Abstract
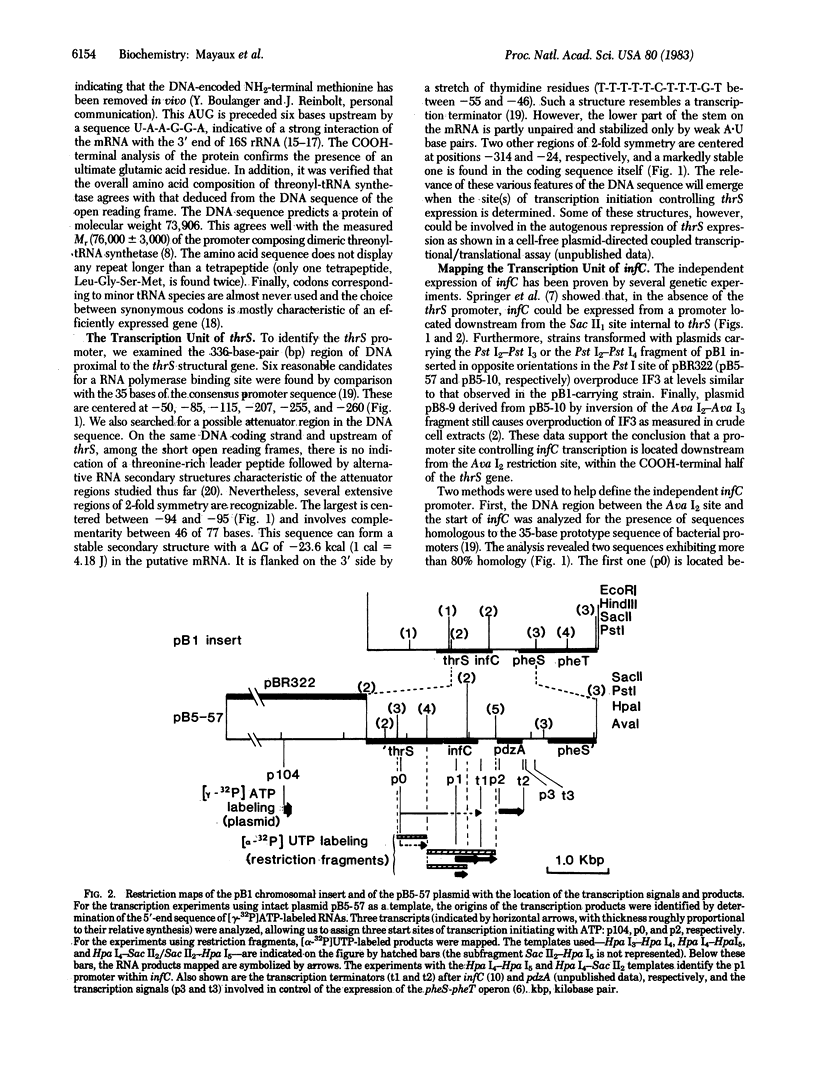
The nucleotide sequence of thrS, the gene encoding dimeric Escherichia coli threonyl-tRNA synthetase [L-threonine:tRNAThr ligase (AMP forming), 6.1.1.3], has been determined. The structural part of the gene is found upstream of and on the same DNA coding strand as infC, the gene for translational initiation factor IF3. thrS is composed of 1,926 base pairs and accounts for a protein of molecular weight 73,906. In addition, a 336-base-pair sequence 5' to the thrS structural gene has been determined. There are only three nucleotides between the stop codon of thrS and the initiator codon of infC. The only potent transcriptional terminator structure is 55 base pairs downstream of the infC coding sequence. This implies that thrS and infC can be expressed from a polycistronic mRNA originating from a promoter upstream to thrS. Although sequence data indicate that thrS and infC are cotranscribed, in vitro transcription and RNA sequence analyses reveal the existence of a promoter within thrS. This promoter can account for the independent expression of infC as reported [Springer, M., Plumbridge, J. A., Trudel, M., Graffe, M. & Grunberg-Manago, M. (1982) Mol. Gen. Genet. 186, 247-252]. A second promoter has been located within infC and could link the expression of infC and that of the next downstream gene, pdzA. Whether these promoters function normally in vivo is an open question.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hennecke H., Böck A., Thomale J., Nass G. Threonyl-transfer ribonucleic acid synthetase from Escherichia coli: subunit structure and genetic analysis of the structural gene by means of a mutated enzyme and of a specialized transducing lambda bacteriophage. J Bacteriol. 1977 Sep;131(3):943–950. doi: 10.1128/jb.131.3.943-950.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J. G., Hershey J. W. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J Biol Chem. 1983 Feb 10;258(3):1954–1959. [PubMed] [Google Scholar]
- Lestienne P., Dondon J., Plumbridge J. A., Howe J. G., Mayaux J. F., Springer M., Blanquet S., Hershey J. W., Grunberg-Manago M. Expression of the gene for Escherichia coli initiation factor IE-3 in vivo and in vitro. Eur J Biochem. 1982 Apr;123(3):483–488. doi: 10.1111/j.1432-1033.1982.tb06556.x. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Neihardt F. C., Parker J., McKeever W. G. Function and regulation of aminoacyl-tRNA synthetases in prokaryotic and eukaryotic cells. Annu Rev Microbiol. 1975;29:215–250. doi: 10.1146/annurev.mi.29.100175.001243. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A., Springer M., Graffe M., Goursot R., Grunberg-Manago M. Physical localisation and cloning of the structural gene for E. coli initiation factor IF3 from a group of genes concerned with translation. Gene. 1980 Oct;11(1-2):33–42. doi: 10.1016/0378-1119(80)90084-0. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A., Springer M. Organization of the Escherichia coli chromosome around the genes for translation initiation factor IF2 (infB) and a transcription termination factor (nusA). J Mol Biol. 1983 Jun 25;167(2):227–243. doi: 10.1016/s0022-2836(83)80333-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Fayat G., Dessen P., Springer M., Plumbridge J. A., Grunberg-Manago M., Blanquet S. Sequence of a 1.26-kb DNA fragment containing the structural gene for E.coli initiation factor IF3: presence of an AUU initiator codon. EMBO J. 1982;1(3):311–315. doi: 10.1002/j.1460-2075.1982.tb01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D., McKenney K., Sobieski D. A., Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982 Oct;30(3):865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Plumbridge J. A., Trudel M., Graffe M., Grunberg-Manago M. Transcription units around the gene for E. coli translation initiation factor IF3 (infC). Mol Gen Genet. 1982;186(2):247–252. doi: 10.1007/BF00331857. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]