C457–74. doi: 10.1152/ajpcell.00380.2002. [DOI] [PubMed] [Google Scholar]
- 37.Murata T, Lin MI, Huang Y, Yu J, Bauer PM, Giordano FJ, et al. Reexpression of caveolin-1 in endothelium rescues the vascular, cardiac, and pulmonary defects in global caveolin-1 knockout mice. J Exp Med. 2007;204:2373–82. doi: 10.1084/jem.20062340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smart EJ, Graf GA, McNiven MA, Sessa WC, Engelman JA, Scherer PE, et al. Caveolins, liquid-ordered domains, and signal transduction. Mol Cell Biol. 1999;19:7289–304. doi: 10.1128/mcb.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Couet J, Lisanti MP. Src Tyrosine Kinases, Galpha Subunits, and H-Ras Share a Common Membrane-anchored Scaffolding Protein, Caveolin. J Biol Chem. 1996;271:29182–90. doi: 10.1074/jbc.271.46.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, Okamoto T, Chun M, Sargiacomo M, Casanova JE, Hansen SH, et al. Evidence for a Regulated Interaction between Heterotrimeric G Proteins and Caveolin. J Biol Chem. 1995;270:15693–701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- 41.Engelman JA, Lee RJ, Karnezis A, Bearss DJ, Webster M, Siegel P, et al. Reciprocal Regulation of Neu Tyrosine Kinase Activity and Caveolin-1 Protein Expression in Vitro and in Vivo. J Biol Chem. 1998;273:20448–55. doi: 10.1074/jbc.273.32.20448. [DOI] [PubMed] [Google Scholar]
- 42.Sowa G, Pypaert M, Sessa WC. Distinction between signaling mechanisms in lipid rafts vs. caveolae. Proc Natl Acad Sci U S A. 2001;98:14072–7. doi: 10.1073/pnas.241409998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engelman JA, Chu C, Lin A, et al. Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo. A role for the caveolin-scaffolding domain. FEBS Lett. 1998;428:205–11. doi: 10.1016/s0014-5793(98)00470-0. [DOI] [PubMed] [Google Scholar]
- 44.Williams TM, Medina F, Badano I, Hazan RB, Hutchinson J, Muller WJ, et al. Caveolin-1 gene disruption promotes mammary tumorigenesis and dramatically enhances lung metastasis in vivo. Role of Cav-1 in cell invasiveness and matrix metalloproteinase (MMP-2/9) secretion. J Biol Chem. 2004;279:51630–46. doi: 10.1074/jbc.M409214200. [DOI] [PubMed] [Google Scholar]
- 45.Wu P, Wang X, Li F, Qi B, Zhu H, Liu S, et al. Growth suppression of MCF-7 cancer cell-derived xenografts in nude mice by caveolin-1. Biochem Biophys Res Commun. 2008;376:215–20. doi: 10.1016/j.bbrc.2008.08.146. [DOI] [PubMed] [Google Scholar]
- 46.Grande-García A, Echarri A, de Rooij J, Alderson NB, Waterman-Storer CM, Valdivielso JM, et al. Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J Cell Biol. 2007;177:683–94. doi: 10.1083/jcb.200701006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–63. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 48.Bartkova J, Horejsí Z, Sehested M, Nesland JM, Rajpert-De Meyts E, Skakkebaek NE, et al. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 2007;26:7414–22. doi: 10.1038/sj.onc.1210553. [DOI] [PubMed] [Google Scholar]
- 49.Racine C, Belanger M, Hirabayashi H, Boucher M, Chakir J, Couet J. Reduction of caveolin 1 gene expression in lung carcinoma cell lines. Biochem Biophys Res Commun. 1999;255:580–6. doi: 10.1006/bbrc.1999.0236. [DOI] [PubMed] [Google Scholar]
- 50.Aldred MA, Ginn-Pease ME, Morrison CD, Popkie AP, Gimm O, Hoang-Vu C, et al. Caveolin-1 and caveolin-2, together with three bone morphogenetic protein-related genes, may encode novel tumor suppressors down-regulated in sporadic follicular thyroid carcinogenesis. Cancer Res. 2003;63:2864–71. [PubMed] [Google Scholar]
- 51.Hayashi K, Matsuda S, Machida K, Yamamoto T, Fukuda Y, Nimura Y, et al. Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res. 2001;61:2361–4. [PubMed] [Google Scholar]
- 52.Koleske AJ, Baltimore D, Lisanti MP. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci U S A. 1995;92:1381–5. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Wong PK. Oxidative stress is linked to ERK1/2-p16 signaling-mediated growth defect in ATM-deficient astrocytes. J Biol Chem. 2009;284:14396–404. doi: 10.1074/jbc.M808116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu N, Stoica G, Yan M, Scofield VL, Qiang W, Lynn WS, et al. ATM deficiency induces oxidative stress and endoplasmic reticulum stress in astrocytes. Lab Invest. 2005;85:1471–80. doi: 10.1038/labinvest.3700354. [DOI] [PubMed] [Google Scholar]
- 55.Razandi M, Pedram A, Rosen EM, Levin ER. BRCA1 inhibits membrane estrogen and growth factor receptor signaling to cell proliferation in breast cancer. Mol Cell Biol. 2004;24:5900–13. doi: 10.1128/MCB.24.13.5900-5913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Yu J, Zhan Q. BRCA1 regulates caveolin-1 expression and inhibits cell invasiveness. Biochem Biophys Res Commun. 2008;370:201–6. doi: 10.1016/j.bbrc.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 57.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–6. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 58.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.