Abstract
ErbB4 is unusual among receptor tyrosine kinases because some isoforms can be efficiently cleaved at the plasma membrane to release a soluble intracellular domain. The cleavage product has high kinase activity and homes to the nucleus. A screen for proteins that associate with the ErbB4 intracellular domain identified candidate interactors including ITCH, WWP2, Nucleolin, and Krab-associated protein 1 (Kap1). Kap1 binds to multiple isoforms of ErbB4 but does not require ErbB4 kinase activity for binding, nor is it an ErbB4 substrate. Kap1 reduces ERBB4 transcription and either directly or indirectly modulates the expression of genes that are themselves regulated by ErbB4. Upregulation of ErbB4 and suppression of MDM2 jointly enhance and accelerate the accumulation of p21CIP1 in response to DNA damage. Overall, these findings further substantiate the role of ErbB4 in conjoint regulation of growth factor signaling and DNA damage responses.
Introduction
Three of the four receptors in the epidermal growth factor family, epidermal growth factor receptor, ErbB2, and ErbB3, are common drivers in human carcinoma, glioblastoma, and other solid tumors. For this reason, they are important targets for new cancer therapeutics. The influence of ErbB4 on human cancer has been more controversial because mutations were rarely reported, and reports on ErbB4 expression as a prognostic marker in carcinoma were contradictory (1). However, the association of ErbB4 with medulloblastoma (2) and the recent identification of candidate ErbB4-activating mutations in lung cancer (3) and melanoma (4) have renewed interest in this problem.
The variable associations of ErbB4 with cancer may be explained by the unusual diversity of ErbB4-regulated signaling processes, which are associated with two different mRNA splice choices. JM-A and JM-B isoforms differ in sequences encoding the extracellular juxtamembrane domain (Fig. 1A; ref. 5). JM-B isoforms function similarly to other receptor tyrosine kinases: they are activated by ligand binding, cross-phosphorylate in dimers, and recruit signaling proteins that bind to the phosphorylation sites and can be further regulated through phosphorylation. Augmenting their “normal” receptor tyrosine kinase signaling capabililty, JM-A isoforms harbor a metalloproteinase cleavage site that is clipped in response to activation by the ErbB4 ligand Neuregulin-1 (NRG1) or the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA; refs. 6, 7). This cleavage releases most of the extracellular domain, leaving a membrane-anchored protein, termed m80, which consists of a small residual extracellular domain fragment and the transmembrane and cytoplasmic domains. The m80 form can then undergo intramembrane cleavage by γ-secretase or a similar enzyme to release the s80 form, consisting of the entire intracellular domain (ICD) and with constitutive tyrosine kinase activity. s80 relocalizes efficiently to the mitochondria and nucleus (7, 8). The nuclear processes regulated by ErbB4 ICD are still emerging, but they include binding of transcriptional coregulators and transcription factors (8).
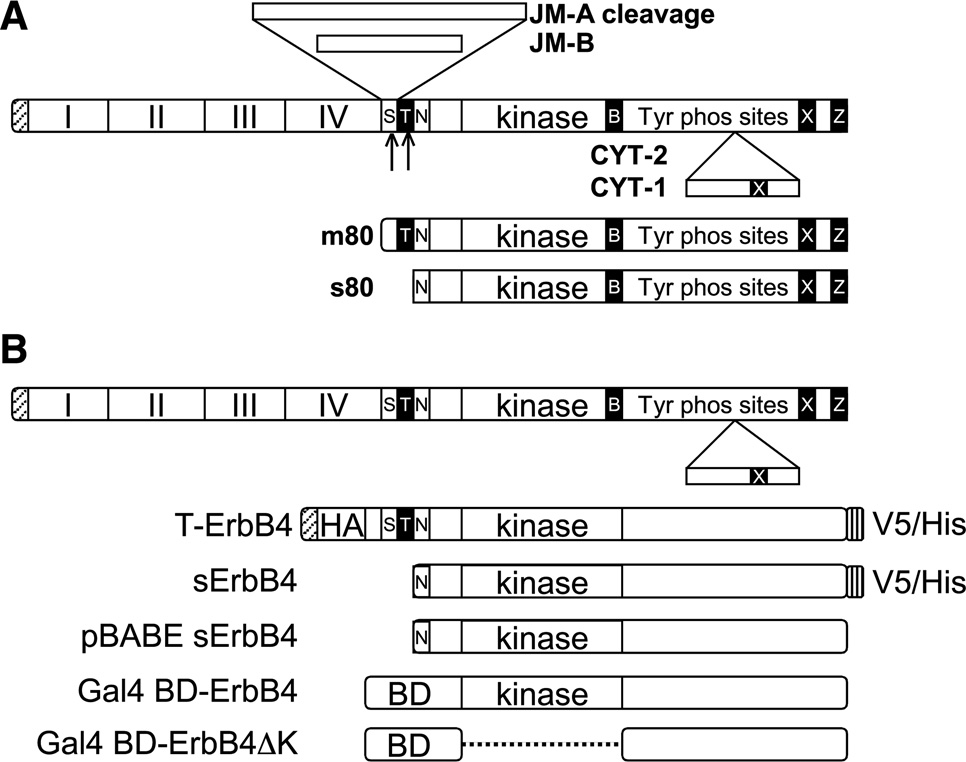
FIGURE 1.
ErbB4 isoforms and constructs. A, ErbB4 splicing and cleavage. Alternative splicing to produce JM-A, JM-B, CYT-1, and CYT-2. Metalloproteinase cleavage of the JM-A stalk releases m80, and then γ-secretase frees s80. S, stalk; T, transmembrane domain; B, BH3 domain; Z, PDZ binding site; N, functionally important nuclear localization signal; X, WW-binding PPXY domain. B, the ErbB4 constructs described in the text.
Another source of diversity in ErbB4 signaling is dictated by a second splice choice in the ICD (Fig. 1A). CYT-1 isoforms include an exon that is absent in CYT-2 (5). Interestingly, in mammary epithelia, the CYT-1 versus CYT-2 choice determines very different biological responses to ErbB4, with CYT-1 associated with differentiation and CYT-2 with growth promotion and transformation (5, 9–11). The mechanisms for these differences have not been uncovered, but they may be linked to the presence of protein binding domains in the CYT-1–specific exon. These include a binding site for the p85 adapter subunit of phosphatidylinositol 3′-kinase and an overlapping PPXY binding site for WW domains (5, 12, 13). The divergence of signaling processes incited by the four different ErbB4 isoforms may explain the discordances in the ErbB4 cancer literature because most research studies fail to consider these isoforms separately. Some recent studies suggest that either the isoform or the subcellular localization of ErbB4 has an effect on prognosis (12, 14).
ErbB4 ICD associates with the transcriptional factors estrogen receptor α and Stat5 (15, 16); with coregulators including Yes-associated protein (YAP; ref. 13), WWOX (12), Eto2 (17), and the TAB2/NcoR complex (18); and with the ubiquitin ligases Itch (19) and MDM2 (20). In an effort to learn more about the global signaling functions of nuclear ErbB4, we have identified proteins that associate with the ICD. This screen identified an association of the ErbB4 ICD with a common transcriptional corepressor, and suggest that ErbB4-containing complexes coordinate cell growth responses to ErbB4 activation and DNA damage.
Materials and Methods
Tissue culture
COS-7, BT-474, MCF-7, T-47D, and MDA-MB452 cell lines were obtained from the American Type Culture Collection and were cultured in RPMI 1640 with glutamate (Gibco) supplemented with 100 units/mL penicillin, 100 µg/mL streptomycin, and 10% fetal bovine serum (FBS; BioWest). HEK293T cells (American Type Culture Collection) were cultured in high-glucose DMEM supplemented with 50 mmol/L HEPES (pH 7.4), penicillin/ streptomycin, and 10% FBS. Transfections in all cell lines were done with FuGENE 6 (Roche) at a 3:1 ratio of DNA to FuGENE according to the manufacturer’s instructions. For some experiments, cells were incubated with NRG (50 ng/mL) and/or TPA (100 ng/mL) in 0.1% FBS for 40 minutes before cell lysis.
Plasmids
Truncated ErbB4 (T-ErbB4) CYT-1 plasmids (Fig. 1B) were gifts from T.W. Kim (College of Physicians and Surgeons, Columbia University, New York, NY). T-ErbB4 CYT-2 was constructed from T-ErbB4 CYT-1 by deletion of nucleotides 3234–3281 using the primer 5′gagcaagaattgcagtttgtatcccg and the Stratagene QuikChange II XL Kit. Kinase-defective mutations were made using the same kit to encode A759R in both isoforms. The soluble s80 CYT-2 plasmid encoding amino acids R675 to V1307 was produced using T-ErbB4 CYT-2 as template for PCR. The primers for the reaction were forward 5′actttaccacaacatgctagactc and reverse 5′cggatccgcccattctgtgtataaa. The product was cloned into the TA cloning vector pcDNA3.1 (Invitrogen). SalI sites were attached to the ends of the insert by PCR. The product was digested and ligated into a SalI-cleaved pBABE retroviral vector to generate s80 pBABE. The plasmids containing the GAL4 DNA binding domain fused with ERBB4 cytoplasmic tail fragment (CTF) and CTF ΔK with deleted kinase domain were kindly provided by Marius Sudol (Geisinger Health System Weis Center for Research, Danville, PA; ref. 13). The human Krab-associated protein 1 (Kap1) plasmid was purchased from Origene. The MDM2 plasmid was a gift from H. Zhang (Peking University Health Science Center, Beijing, China; ref. 21). The shMDM2 plasmid and virus were produced using the pSiren system (BD Biosciences) according to the manufacturer’s instructions, with the target sequence sense strand 5′ggatcccaggtgtcaccttgaagtgaattctac. The shTp53 plasmid was a gift from Kristin Yates (Yale University, New Haven, CT; ref. 22). The shKap plasmid was a gift from A. Ivanov (West Virginia University School of Medicine, Morgantown, WV; ref. 23).
Immunoprecipitation and immunoblotting
For total cell extracts, cells were lysed in NP40 buffer [1% NP40, 150 mmol/L NaCl, 50 mmol/L Tris-HCl (pH 7.4)] with phosphatase inhibitor cocktails 1 and 2 (Sigma). Nuclear and cytoplasmic extracts were prepared using the NE-PER kit (Thermo Pierce) according to the manufacturer’s instructions. Protein concentrations were determined using detergent-compatible protein assay (Bio-Rad). For immunoprecipitations, portions of lysates were reacted with 2 µg of specific antibody for 3 hours in the presence of protein A/G beads (Thermo) at 4°C. ErbB4 was immunoprecipitated with sc-8050 (Santa Cruz), Kap1 with A300-274A (Bethyl Laboratories), V5 with 46–0705 (Invitrogen), and Flag with A8592 (Sigma). The beads were harvested by centrifugation and washed five times in lysis buffer. Bound protein was solubilized by heating at 100°C in 2× Laemmli sample buffer. Immunoblots on polyvinylidene difluoride membranes were blocked with 5% nonfat dry milk in PBST (Dulbecco’s PBS with 0.1% Tween 20). The antibodies against the following proteins were incubated with the membranes in 5% nonfat milk in PBST: Kap1 (A300-274A) and phospho-Kap1 (A300–767A; Bethyl Laboratories), ErbB4 (sc-283, sc-8050), p21 (sc-397), p53 (sc-126), MDM2 (sc-965), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; c-25778, Santa Cruz). Membranes were washed in PBST and incubated with horseradish peroxidase–conjugated secondary antibody in 5% nonfat dry milk in PBST.
Preparative immunoprecipitation and tandem mass spectrometry
Ten 100-mm plates of BT-474 cells were incubated in low serum overnight and stimulated with 100 ng/mL TPA. Cells were harvested and washed in PBS. They were lysed in a hypotonic buffer [10 mmol/L HEPES (pH 7.9), 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.2 mmol/L phenylmethylsulfonyl fluoride, 0.5 mmol/L DTT] with Dounce homogenization. The nuclei were collected by centrifugation and the supernatant was removed. The nuclei were washed and resuspended in half the pellet volume in low-salt buffer [20 mmol/L HEPES (pH 7.9), 20 mmol/L KCl, 1.5 mmol/L MgCl2 25% glycerol (v/v), 0.2 mmol/L EDTA, 0.2 mmol/L phenylmethylsulfonyl fluoride, 0.5 mmol/L DTT]. The nuclei were extracted by rocking for 30 minutes at 4°C in half the pellet volume in high-salt buffer (1.2 mol/L KCl in low-salt buffer). The nuclei were pelleted by centrifugation and the supernatant was used for preparative immunoprecipitation. Samples were resolved on a 4% to 12% acrylamide Bis-Tris gel and stained with Simply Blue (Invitrogen) colloidal Coomassie blue stain. A band in the 110 kDa range was excised and analyzed by the Taplin Biomedical Mass Spectrometry Facility of Harvard Medical School, by tandem mass spectrometry (MS/MS). The Taplin Facility conducted in-gel digestion of the gel band, microcapillary liquid chromatography-MS/MS analysis, protein database searching, data analysis, and web-based reporting of the data.
RNA extraction and real-time PCR
Total RNA was extracted from cells using the Qiagen RNeasy Mini Kit according to the manufacturer’s instructions. One hundred nanograms of RNA were converted to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). cDNA was analyzed by TaqMan (Applied Biosystems) quantitative PCR reactions for TP53 Hs00153349_m1, PGR Hs00172183_m1, Trim28 Hs01076234_m1, MDM2 Hs016066930_m1, CDKN1A Hs00355782_m1, and GAPDH Hs9999905_m1.
Luciferase assay
Luciferase assays were conducted using the dual reporter system (Promega) according to the manufacturer’s instructions and analyzed on a 20/20n Single Tube Luminometer (Turner Biosystems).
Results
BT-474 breast tumor cells express high levels of endogenous ErbB4. TPA induces ErbB4 cleavage in these cells, and we verified that the s80 formed under these conditions is indeed soluble (m80 and s80 cannot be differentiated by size) and localized to the nucleus (data not shown). In an effort to identify functional partners for the ErbB4 ICD, we performed a preparative immunoprecipitation of the ErbB4 ICD from BT-474 cells for analysis of associated proteins by MS/MS. Nuclear extracts from TPA-treated BT-474 cells were immunoprecipitated with anti-ErbB4 endodomain antibody or IgG control. Criteria for ICD-associated proteins were that they be precipitated with anti-ErbB4 but not the IgG control and that they be immunoprecipitated only from TPA-treated cells. A prominent band of ~110 kDa was excised and analyzed by MS/MS. The resulting candidate proteins (Table 1) include the heterogeneous nRNA binding proteins HnRNPU, Nucleolin, RMB15, and RMB25; the splicing factor U5S1; the RNA helicase DDX23; poly(ADP-ribose) polymerase 1; the ubiquitin ligases Itch and WWP2; and the transcriptional corepressor known variously as Trim28/Kap1/Tif1β and referred to herein as Kap1.
Table 1.
MS/MS analysis of proteins that coimmunoprecipitate with ErbB4
| Gene | Protein | Peptide matches to sequence |
|---|---|---|
| PRPU | U5 snRNP protein | 33/33 |
| DDX23 | RNA helicase | 29/29 |
| MATR3 | Matrin 3 | 19/19 |
| RBM15 | RNA binding protein 15 | 13/13 |
| ILF3 | Interleukin-binding protein | 11/11 |
| KAP1 | Krab-associated protein 1 | 7/7 |
| U5S1 | U5 snRNP | 5/5 |
| Q9BR70 | Unknown | 5/5 |
| ITCH | E3 ligase | 4/4 |
| HNRPU | HnRNA protein U | 4/4 |
| AP2A1 | Alpha-adaptin 1 | 4/4 |
| Nulc | Nucleolin | 4/4 |
| Q8WVCO | Paf1 complex protein | 3/3 |
| WWP2 | NEDD-like E3 ligase | 3/3 |
| IGHG1 | IgG-like protein | 3/3 |
| HXK1 | Brain hexokinase | 2/2 |
| ARS2 | Arsenite binding protein | 2/2 |
Abbreviations: snRNP, small nuclear ribonucleoprotein; hnRNA, heterogeneous nRNA.
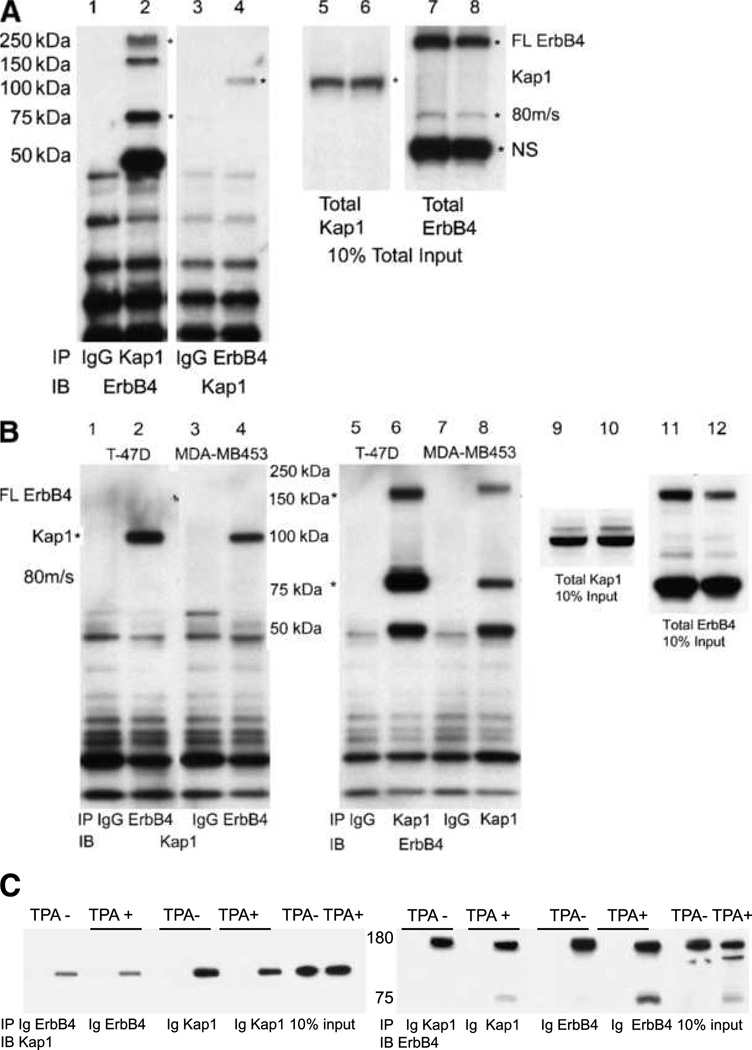
While this work was in progress, another laboratory validated Itch as an ErbB4 binding protein and has linked Itch to downregulation of ErbB4 in vivo (19, 24). Nucleolin has now been verified as an ErbB4-interacting protein (25). The ability of Kap1 and ErbB4 ICD to form a complex was further evaluated in BT-474 cells. The cells were stimulated with NRG to induce ErbB4 cleavage, followed by immunoprecipitation of either ErbB4 or Kap1 (Fig. 2A). Although both full-length and cleaved forms of ErbB4 were present in anti-Kap1 immunoprecipitates (Fig. 2A, lane 2, asterisks), anti-Kap1 enriched the ~80-kDa cleaved form (Fig. 2A, compare lane 2 with lane 8). Likewise, anti-ErbB4 immunoprecipitated Kap1 (lane 4). Similarly, Kap1 and ErbB4 cross-immunoprecipitated after TPA treatment to induce ErbB4 cleavage in two other breast tumor cell lines, T-47D and MDA-MB453, which expressed lower levels of endogenous ErbB4 (Fig. 2B). Hence, ErbB4 and Kap1 can be isolated in a single complex with endogenous levels of expression in these three tumor cell lines.
FIGURE 2.
Reciprocal coimmunoprecipitation of ErbB4 and Kap1 in breast cancer cell lines. A, coimmunoprecipitation from NRG-treated BT-474 cells. Five hundred micrograms of BT-474 cell lysate were immunoprecipitated with 2 µg of total rabbit IgG, anti-Kap1, or anti-ErbB4. Immune complexes were isolated using protein A/G beads and analyzed on 4% to 12% NuPAGE Bis-Tris gels. Portions of the lysates were analyzed directly in lanes 5 to 8. Immunoblots were probed with the reciprocal antibodies to those used for immunoprecipitation. Asterisks mark the full-length and 80-kDa cleaved forms of ErbB4 (lanes 2, 7, and 8) or Kap1 (lanes 4–6). NS, nonspecific band on ErbB4 blots. B, coimmunoprecipitation of ErbB4 and Kap1 from NRG-treated T-47D and MDA-MB453 cells. Portions of the lysates were analyzed directly without immunoprecipitation in lanes 9 to 12. C, HEK293T cells were transfected with 1 µg each of the plasmids encoding full-length ErbB4 and Flag-Kap1. After 24 h, the cells were switched to starvation medium (0.1% FBS in DMEM) for 16 h and then mock stimulated with DMSO vehicle (TPA−) or with 100 ng/mL TPA (TPA+) in DMSO. Lysates were immunoprecipitated with anti-Kap1, anti-ErbB4, or control IgG (Ig). Kap1 and ErbB4 were identified by immunoblotting, with ~100-kDa Kap1 and full-length and cleaved ErbB4 forms. Blotting for GAPDH (not shown) verified even loads for 10% input tracks. Data represent at least two biological replicates in identical format plus two similar biological replicates (A and B).
Kap1 is a generalized transcriptional corepressor that works together with KRAB-family transcription factors. Kap1 recruitment to chromatin through these transcription factors and heterochromatin protein 1 brings in Kap1 and the Kap1-binding histone methyltransferase SETDB1. The whole complex induces heterochromatinization (26). Interestingly, Kap1-dependent chromatin compaction is relieved by ATM phosphorylation of Kap1 after DNA damage, thereby linking a DNA damage response to Kap1-dependent regulation (27, 28).
Requirements for Kap1/ErbB4 interaction
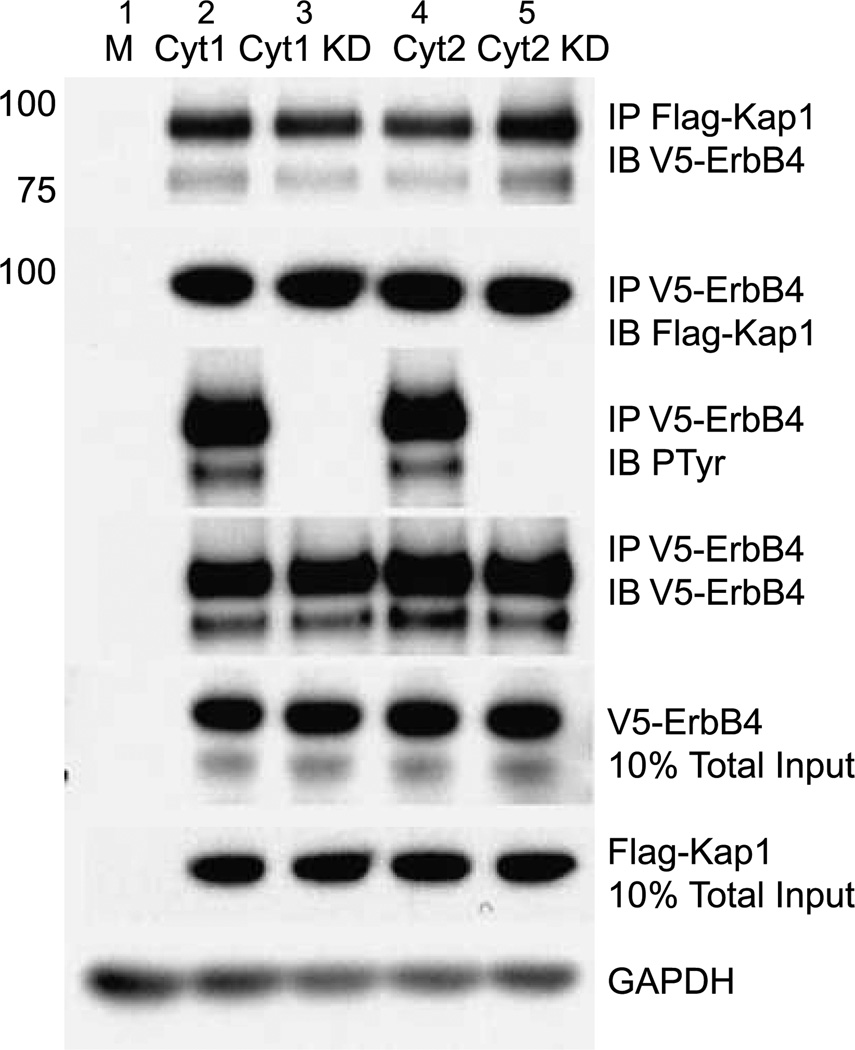
Endogenous Kap1 coimmunoprecipitates with anti-ErbB4 in BT-474, T-47D, and MDA-MB453 breast cancer cells (Fig. 2). Because the biological activities of ErbB4 CYT-1 and CYT-2 can be quite different (9), we determined whether Kap1 binds both isoforms. To facilitate recovery of the m80 and s80 forms of ErbB4, for these and the subsequent experiments, we expressed tagged NH2-terminally truncated forms of ErbB4 that either have the transmembrane domain (T-ErbB4, Fig. 1B) or lack the transmembrane domain and are soluble (V/His sErbB4, Fig. 1B). Cleavage of T-ErbB4 to release the ICD was triggered by treatment with TPA. Both the CYT-1 and CYT-2 forms of V5-tagged T-ErbB4 coimmunoprecipitated with Flag-tagged Kap1 when both were expressed in COS-7 cells (Fig. 3). Thus, Kap1 binding is CYT isoform independent. Likewise, because these forms lack most of the ectodomain, most of this domain is not required for the interaction. These data indicate that a subpopulation of Kap1 interacts with a subpopulation of full-length ErbB4, m80, and s80. Because Kap1 interacts with both cleaved and noncleaved forms of ErbB4, we determined the extent to which cleavage modulates coimmunoprecipitation. TPA-induced cleavage of ErbB4 did not significantly shift the total amount 8of Kap1 immunoprecipitated with anti-Kap1 (Fig. 2C, left), and the amounts of ErbB4 that coprecipitated with anti-Kap1 roughly paralleled the relative proportions of full-length and truncated ErbB4 (Fig. 2C, right, compare anti-Kap1 and anti-ErbB4 immunoprecipitates). Despite the association of Kap1 with ErbB4, Kap1 did not react with anti-phosphotyrosine, whether basally or after exposure of cells to TPA or NRG. Thus, Kap1 is not a substrate for the ErbB4 kinase (data not shown). Furthermore, kinase-defective forms of ErbB4 as well as kinase-active ErbB4 coprecipitated with Kap1; thus, the kinase activity of ErbB4 is not required for the association of CYT-1 or CYT-2 isoforms with Kap1 (Fig. 3, lanes “KD”).
FIGURE 3.
Effect of ErbB4 kinase activity and ICD isoform on interaction with Kap1. Truncated V5-tagged ErbB4 isoforms CYT-1, kinase-deficient CYT-1 (CYT-1 KD), CYT-2, or CYT-2 KD were transiently coexpressed with Flag-tagged Kap1 in COS-7 cells that were then stimulated with 100 ng/mL TPA. Protein extracts were immunoprecipitated using antibodies against the epitope tags and immunoblotted as marked. Portions of the lysates were analyzed directly by immunoblotting for V5, Flag, and GAPDH without immunoprecipitation. Data represent two biological replicates in identical format plus two similar biological replicates.
MDM2
ErbB4 CYT-2 s80 interacts with human MDM2 (20). Coexpression of s80 ICD and MDM2 enhances MDM2 Tyr phosphorylation, which requires s80 kinase activity, enhances MDM2 ubiquitination, and reduces the levels of MDM2 with concomitant upregulation of p53 protein and transcriptional activity on the p21CIP promoter. Because Kap1 also interacts with MDM2 (23), it is possible that a ternary complex of Kap1, MDM2, and ErbB4 is formed.
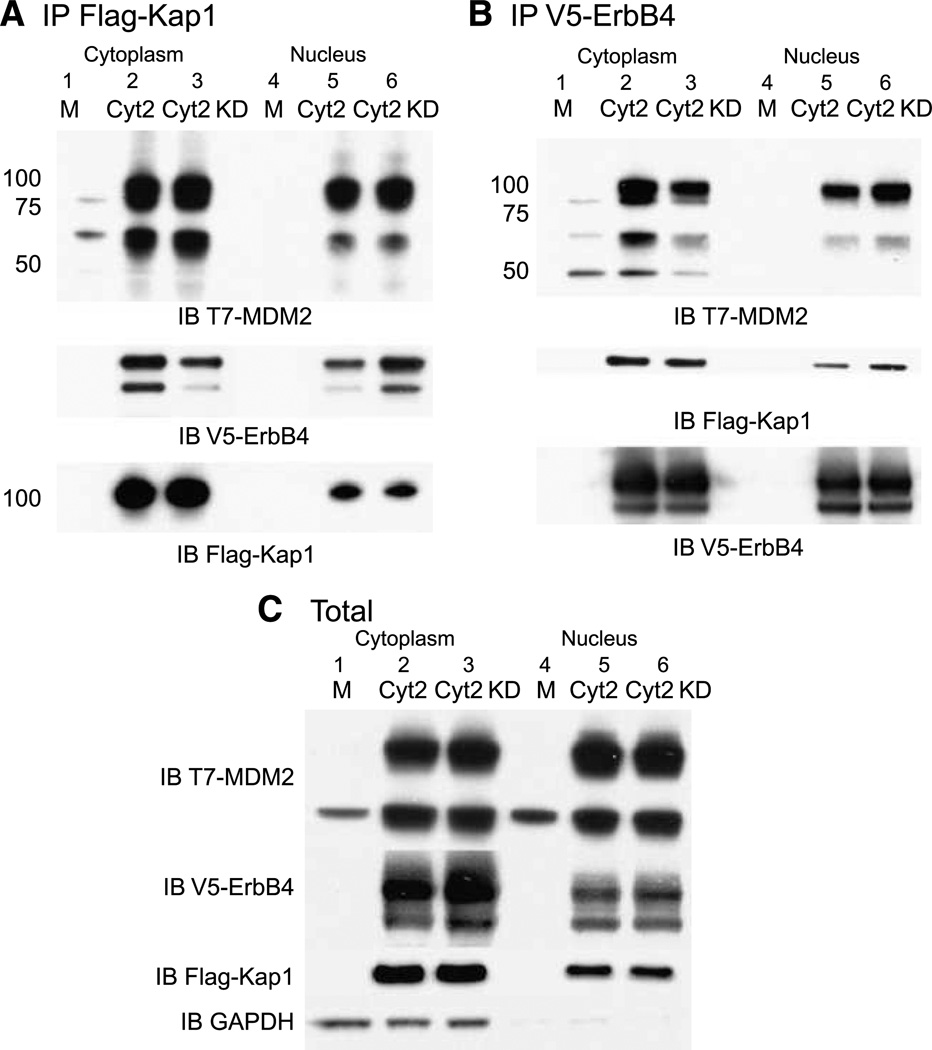
Truncated forms of ErbB4 CYT-2 coprecipitated with Kap1 when expressed in COS-7 cells (Fig. 4A), and Kap1 was cross-immunoprecipitated by anti-ErbB4 (Fig. 4B). Similarly, exogenous MDM2 coprecipitated with anti-Kap1 (Fig. 4A) and with anti-ErbB4 (Fig. 4B). s80 CYT-2 ErbB4 coprecipitated with Kap1 and with MDM2, even when ErbB4 was kinase-deficient because of a K795R substitution (Fig. 4B).
FIGURE 4.
ErbB4 and Kap1 interact with MDM2. COS-7 cells were transiently transfected with vector alone (M) or vectors encoding truncated CYT-2 or CYT-2 KD, Flag-tagged Kap1, and T7-tagged MDM2. Nuclear and cytoplasmic fractions were immunoprecipitated with anti-Kap1 (A) or anti-ErbB4 (B) and immunoblotted with anti-ErbB4, Kap1, or MDM2 antibodies. The blot in A was stripped and reprobed with anti-Kap antibody. The blot in B was stripped and reprobed with anti-ErbB4 antibodies. A separate blot of the same immunoprecipitations was probed with anti-ErbB4 antibody (A) to confirm the Kap1/ErbB4 interaction. Similarly, the blot in B was probed with anti-Kap antibody to confirm the ErbB4/Kap1 interaction. C, portions of the nuclear and cytoplasmic extracts were analyzed without immunoprecipitation, as marked. Data represent two biological replicates in identical format plus two similar biological replicates.
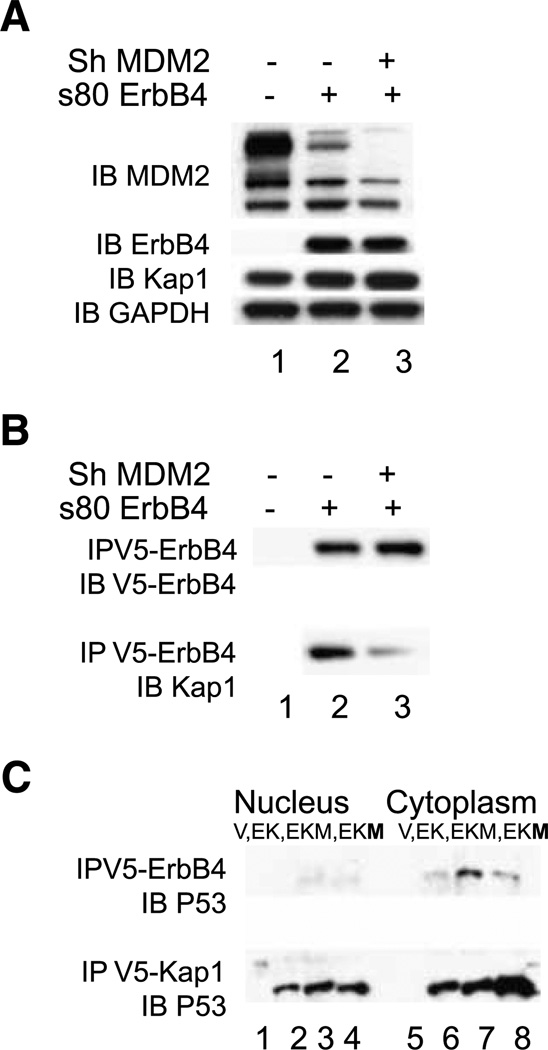
Because MDM2 binds both Kap1 and ErbB4, it was possible that the coprecipitation of ErbB4 and Kap1 was mediated through bridging by MDM2. ShRNA knockdown of MDM2 (Fig. 5A, lane 3) reduced the coimmunoprecipitation of s80-V5 ErbB4 with Kap1 (Fig. 5B, lane 3), suggesting that MDM2 participated in the interaction.
FIGURE 5.
Effect of MDM2 expression on ErbB4/Kap1/p53 interactions. A and B, ErbB4 and Kap1. HEK293T cells were infected with pSiren or pSiren shMDM2 virus. After 24 h, the cells were transfected with pcDNA3.1 V5/His or s80 Cyt2 in the same vector. At 72 h after infection, the cells were harvested and total protein was extracted. A, immunoblots for endogenous MDM2, Kap1, and GAPDH and ectopic s80 ErbB4 in cell lysates. B, anti-V5 immunoprecipitation, with ErbB4 or Kap1 immunoblot from the samples analyzed in A. C, ErbB4, Kap1, and p53. COS-7 cells were transfected with vector (V; lanes 1 and 5); with 2 µg of truncated CYT-1 ErbB4 and 2 µg of Flag-tagged Kap1 (EK; lanes 2 and 6); with2 µg of truncated CYT-1 ErbB4, 2 µg of Flag-tagged Kap1, and 2 µg of MDM2 (EKM; lanes 3 and 7); and with 2 µg of truncated CYT-1 ErbB4, 2 µg of Flag-tagged Kap1, and 4 µg of MDM2 (EKM; lanes 4 and 8). Cells were treated with 10 µmol/L MG132 for 3.5 h and stimulated with TPA for 30 min before harvest at 48 h. Data represent two biological replicates in identical format plus two similar biological replicates (A and B).
p53
MDM2 regulates p53 through formation of a complex in which MDM2 ubiquitinates p53. The Kap1-MDM2 complex further modulates p53 through recruitment of Kap1 binding proteins that themselves deacetylate p53 (23). Hence, we investigated whether ErbB4 associates with p53. With MDM2 and p53 stabilized using the proteasome inhibitor MG132, p53 was readily precipitated with anti-ErbB4 or Flag-Kap1 (Fig. 5C). Kap1 interactions with p53 were enhanced with MDM2 (Fig. 5C, bottom, lanes 3, 4, 7, and 8), consistent with MDM2 bridging p53 and Kap1. Associations of p53 with ErbB4 increased with the presence of MDM2 in the cytoplasm (top, lanes 7 and 8).
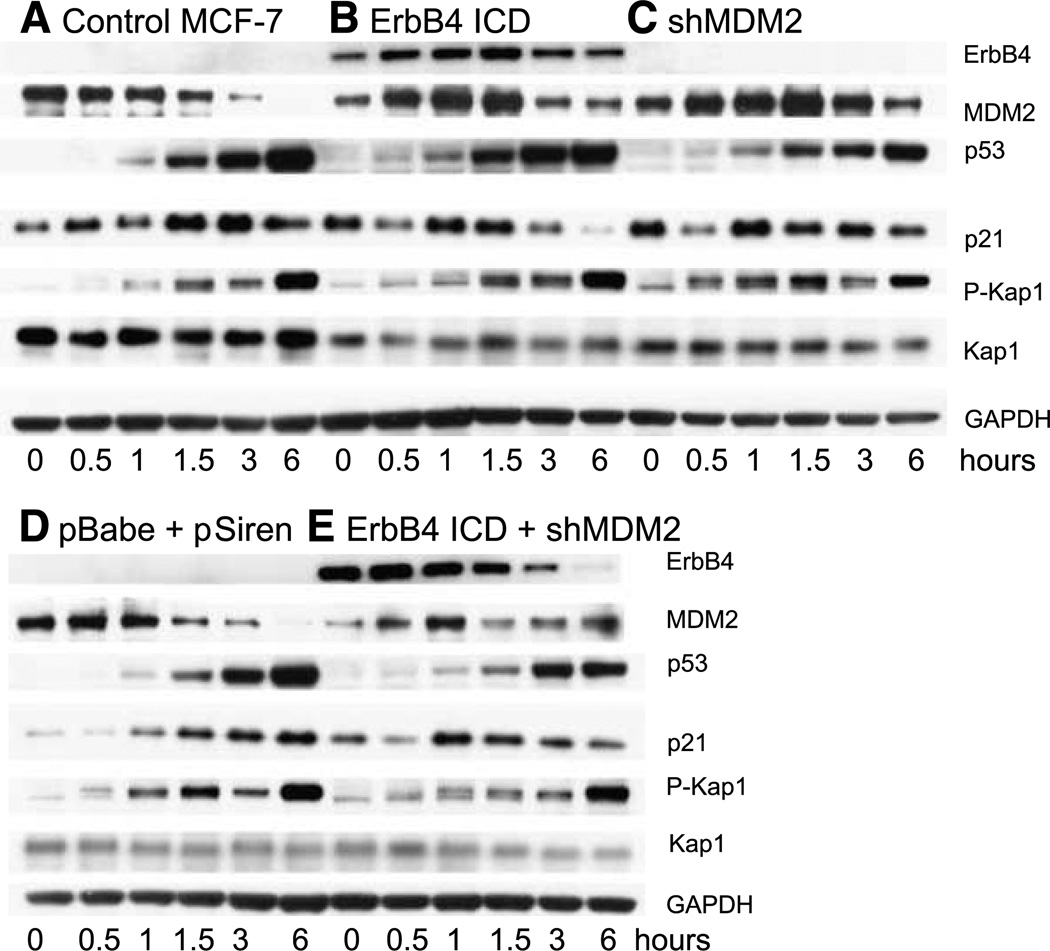
The interactions under investigation here include a direct transcriptional regulator, p53, and coregulators ErbB4 and Kap1. Furthermore, ErbB4 enhances p53 accumulation through a reduction of MDM2 (24). Because p53 is a DNA damage response protein, we next evaluated the effect of DNA damage on these interactions in MCF-7 cells, which have wild-type TP53. In MCF-7 controls (Fig. 6A and D), camptothecin induced accumulation of p53 (within 1 hour, and still increasing between 3 and 6 hours). Upregulation of the p53 transcriptional target p21 occurred within 1 hour and plateaued by 3 hours. Either overexpression of ErbB4 ICD (Fig. 6B) or knockdown of MDM2 (Fig. 6C) or the combination (Fig. 6E) induced baseline accumulation of p21 (0-hour time points), which dipped 30 minutes after camptothecin treatment. In camptothecin-treated cells, either ErbB4 ICD expression (Fig. 6B) or MDM2 shRNA (Fig. 6C) was associated with a more rapid increase and then decrease in p21CIP expression after DNA damage. Expression of the ErbB4 ICD was itself upregulated and then reduced over time with DNA damage (Fig. 6B), with the reduction more rapid in the presence of MDM2 shRNA (Fig. 6, B versus E).
FIGURE 6.
MDM2 and ErbB4 s80 affect DNA damage response. MCF-7 control cells (A), MCF-7 cells stably infected with pBabe CYT-2 s80 (B), MCF-7 cells transiently infected with pSiren shMDM2 (C), MCF-7 cells stably with pBabe and transiently infected with pSiren (D), and MCF-7 stably infected with pBABE CYT-2 s80 and transiently infected with pSiren shMDM2 (E) were treated with 10 µmol/L camptothecin for the times indicated (hours). Total protein was extracted using 2× Laemmli sample buffer and proteins were detected by immunoblotting filter strips as indicated. The strips used for blotting of phospho-Kap1 were stripped once and reprobed for MDM2 and then restripped and probed for Kap1. Data represent three biological replicates in identical format plus two similar biological replicates.
Kap1 phosphorylation
The phosphatidylinositol 3′-kinase–like kinase ATM operates near the apex of DNA checkpoint signal cascades initiated by double-strand DNA breaks that lead directly and through Chk2 to phosphorylation of MDM2 and p53. In addition to the Kap1-dependent regulation of p53 activity through binding of MDM2, an entirely different connection of Kap1 with DNA damage responses has been uncovered (27, 28). ATM activation at double-strand breaks leads to phosphorylation of Kap1 at S824. This phosphorylation is required for a Kap1-dependent global relaxation of chromatin, which is hypothesized to facilitate DNA repair. S824 phosphorylation opens up the Kap1-repressed DNA damage response genes p21 and puma for transcription by p53 (29).
Because ErbB4 is also involved in p53-regulated processes and interacts with Kap1, we wondered whether ErbB4 affects Kap1 phosphorylation. We did not detect any Tyr phosphorylation of Kap1 in immunoblotting experiments, and thus it seems unlikely that Kap1 is itself a substrate for ErbB4 (data not shown). In camptothecin-treated MCF-7 cells, Kap phosphorylated at S824 accumulated over a period of hours, without major changes in the levels of Kap1 (Fig. 6A and D). Expression of ErbB4 s80 or knockdown of MDM2 had variable effects on early Kap1 S824 phosphorylation (Fig. 6).
Interestingly, anti–P-Kap1 detected multiple isoforms of Kap1 with slightly different electrophoretic mobilities (Fig. 6), and mobility shifts were also evident in published work (27). A form with slower mobility was evident within 30 minutes of camptothecin treatment in controls (Fig. 6A and D), then diminished by 1 hour, and later returned with further accumulation of P-Kap1. Although this form was readily detected with anti–P-S824 Kap1, it did not correlate with the intensity of P-Kap1, and the most intense P-Kap1 bands seemed to be a mixture of forms. The simplest interpretation is that the shift marks a subset of P-S824 Kap1 that is further modified covalently, either by phosphorylation at a second site or by other modifications such as sumoylation, ubiquitination, or acetylation.
ErbB4 transactivation activity
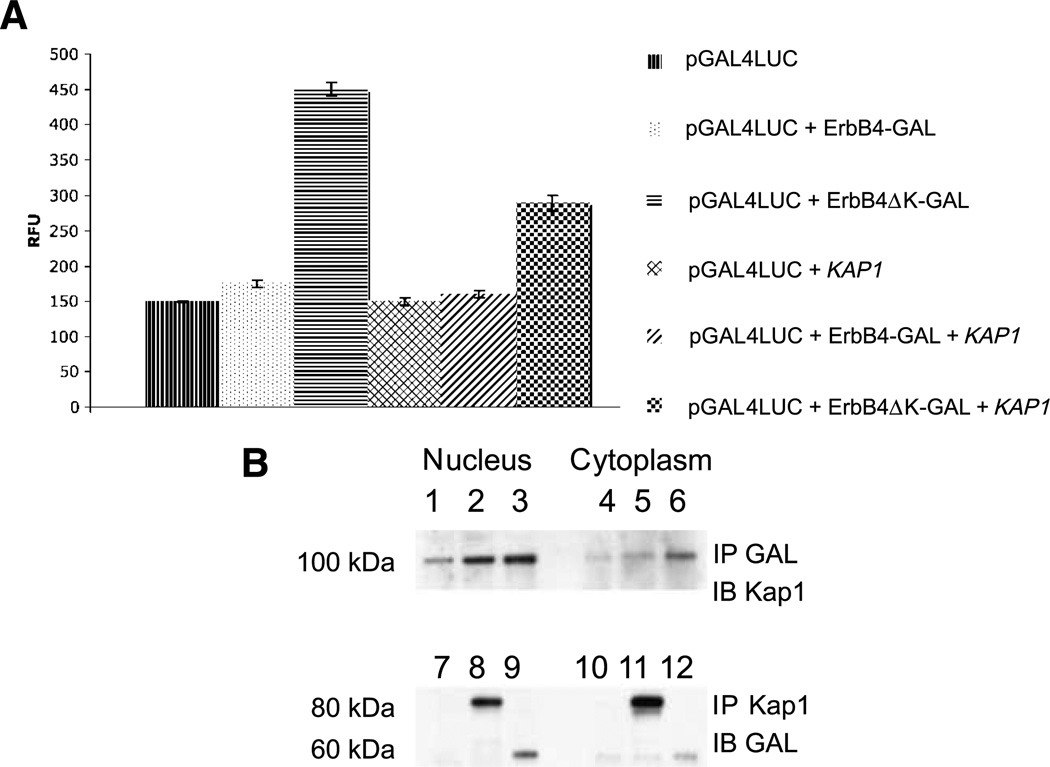
ErbB4 enhances gene transcription through association with sequence-specific DNA binding proteins, including Stat5a and estrogen receptor, and with the transcriptional regulators NCoR and Eto2 (30). ErbB4 itself has intrinsic transactivating activity (7, 13). Binding of YAP enhances ErbB4 transactivation activity, as does deletion of the ErbB4 kinase domain (13). Hence, ErbB4 ICD transactivation activity may exist basally in a cryptic state that is unmasked through conformational changes induced by YAP binding or deletion. In coimmunoprecipitation experiments, kinase-deleted GAL4-ErbB4ΔK bound Kap1 better than did ErbB4 containing the kinase domain (Fig. 7B). This is consistent with a conformational change or exposure of a binding domain induced by deletion of this domain. The same constructs were used to assay the ErbB4 transactivation activity of the GAL4-luciferase reporter. As reported (13), GAL4-ErbB4 has a weak transactivating activity relative to the reporter control, and GAL4-ErbB4ΔK is much stronger (Fig. 7A). In the presence of exogenous Kap1, GAL4-ErbB4 activity is similar, but the relative enhancement with ErbB4ΔK is reduced (Fig. 7A). These results support a model in which ErbB4 transactivation activity is reduced by Kap1 directly or in complex with other protein(s).
FIGURE 7.
Kap1 inhibits ErbB4 transactivation. A plasmid containing the luciferase gene driven by the GAL4 promoter was introduced into COS-7 cells, together with vectors encoding the GAL4 DNA binding domain with the entire ErbB4 ICD (amino acid residues 676–1292; Fig. 1B) or with a kinase domain deletion [amino acid residues 988–1292, Delta kinase (ΔK)] in the absence or presence of KAP1. All transfections included a plasmid carrying the Renilla luciferase gene for normalization of transfection efficiency. B, mock-transfected cells (lanes 1,4, 7, and 9) or cells transfected with ErbB4-GAL (lanes 2, 5, 8, and 11) or GAL-ErbB4ΔK (lanes 3, 6, 9, and 12) were immunoprecipitated with anti-GAL (lanes 1–6) or anti-Kap1 (lanes 7–12) and immunoblotted with the complementary antibody. A, each analysis was done in triplicate with three biological replicates. B, data represent three biological replicates.
Effects of Kap1 on ErbB4 transcription
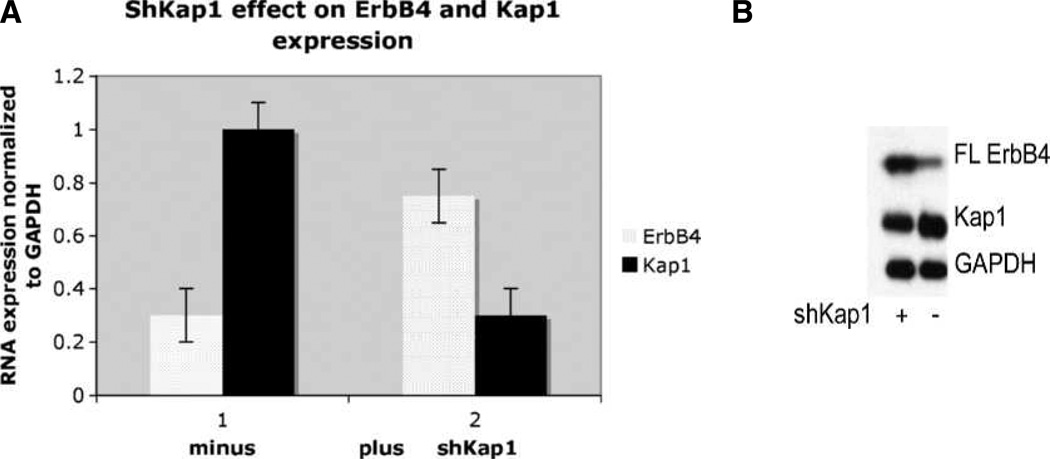
Because the ERBB4 promoter was identified in a genome-wide ChIP-chip screen for Kap1 targets in which DNA isolated by Kap1 chromatin immunoprecipitation (ChIP) was analyzed on solid-phase arrays (31), we determined the effect of Kap1 on ERBB4 expression. shRNA knockdown of Kap1 consistently enhanced the levels of ERBB4 mRNA (Fig. 8A) and protein (Fig. 8B). Because Kap1 has been localized to the ErbB4 promoter by ChIP-chip, these data provide further evidence that Kap1 is an ERBB4 corepressor.
FIGURE 8.
Kap1 knockdown increases ErbB4 RNA and protein expression. MCF-7 cells were infected with shKap1 virus or control virus for 3 d. Total RNA and protein were extracted using the Qiagen RNeasy Mini Kit. A, RNA was analyzed for the presence of Kap1 and ErbB4 RNA using the TaqMan assay system and normalized to GAPDH expression, which was constant. Bars, SD. A, each analysis was done in triplicate with three biological replicates. B, expression of Kap1 and ErbB4 with Kap1 knockdown (left) or control (right). Data in B represent three biological replicates in identical format.
Discussion
The soluble ErbB4 ICD has been linked to a number of functions associated with transcriptional regulation and with DNA damage responses. We have identified several proteins associated with ErbB4, some of which have now been confirmed in other laboratories. Here, we describe the functional implications of ErbB4 association with the pleiotropic transcriptional corepressor Kap1. Both CYT-1 and CYT-2 ErbB4 isoforms bind Kap1, and binding does not require ErbB4 kinase activity nor a significant portion of the kinase domain. ErbB4 alters Kap1-related processes, but ErbB4 does not seem to phosphorylate Kap1.
Our data linking ErbB4 to Kap1 add to a growing list of ErbB4 interactions with transcriptional coregulators. ErbB4 binds to the corepressor Eto2, which restrains erythropoietic differentiation and has unknown functions in mammary tissue (30). ErbB4 reduces Eto2 corepression in reporter assays. In another context, ErbB4 ICD is linked to the corepressor NCoR through an interaction with the adapter TAB2 (18). The ternary complex moves to the nucleus and represses glial-specific genes to maintain a neurogenic phenotype. The NRG1-dependent association of ErbB4 ICD with the GFAP and S100β promoters was confirmed by ChIP. Estrogen-dependent upregulation of estrogen response elements in the ERBB4 promoter indicated a positive feedback between ErbB4 and estrogenic growth functions (32). Taken together with the known interactions of ErbB4 with YAP and with the PR gene, these findings implicate ErbB4 as a transcriptional modulator working through interaction with transcription factors and transcriptional coregulators. Recent work documents the reactivation of the ErbB4 gene in BT-20 cells by histone deacetylase, supporting the epigenetic control of ErbB4 in breast cancer cells (33), as well as the role of the ICD as a coactivator of progesterone receptor regulation (34).
Kap1 and ErbB4 also form complexes with MDM2. This means that the interactions detected may be indirect and mediated by an MDM2 bridge or other larger complexes. Our data are consistent with the conclusion that MDM2 bridges Kap1/ErbB4 interactions. These multiple interactions affect the transcriptional regulation of p53-and ErbB4-dependent genes. MDM2 interacts directly with p300/CBP and inhibits the acetylation of p53. The acetylation of p53 stabilizes the protein. Because acetylation and ubiquitination occur on the same lysine residues, inhibition of acetylation favors p53 ubiquination by Mdm2 and turnover of p53 (30). MDM2 also recruits histone deacetylase 1 to deacetylate lysine residues on p53, thereby freeing them for ubiquitination (35). These processes can be inhibited by the removal of Mdm2 from the nucleus to the nucleolus by p19ARF. Kap1 also interacts with Mdm2 and p53 (23). Kap1 binds directly to Mdm2 and enhances Mdm2 nuclear localization by competing with p19ARF. Kap1 binds histone deacetylase 1 to the Mdm2/p53 complex, causing the deacetylation of p53. Kap1 functions as a cofactor in p53 protein turnover.
Ectopically expressed ErbB4 soluble ICD phosphorylates MDM2, resulting in self-ubiquitination and degradation of Mdm2, which in turn disrupts the Kap1/p53/ Mdm2 complex (20). In this process, the total amount of Mdm2 is reduced by 50% to 70%, but p53 levels are not elevated and apoptosis is not induced. Under these conditions, Kap1 protein levels remain unchanged. It is noteworthy that the baseline p21 accumulations with high ErbB4 ICD or MDM2 knockdown occurred despite the low levels of p53. p53-independent transcriptional regulation of p21 has been reported (36). We found that ErbB4 ICD accelerates DNA damage–induced accumulation of p21 in MCF-7 cells. Knockdown of MDM2 in MCF-7 cells gives similar results, suggesting that the effects seen in the ErbB4 ICD cells may be mediated by the degradation of MDM2. Overall, the presence of ErbB4 CYT-2 s80 may stress cells and prime them for the DNA damage response.
In summary, we have shown a series of complex interactions between ErbB4 and Kap1. Kap1 is a corepressor of ErbB4 gene transcription. ErbB4 ICD can promote MDM2 phosphorylation and thereby indirectly stabilize p53 (20). The ICD of ErbB4 destabilizes Mdm2 by triggering its autoubiquitination and turnover, thereby stabilizing p53. As ErbB4 may also activate mitochondrial apoptotic pathways (37), these findings illuminate a web of processes through which ErbB4 ICD can impede cell cycle progression and promote apoptosis. They represent a point of direct communication between growth factor–regulated receptor tyrosine kinase signaling processes and a major component of the DNA damage response system. The extent to which these processes contribute to the maintenance of genome stability in cells confronted with DNA damage remains to be determined. Both Kap1 and ErbB4 ICD bind Mdm2, but their effects are mutually antagonistic. Kap1 stabilizes Mdm2 and its interactions with p53, which lead to p53 turnover. Interestingly, the ErbB4 ICD can promote dissimilar processes (growth and transformation versus differentiation and apoptosis), perhaps depending on the proteins recruited to the exon uniquely present in the CYT-1 isoform. It is possible that the Kap1-containing complexes are a switch point for channeling these responses to two different directions.
Acknowledgments
We thank Marius Sudol, Kristin Welage, T.W. Kim, A. Ivanov, and H. Zwang for providing constructs used in this work; Sabine Lang and Robert Means for suggestions about the experimental work; and JoAnn Falato for administrative support.
Grant Support
USPHS grant R01 CA80065 from the National Cancer Institute.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Sundvall M, Iljin K, Kilpinen S, Sara H, Kallioniemi OP, Elenius K. Role of ErbB4 in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:259–268. doi: 10.1007/s10911-008-9079-3. [DOI] [PubMed] [Google Scholar]
- 2.Ferretti E, Di Marcotullio L, Gessi M, et al. Alternative splicing of the ErbB-4 cytoplasmic domain and its regulation by hedgehog signaling identify distinct medulloblastoma subsets. Oncogene. 2006;25:7267–7273. doi: 10.1038/sj.onc.1209716. [DOI] [PubMed] [Google Scholar]
- 3.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prickett TD, Agrawal NS, Wei X, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kainulainen V, Sundvall M, Maatta JA, Santiestevan E, Klagsbrun M, Elenius K. A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survivalorchemotaxis. J Biol Chem. 2000;275:8641–8649. doi: 10.1074/jbc.275.12.8641. [DOI] [PubMed] [Google Scholar]
- 6.Ni CY, Yuan H, Carpenter G. Role of the ErbB-4 carboxyl terminus in γ-secretase cleavage. J Biol Chem. 2003;278:4561–4565. doi: 10.1074/jbc.M210504200. [DOI] [PubMed] [Google Scholar]
- 7.Ni CY, Murphy MP, Golde TE, Carpenter G. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 8.Schlessinger J, Lemmon MA. Nuclear signaling by receptor tyrosine kinases: the first robin of spring. Cell. 2006;127:45–48. doi: 10.1016/j.cell.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Sundvall M, Peri L, Maatta JA, et al. Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene. 2007;26:6905–6914. doi: 10.1038/sj.onc.1210501. [DOI] [PubMed] [Google Scholar]
- 10.Muraoka-Cook RS, Sandahl MA, Strunk KE, et al. ErbB4 splice variants Cyt1 and Cyt2 differ by 16 amino acids and exert opposing effects on the mammary epithelium in vivo. Mol Cell Biol. 2009;29:4935–4948. doi: 10.1128/MCB.01705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muraoka-Cook RS, Feng SM, Strunk KE, Earp HS., III ErbB4/HER4: role in mammary gland development, differentiation and growth inhibition. J Mammary Gland Biol Neoplasia. 2008;13:235–246. doi: 10.1007/s10911-008-9080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aqeilan RI, Donati V, Gaudio E, et al. Association of Wwox with ErbB4 in breast cancer. Cancer Res. 2007;67:9330–9336. doi: 10.1158/0008-5472.CAN-07-2147. [DOI] [PubMed] [Google Scholar]
- 13.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 14.Thor AD, Edgerton SM, Jones FE. Subcellular localization of the HER4 intracellular domain, 4ICD, identifies distinct prognostic outcomes for breast cancer patients. Am J Pathol. 2009;175:1802–1809. doi: 10.2353/ajpath.2009.090204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones FE, Welte T, Fu XY, Stern DF. ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J Cell Biol. 1999;147:77–88. doi: 10.1083/jcb.147.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams CC, Allison JG, Vidal GA, et al. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J Cell Biol. 2004;167:469–478. doi: 10.1083/jcb.200403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linggi B, Cheng QC, Rao AR, Carpenter G. The ErbB-4 s80 intracellular domain is a constitutively active tyrosine kinase. Oncogene. 2006;25:160–163. doi: 10.1038/sj.onc.1209003. [DOI] [PubMed] [Google Scholar]
- 18.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006;127:185–197. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 19.Omerovic J, Santangelo L, Puggioni EM, et al. The E3 ligase Aip4/ Itch ubiquitinates and targets ErbB-4 for degradation. FASEB J. 2007;21:2849–2862. doi: 10.1096/fj.06-7925com. [DOI] [PubMed] [Google Scholar]
- 20.Arasada RR, Carpenter G. Secretase-dependent tyrosine phosphorylation of Mdm2 by the ErbB-4 intracellular domain fragment. J Biol Chem. 2005;280:30783–30787. doi: 10.1074/jbc.M506057200. [DOI] [PubMed] [Google Scholar]
- 21.Banks D, Wu M, Higa LA, et al. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle. 2006;5:1719–1729. doi: 10.4161/cc.5.15.3150. [DOI] [PubMed] [Google Scholar]
- 22.Yates KE, Korbel GA, Shtutman M, Roninson IB, DiMaio D. Repression of the SUMO-specific protease Senp1 induces p53-dependent premature senescence in normal human fibroblasts. Aging Cell. 2008;7:609–621. doi: 10.1111/j.1474-9726.2008.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Ivanov A, Chen L, et al. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundvall M, Korhonen A, Paatero I, et al. Isoform-specific monoubiquitination, endocytosis, and degradation of alternatively spliced ErbB4 isoforms. Proc Natl Acad Sci U S A. 2008;105:4162–4167. doi: 10.1073/pnas.0708333105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farin K, Di Segni A, Mor A, Pinkas-Kramarski R. Structure-function analysis of nucleolin and ErbB receptors interactions. PLoS ONE. 2009;4:e6128. doi: 10.1371/journal.pone.0006128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziv Y, Bielopolski D, Galanty Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 28.White DE, Negorev D, Peng H, Ivanov AV, Maul GG, Rauscher FJ., III KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 2006;66:11594–11599. doi: 10.1158/0008-5472.CAN-06-4138. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Lee YK, Jeng JC, et al. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 30.Linggi B, Carpenter G. ErbB-4 s80 intracellular domain abrogates ETO2-dependent transcriptional repression. J Biol Chem. 2006;281:25373–25380. doi: 10.1074/jbc.M603998200. [DOI] [PubMed] [Google Scholar]
- 31.O’Geen H, Squazzo SL, Iyengar S, et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y, Sullivan LL, Nair SS, et al. Coregulation of estrogen receptor by ERBB4/HER4 establishes a growth-promoting autocrine signal in breast tumor cells. Cancer Res. 2006;66:7991–8. doi: 10.1158/0008-5472.CAN-05-4397. [DOI] [PubMed] [Google Scholar]
- 33.Das PM, Thor AD, Edgerton SM, Barry SK, Chen DF, Jones FE. Reactivation of epigenetically silenced HER4/ERBB4 results in apoptosis of breast tumor cells. Oncogene Epub. 2010 Jul 5; doi: 10.1038/onc.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rokicki J, Das PM, Giltnane JM, et al. The ERα coactivator, HER4/4-ICD, regulates progesterone receptor expression in normal and malignant breast epithelium. Mol Cancer. 2010;9:150. doi: 10.1186/1476-4598-9-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitazono M, Bates S, Fok P, Fojo T, Blagosklonny MV. The histone deacetylase inhibitor FR901228 (desipeptide) restores expression and function of pseudo-null p53. Cancer Biol Ther. 2002;1:665–668. doi: 10.4161/cbt.317. [DOI] [PubMed] [Google Scholar]
- 36.Lee YK, Thomas SN, Yang AJ, Ann DK. Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J Biol Chem. 2007;282:1595–1606. doi: 10.1074/jbc.M606306200. [DOI] [PubMed] [Google Scholar]
- 37.Naresh A, Long W, Vidal GA, et al. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;66:6412–6420. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]