102.1 Introduction
PEDF-R is a PNPLA2 protein with demonstrable triglyceride lipase, triacylglycerol transacylase, and phospholipase activities. It has an N-terminal acyltransferase/lysophospholipase domain (human amino acid sequence numbers 3–178) and a patatin domain (human amino acid sequence numbers 10–179). PEDF-R is also known as TTS-2.2, iPLA2ζ, ATGL and desnutrin (Notari et al. 2006). Among human, rat, and mouse species, PEDF-R is highly conserved (87% identity for both human/mouse and for human/rat and 96% for mouse/rat).
The gene of PEDF-R (pnpla2) has been identified in the retina (Notari et al. 2006). Although PEDF-R transcripts are abundantly identified in adipose tissues, the ARPE-19 cell line and the retina precursor R28 and RGC-5 cell lines also express PEDF-R transcripts (Notari et al. 2006). In the native retina, the PEDF-R protein is distributed in the RPE and in the inner segments of the photoreceptors, and at lower levels, in the inner nuclear and retinal ganglion cell layer (Notari et al. 2006). Several lines of evidence point to the subcellular localization of PEDF-R to plasma membranes having four transmembrane domains, two extracellular loops, one intracellular loop, and intracellular N-end and C-end tails (Notari et al. 2006).
PEDF-R is an enzyme with phospholipase A activity that hydrolyzes phospholipids into fatty acids and lysophospholipids, In particular, phospholipase A2 can specifically hydrolyze the sn-2 acyl bond of phospholipids releasing fatty acid, like arachidonic acid or docosahexaenoic acid. These products can act as lipid second messengers and cause further downstream signaling. Thus, regulation of this enzyme can result in important downstream biological events. In this regard, we have demonstrated that PEDF-R has high affinity binding for PEDF (Notari et al. 2006), a multifunctional protein involved in retinal neuronal survival and differentiation, and in preventing angiogenesis and the growth and invasion of tumor cells and has anti-inflammatory properties (Crawford et at. 2001; Bouck 2002; Wang et al. 2003; Barnstable and Tombran-Tink 2004; Garcia et al. 2004). More interestingly, PEDF can stimulate the in vitro PLA activity of PEDF-R (Notari et al. 2006) and it can enhance the liberation of a DHA derivative termed neuroprotectin D1 (Bazan et al. 2005), which is a neuronal survival and anti-inflammatory agent (Bazan 2005) like PEDF. Therefore, it has been proposed that the signaling activated by PEDF is mediated by the interactions between PEDF and PEDF-R to enhance retina cell survival.
Given that understanding the interactions between PEDF-R and PEDF are of interest to elucidate mechanisms of action of PEDF, it is important to have well-characterized tools for studying PEDF-R. In this study, we have characterized an antibody for PEDF-R available through commercial source (R&D systems) that can be used to detect PEDF-R in samples from human, mouse, and rat. We have explored the antibody-binding site(s) on PEDF-R using recombinant PEDF-R polypeptides and peptides. We have also used rat retina R28 cells as native source, because recent studies have shown that PEDF is a survival factor for R28 cells in response to serum starvation (Notari et al. 2005; Murakami et al. 2008). We provide information for an epitope and blocking peptides for the anti-PEDF-R as tools for further PEDF-R studies.
102.2 Materials and Methods
102.2.1 Peptides, Proteins, and Antibodies
Peptides were designed from exons 4, 5, 6, 7, and 8 of human PEDF-R and were chemically synthesized by a commercial source (Aves labs). Expression vectors for PEDF-R and PEDF-R4 were constructed into pEXP1-DEST vector with N-terminal epitope-tags (Xpress and His) as described (Notari et al. 2006). Recombinant proteins were expressed by cell-free in vitro protein synthesis using the pEXP-based vectors and Escherichia coli extracts from IVPS™ (Invitrogen). Recombinant proteins were purified using His tag affinity column chromatography with Ni-NTA resin (Invitrogen). Sheep polyclonal anti-PEDF-R was from R&D systems (Cat# AF5365); Secondary antibody HRP-conjugated donkey anti-sheep IgG was from SIGMA.
102.2.2 Slot Blot
Solutions of synthetic peptides (1 μg) were applied to wells in a manifold (Life Technologies) containing a nitrocellulose membrane (Bio-rad, Cat# 162-0116, 0.45 μm) presoaked in transfer buffer (Tris/Glycine/methanol). Peptides were transferred to membranes using vacuum as a driving force and the membrane subjected to immuno-blot.
102.2.3 Membrane Fractionation
R28 cells (kind gift of Dr. Gail Seigel, University of Buffalo) were cultured in DMEM media with 10% of fetal calf serum (FCS) and 1% of Penicillin/Streptomycin (P/S) at 37°C with 5% CO2. Confluent cells (90%) were harvested and separation of cytosolic and membrane fraction was obtained by centrifugation at 80,000 × g as described previously (Notari et al.). Protein concentration was determined with Protein Assay (Bio-Rad).
102.2.4 Polyacrylamide Gel Electrophoresis
Protein samples were resolved using NuPAGE 4–12% polyacrylamide gel in Bis-Tris buffer with NuPAGE MOPS-SDS as running buffer (Invitrogen). After electrophoresis, proteins from gel were then transferred to nitrocellulose membranes using the iBlot Gel Transfer system (Invitrogen) for immuno-blot. Prestained markers were from Bio-Rad (Cat# 161-0305).
102.2.5 Immuno-Blot
The membrane was incubated in blocking solution (1 % BSA in Tris buffered saline plus 0.1% Tween-20, TBS-T) for 1 h at room temperature. The primary antibody was 0.25 mg/mL anti-PEDF-R in 1% BSA/TBS-T, and the secondary was HRP-conjugated donkey anti-sheep IgG (diluted 1:20,000 in 1% BSA/TBS-T). To block the binding of anti-PEDF-R, the antibody was preincubated with E4a and E4b peptides (at 1 mg/mL each) for 1 h at room temperature, followed by 16 h at 4°C before addition to the blot. Washes between primary and secondary antibody incubations were with TBS-T for 5 min each and 3 times. For immunodetection, SuperSignal West Dura Extended Duration Substrate (Pierce) was used following the manufacturer’s protocol. The blot was exposed to an X-ray film to visualize the immunoreactive signal by chemiluminescence.
102.3 Results
102.3.1 Immunoreactivity to Recombinant PEDF-R Polypeptide Fragments
We tested the immunoreactivity of anti-PEDF-R to recombinant PEDF-R poly-peptide fragments fused to Xpress and His tags. We expressed full-length PEDF-R and PEDF-R4, a C-terminal truncated version that is derived from the first four exons of PEDF-R. Given that these recombinant polypeptides have the Xpress tag, we used anti-Xpress antibody to confirm their expression (Fig. 102.1a). The PEDF-R antibody recognized both recombinant proteins of apparent molecular weights ~81 kDa for the full-length PEDF-R and ~40 kDa for the truncated PEDF-R4 version (Fig. 102.1b). The results suggest that the antibody recognition site does not require the C-terminal half of PEDF-R, and it could be located within the first four exons 1–4.
Fig. 102.1.

Western blot of recombinant PEDF-R polypeptides. Full-length PEDF-R and C-terminal truncated PEDF-R4 were expressed using in vitro cell-free Escherichia coli expression system. Purified proteins were resolved by SDS-PAGE and electrotransfered to a membrane for immunostaining. Photographs of blots immunostained with anti-Xpress (a) and anti-PEDF-R (b) are shown. Lanes 1 and 3 were PEDF-R, lanes 2 and 4 were PEDF-R4. Migration positions of PEDF-R and PEDF-R4 are indicated with arrows, and of molecular weight markers are in between the two blots
102.3.2 Immunoreactivity to Synthetic PEDF-R Peptides
Ten peptides spanning exons 4, 5, 6, 7, and 8 (Fig. 102.2) were generated synthetically. With equal amounts of each peptide transferred onto the membrane immunoreactions with anti-PEDF-R antibody revealed that only three peptides E4a, E4b, and P1 were detected (Fig. 102.3). Anti-PEDF-R bound with highest affinity to E4a followed by E4b and then P1. The results demonstrate that the exon 4 region contains the epitope for anti-PEDF-R, the E4a region being more antigenic than E4b.
Fig. 102.2.
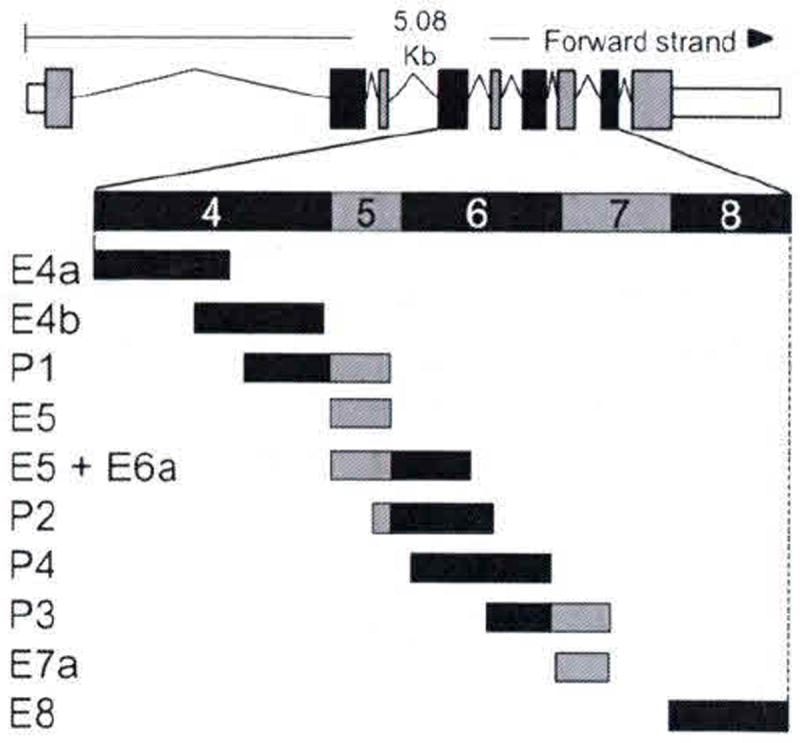
Schematic of rat PEDF-R transcript. Transcript summary information for rat PEDF-R was obtained from http://www.ensembl.org for ENSRNOT00000025319. Exons are illustrated by boxes; coding regions are black and gray; introns are the lines flanking the boxes. Expanded region illustrates the design of synthetic peptides
Fig. 102.3.
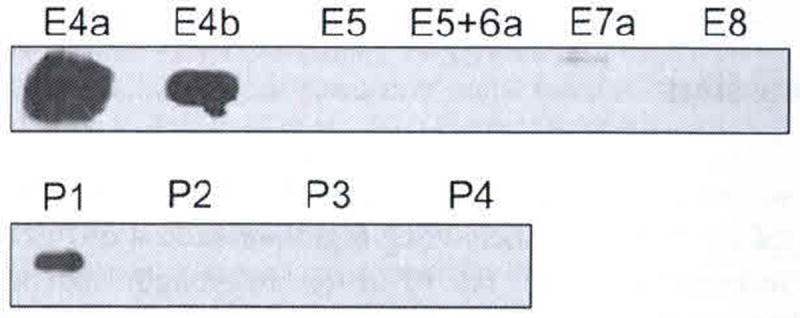
Slot blot of PEDF-R peptides. Peptides (1 mg) were applied to a nitrocellulose membrane using a slot-blot technique and immunostained with anti-PEDF-R. Peptides are indicated to the top of the photograph
102.3.3 Immunoreactivity to Native Rat PEDF-R
Western blots of R28 cell membrane proteins with anti-PEDF-R antibody revealed three distinct immunoreactive protein bands (Fig. 102.4). The molecular sizes for these proteins were estimated to be 81, 70, and 65 kDa, relative to the migration pattern of the prestained markers. The signal for the three bands decreased when anti-PEDF-R was preincubated with a mixture of E4a and E4b peptides (Fig. 102.4), indicating that the immunoreactivity was blocked with E4, the antibody-binding region. This demonstrated that these three rat R28 proteins were specifically recognized by anti-PEDF-R.
Fig. 102.4.
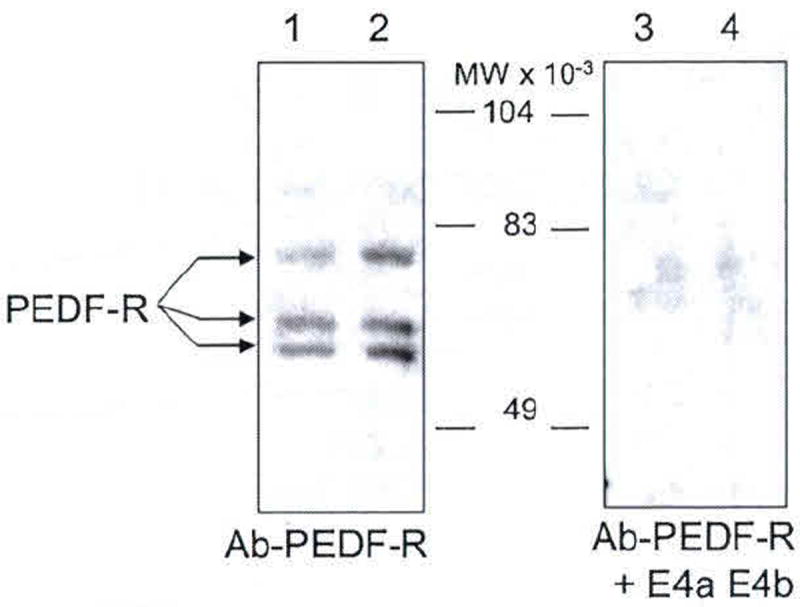
Western blot of native PEDF-R from retina R28 cells. Membrane fractions obtained from R28 cells were resolved by SDS-PAGE. Total protein loaded in lanes 1–4 was 6 μg each. Lanes 1 and 2, and lanes 3 and 4 were replicates. Immunoreactions with anti-PEDF-R were for lanes 1 and 2, and with anti-PEDF-R preincubated with peptides E4a and E4b were for lanes 3 and 4. Migration positions of PEDF-R isoforms are indicated with arrows, and molecular weight markers are in the center
102.4 Discussion
In this study, we have identified the epitope of the antibody that detects PEDF-R. We have mapped the epitope to the coding region of exon 4 of PEDF-R. Antibody anti-PEDF-R recognizes native rat PEDF-R, full-length recombinant human PEDF-R and the terminal-truncated PEDF-R4. PEDF-R4 terminates after exon 4, thereby retaining the antibody epitope. Given that the intensity of PEDF-R immunoreactivity decreased in this order E4a>E4b>P1, and that peptide E5 is not immunostained with anti-PEDF-R, the signal with P1 was most likely due to detection of its overlap with the E4b region. The antibody did not recognize any other region of PEDF-R. These results lead us to conclude that the epitope is located on the coding region of exon 4 of PEDF-R.
The rat PEDF-R gene contains nine exons with a coding capacity of 478 amino acids, in contrast to the 504 amino acid for the human product. Immunoreactions of rat R28 cell membrane fractions with anti-PEDF-R revealed three proteins with apparent molecular weights of 81, 70, and 65 kDa that contain the E4 region of PEDF-R. These proteins were specifically recognized to be PEDF-R as they were blocked upon preincubation of the antibody with the peptides E4a and E4b. While the 81-kDa protein corresponds to the full-length PEDF-R, the smaller proteins may result from alternative splice transcripts of PEDF-R. Ensembl reveals a splice variant lacking exon 6 in rat PEDF-R, thus resulting in a shorter polypeptide. R&D Systems reports three alternative splice mouse mRNAs with coding regions for three polypeptides of molecular weights similar to those in the present study. Another possibility is that the 70- and 65-kDa protein versions result after posttranslational modifications. Previous studies demonstrated a single PEDF-R immunoreactive band of ~83-kDa protein in R28 cells with an antibody to RA peptide derived from the N-terminal half of E4b (Subramanian et al. 2010). This suggests that the RA region might be missing in the 71- and 65-kDa PEDF-R proteins. More importantly, our results demonstrate that R28 cells contain PEDF-R protein versions with the E4 region, which contains the PEDF-binding region (Locatelli-Hoops et al. 2008) important for stimulating the PLA activity of PEDF-R.
In summary, the antibody for PEDF-R used in this study is a useful tool to identify PEDF-R protein, and the E4a and E4b peptides are excellent blocking peptides for this antibody, which will prove useful in characterization of PEDF-R isoforms.
Contributor Information
Preeti Subramanian, Email: subramanianp@nei.nih.gov, National Eye Institute, National Institutes of Health, Building 6, Room 134, 6 Center Drive, MSC 0608, Bethesda, MD 20892-0608, USA.
Matthew Rapp, University of Maryland Baltimore County, Baltimore, MD 21250, USA.
S. Patricia Becerra, National Eye Institute, National Institutes of Health, Building 6, Room 134, 6 Center Drive, MSC 0608, Bethesda, MD 20892-0608, USA.
References
- Barnstable CJ, Tombran-Tink J. Neuroprotective and antiangiogenic actions of PEDF in the eye: molecular targets and therapeutic potential. Prog Retin Eye Res. 2004;23:561–577. doi: 10.1016/j.preteyeres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Bazan NG. Neuroprotectin D1 (NPD1): a DHA-derived mediator that protects brain and retina against cell injury-induced oxidative stress. Brain Pathol. 2005;15:159–166. doi: 10.1111/j.1750-3639.2005.tb00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan NG, Marcheselli VL, Hu J, et al. Pigment epithelium-derived growth factor (PEDF) selectively up-regulates NPD1 synthesis and release through the apical side of human RPE cells in primary cultures. Invest Ophthalmol Vis Sci. 2005;46:167. [Google Scholar]
- Bouck N. PEDF: anti-angiogenic guardian of ocular function. Trends Mol Med. 2002;8:330–334. doi: 10.1016/s1471-4914(02)02362-6. [DOI] [PubMed] [Google Scholar]
- Crawford SE, Stellmach V, Ranalli M, et al. Pigment epithelium-derived factor (PEDF) in neuroblastoma: a multifunctional mediator of Schwann cell antitumor activity. J Cell Sci. 2001;114:4421–4428. doi: 10.1242/jcs.114.24.4421. [DOI] [PubMed] [Google Scholar]
- Garcia M, Fernandez-Garcia NI, Rivas V, et al. Inhibition of xenografted human melanoma growth and prevention of metastasis development by dual antiangiogenic/antitumor activities of pigment epithelium-derived factor. Cancer Res. 2004;64:5632–5642. doi: 10.1158/0008-5472.CAN-04-0230. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Ikeda Y, Yonemitsu Y, et al. Inhibition of nuclear translocation of apoptosis-inducing factor is an essential mechanism of the neuroprotective activity of pigment epithelium-derived factor in a rat model of retinal degeneration. Am J Pathol. 2008;173:1326–1338. doi: 10.2353/ajpath.2008.080466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notari L, Baladron V, Aroca-Aguilar JD, Balko N, et al. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem. 2006;281:38022–38037. doi: 10.1074/jbc.M600353200. [DOI] [PubMed] [Google Scholar]
- Notari L, Miller A, Martinez A, et al. Pigment epithelium-derived factor is a substrate for matrix metalloproteinase type 2 and type 9: implications for downregulation in hypoxia. Invest Ophthalmol Vis Sci. 2005;46:2736–2747. doi: 10.1167/iovs.04-1489. [DOI] [PubMed] [Google Scholar]
- Subramanian P, Notario PM, Becerra SP. Pigment Epithelium-derived Factor Receptor (PEDF-R): A Plasma Membrane-linked Phospholipase with PEDF Binding Affinity. Adv Exp Med Biol. 2010;664:29–37. doi: 10.1007/978-1-4419-1399-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Schmitz V, Perez-Mediavilla A, et al. Suppression of angiogenesis and tumor growth by adenoviral-mediated gene transfer of pigment epithelium-derived factor. Mol Ther. 2003;8:72–79. doi: 10.1016/s1525-0016(03)00128-x. [DOI] [PubMed] [Google Scholar]
- Locatelli-Hoops S, Notari L, Becerra SP. Identification of Structural Determinants on PEDF-R Responsible for Binding PEDF. Invest Ophthalmol Vis Sci; ARVO Meeting Abstracts; 2008. p. 5768. [Google Scholar]


