Abstract
The leucine-rich repeats (LRR)-containing domain is evolutionarily conserved in many proteins associated with innate immunity in plants, invertebrates and vertebrates. Serving as a first line of defense, the innate immune response is initiated through the sensing of pathogen-associated molecular patterns (PAMPs). In plants, NBS (nucleotide-binding site)-LRR proteins provide recognition of pathogen products of avirulence (AVR) genes. LRRs also promote interaction between LRR proteins as observed in receptor-coreceptor complexes. In mammals, toll-like receptors (TLRs) and NOD-like receptors (NLRs) through their LRR domain, sense molecular determinants from a structurally diverse set of bacterial, fungal, parasite and viral-derived components. In humans, at least 34 LRR proteins are implicated in diseases. Most LRR domains consist of 2–45 leucine-rich repeats, with each repeat about 20–30 residues long. Structurally, LRR domains adopt an arc or horseshoe shape, with the concave face consisting of parallel β-strands and the convex face representing a more variable region of secondary structures including helices. Apart from the TLRs and NLRs, most of the 375 human LRR proteins remain uncharacterized functionally. We incorporated computational and functional analyses to facilitate multifaceted insights into human LRR proteins and outline a few approaches here.
Key words: systems biology, pathogen sensors, pathogen response, antibacterial, inflammation, autophagy, bioinformatics, computational biology
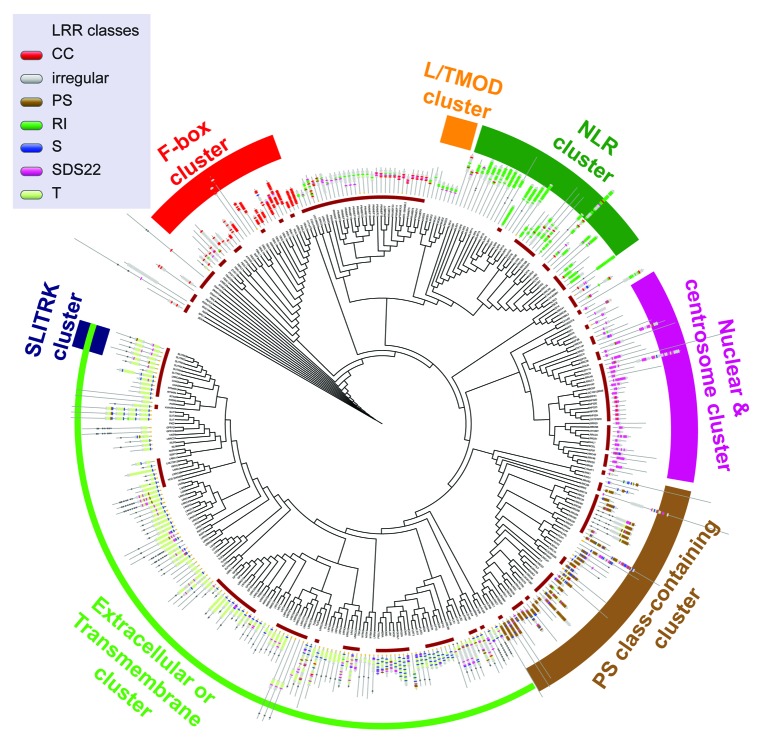
Given that the LRR domain provides the important structural framework required for molecular interactions, and also for PAMP recognition by TLRs and NLRs, we reasoned that functionally related LRR proteins can be grouped together based on the similarity of their LRR domains, and by extension, additional functional inference can be made about poorly characterized LRR proteins that cluster alongside well-studied LRR proteins. We devised semiautomated methods to cluster or group LRR proteins on the basis of LRR classes. By constructing hidden Markov models (HMMs), we identified and classified each repeat sequence within each LRR domain into the 7 ‘regular’ classes of leucine-rich repeats: S, RI, CC, SDS22, PS, T and Tp. In addition, we also used pattern-matching algorithms to find predicted secondary structures and atypical leucine-rich repeat sequences (termed ‘irregular LRRs’) adjacent to regular LRRs. We next grouped the human LRR proteins based on the LRR class assignments. Two methods were used to cluster LRR proteins. The first was based on the sequence similarity of LRRs within each class and, with this, we constructed a comprehensive map of the clustered LRR proteins (Fig. 1; http://xavierlab.mgh.harvard.edu/LrrProteins/). This allowed us to not only group functionally related LRR proteins together, such as members of the NLR, SLITRK and F-box families, but also less well-characterized LRR proteins on the basis of LRR class composition. From this, we found CEP72 (an LRR protein that was recently implicated in ulcerative colitis from genome-wide association studies) clustered with LRRC36, an RORγ-binding protein, suggesting a gene-regulatory interaction. The second clustering approach partitioned LRR proteins into categories based on the class to which the majority of LRRs in each protein belong. We observed a striking co-occurrence of certain non-LRR functional domains with a large number of LRR proteins in several of the resulting clusters. In particular, a statistically significant number of LRR proteins in the T and
Figure 1. LRR proteins were clustered using LRR classes. Colored bands on the periphery identify clusters containing certain features observed among LRR protein members in the respective clusters. Adapted from Ng AC, et al. Proc Natl Acad Sci USA 2011; 108:4631–8; PMID:20616063; 10.1073/pnas.1000093107 with permission of the publisher. A high-resolution version of this figure is available (see Fig. S3 of the original manuscript).
S + T categories have transmembrane (TM) domains, whereas most of the LRR proteins in the RI and CC categories contain a NACHT NTPase domain and an F-box domain which mediates interactions with the E3 ubiquitin ligase complex. Among proteins containing S and T LRR classes (which are the most commonly occurring regular LRR classes), we found a statistical enrichment of processes associated with receptor-mediated signaling, including cytokine- and chemokine-mediated signaling, immunity and defense, suggesting S and T LRRs might play a role in these processes.
We inspected LRR proteins in terms of their evolutionary characteristics and compared general trends with members in the SH2 and PDZ family of proteins. In contrast to SH2 and PDZ domain-containing proteins, a significantly large number of LRR proteins have orthologs primarily in mammals. Also of interest, a subset of LRR proteins exhibiting strong conservation in fungi is enriched for nucleic acid binding function. One of these, NXF1, a nuclear RNA-export factor, was recently identified in two genome-wide siRNA screens as a key host-dependency factor for influenza virus. By combining analyses of class-based LRR categories and LRR evolutionary profiles, there appears to be an intriguing connection between the former and the extent to which their members are conserved. For example, the SDS22, RI and mixed LRR categories contain very few proteins with high degrees of conservation relative to the other LRRs. We also note that almost all the proteins in the T and S + T categories have orthologs only in metazoans.
To uncover candidate LRR genes that might be involved in host-pathogen innate immune response, but have not been previously identified to play a role in pathogen response, we examined time-series expression profiles of LRR genes in primary human macrophages infected with clinically important bacterial pathogens (Staphylococcus aureus, Mycobacterium tuberculosis, Listeria monocytogenes, enterohemorrhagic Escherichia coli and Salmonella enterica serovar typhi). We observed a number of clusters of LRR genes that exhibited strong transcriptional responses to the pathogens. In particular, MFHAS1, a gene that exhibits enriched expression in immune cells, was identified as a candidate from its strong transcriptional induction response to four of the five pathogens examined. RNAi knockdown in RAW macrophages strongly enhances IL-6 production following LPS and polyI:C stimulation. This places MFHAS1 in the signaling network downstream of TLR signaling and highlights its potential role as a negative modulator of the inflammatory response.
We were also interested to explore if LRR proteins might be involved in autophagy. Through literature mining, we note that the only previously known LRR protein to be implicated in autophagy is WDFY3. By extending this search using protein-protein interaction (PPI) network data, we uncovered an additional LRR protein, LRSAM1. We observed that LRSAM1 is induced transcriptionally by Salmonella, and as it also has a RING (E3-ubiquitin ligase) domain, which regulates TSG101’s ability to sort endo- and exocytic cargoes, we hypothesized and validated its involvement in anti-Salmonella autophagy using a functional RNAi approach.
Using information from orthogonal sources can be useful for inferring function of uncharacterized LRR proteins. With this framework, we have employed LRR class composition to facilitate the functional classification of novel LRR proteins alongside those of known function, transcriptional profiling to identify candidates with roles in pathogen sensing and response, as well as PPI networks and functional RNAi to uncover molecular pathways associated with LRR proteins.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/16464