Abstract
Fank1 is exclusively expressed in the testis from the meiosis phase to the haploid phase of spermatogenesis. In this study, we examined the function of Fank1 by establishing a Fank1-knockdown transgenic mouse model. The apoptotic statuses of the testes of the transgenic mice were tested using the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) method. The FANK1 consensus DNA-binding sequence was identified using cyclic amplification of sequence target (CAST) analysis. Differentially expressed genes were examined using microarray analysis. A reduction in sperm number and an increase in apoptotic spermatocytes were observed in Fank1-knockdown mice, and the apoptotic cells were found to be primarily spermatogonia and spermatocytes. The CAST results demonstrated that the consensus DNA-binding sequence was AAAAAG, in which the percentage occurrence of each base at each position ranged from 55 to 86%. This sequence was present in the promoter regions of 10 differentially expressed genes that were examined using microarray analysis. In total, 17 genes were differentially expressed with changes in their expression levels greater than twofold. The abnormal expression of Fank1 target genes that were regulated directly or indirectly by Fank1 reduced the number of sperm in the knockdown mice. Thus, FANK1 may play a pivotal role in spermatogenesis as a transcription factor.
Keywords: Fank1, knockdown mice, spermatogenesis
INTRODUCTION
Spermatogenesis is a complex cellular process in which male germ cells proliferate and differentiate into functional highly specialized male gametes called spermatozoa. Spermatogenesis is divided into three phases: the mitotic phase (multiplication and differentiation of spermatogonia), the meiotic phase (meiotic cell division of spermatocytes) and the haploid phase (differentiation of spermatids into spermatozoa).1,2 In this process, testis-specific gene expression occurs, which produces several enzymes and proteins that regulate spermatogenesis.3 When testis-specific gene abnormalities cause oligozoospermia, asthenozoospermia or sperm abnormalities occur. Therefore, the identification of expressed testis-specific genes is necessary to determine the molecular pathways and signaling systems that control male gamete formation.
In our previous studies,4 we analyzed the evolutionary conservation, expression and localization of Fank1 mRNA and protein. Fank1 was identified as a testis-expressed gene based on a bioinformatics approach and was subsequently shown to be expressed uniquely in the testis, beginning in mid-pachytene germ cells and extending into spermatids. The sequence revealed both ankyrin and fibronectin III homology repeats, which were combined to generate its name. The fibronectin III homology domain indicates a DNA-binding domain, which suggests that Fank1 is a transcription factor, a view supported by the fact that the protein is found largely or exclusively in the nucleus, as shown by immunohistology. Here, we studied the effects of in vivo knockdown using transgenic mice expressing hairpin RNAi sequences. The mice carrying the knockdown mutation were subfertile with low sperm counts. A gene-chip screen of the mutant testis RNA identified several downregulated transcripts and a lesser number of upregulated transcripts. In addition, several of these transcripts possessed high affinity Fank1-binding sites in the general region of the promoter. Our observations indicate that FANK1 is essential for spermatogenesis as a transcription factor. These results provide several clues regarding the biology and molecular function of Fank1 during spermatogenesis.
MATERIALS AND METHODS
Expression and Purification of GST-FANK1
The pGEX-4T-2-FANK1 recombinant protein expression plasmid was constructed and expressed in E. coli BL21 (DE3) bacterial cells. The FnIII domain of Fank1 was amplified by polymerase chain reaction (PCR) using the pds RED-Fank1 vector as the template, which coexpressed the red fluorescence protein (RFP) and the Fank1 domain. The forward primer was 5’-CCGGGATCCATGGAGCCCCACAAAGTCG-3’, containing an Sal I site, and the reverse primer was 5’-ACGCGTCGACCCTTTCTGAGCAGCAACC-3’, with a Bam HI site. The PCR conditions used were as follows: 95 °C for 5 min, followed by 30 cycles of 30 s at 95 °C, 30 s at 55 °C and 30 s at 72 °C, and a final cycle of PCR extension at 72 °C for 5 min (TaKaRa Biotechnology, Dalian, China). The target fragment was inserted downstream of the GST gene in the pGEX-4T-2 vector. Competent E. coli BL21 (DE3) cells were transformed with pGEX-4T-2-FANK1. The recombinant plasmid was confirmed by restriction enzyme digestion and sequencing.
The vector-containing bacteria were grown in Luria-Bertani broth at 37 °C overnight (A600 = 0.5). Overnight cultures of bacterial cells were diluted 20-fold with Luria-Bertani broth (A600 = 1.0) and were then induced to express the recombinant protein for 3 h by adding 0.5 mmol l−1 IPTG at 30 °C. Purified protein was obtained using GST-bound resin affinity chromatography with the dissolved protein, and the recombinant protein was obtained after lysing (GST-Bind Kits, Novagen, Darmstadt, Germany), dialyzing and renaturation (Protein Refolding Kit, Novagen, Darmstadt, Germany) according to the manufacturer's instructions. The purity of the recombinant protein was verified by Western blotting, and the purified GST-FANK1 proteins were stored at -80 °C for later cyclic amplification of sequence target (CAST) analysis.
Cyclic amplification of sequence targetAnalysis
CAST was performed as previously described with several modifications.5 The forward primer (5’-GCTGCAGTTGCACTGAATTCGCCTC (N)26CGACAGGATCCGCTGA ACTGACCTG-3’) and reverse primer (5’-CAGGTCAGTTCAGCGGATCCTGTCG (N)26GAGGCGAATTCAGTGCAACTGCAGC-3’) contained flanking PCR primer sequences and a central core of 26 random bases. The oligonucleotides were annealed to short double-stranded DNA that was dissolved in DNA-binding buffer (10 mmol l−1 Tris pH 7.5, 50 mmol l−1 NaCl, 7.5 mmol l−1 MgCl2, 1 mmol l−1 ethylenediaminetetraacetic acid, 5% (v/v) glycerol, 5% (w/v) sucrose, 0.1% (v/v) Nonidet P-40 and 5 mg ml−1 bovine serum albumin) with the GST-FANK1 fusion protein. After a 20-min incubation period at room temperature on a shaking platform, the complexes were washed three times with the binding buffer.
The mixture was amplified by PCR (TaKaRa Biotechnology, Dalian, China), and the PCR conditions used were as follows: 95 °C for 5 min followed by 10 cycles of 1 min at 94 °C, 1 min at 62 °C and 30 s at 72 °C, and a final cycle of PCR extension at 72 °C for 5 min. Next, the product was used as the template to initiate the next CAST cycle. A total of five cycles of CASTs were performed. The final PCR products were cloned into the pGEM-T easy vector (Promega, Madison, WI, USA). The data were analyzed using LaserGene 7.10 software (DNAStar) after sequencing.
pSUPER-shFank1 and pDsRed-Fank1 Construction and In Vivo Verification
According to the Fank1 cDNA sequence (NM_025850) from GenBank and siRNA design principles,6 two siRNA sequences that were able to specifically knock down the expression of Fank1 and a negative control sequence were obtained using software available on the manufacturer's website (Thermo Scientific/Dharmacon; http://www. dharmacon. com/). The sequences were inserted into the Bgl II and Xho I sites (Table 1). After annealing, the ds oligo was digested with Bgl II and Xho I restriction endonucleases and inserted between the Bgl II and Xho I sites of the pSUPER-neo -green fluorescence protein vector downstream of the H1 promoter.
Table 1.
siRNA sequences of Fank1 and the negative control

The Fank1 cDNA coding sequence was amplified by PCR from the testes of adult C57BL/6 mice. The specific primer for the Fank1 ORF was designed with a start codon (ATG) but no termination codon. The forward primer was 5’-CGAAGACAGGCGTTAGGAG-3’, and the reverse primer was 5’-GTGGACAGAGGACTTCCTT-3’. The PCR product was ligated with pDsRed-N1 to create the pDsRed-Fank1 vector, resulting in a recombinant plasmid containing the Fank1 gene and the red fluorescent protein sequence.
Thirty-six hours after co-transfection of the 293-T cells with the pSUPER-shFank1 and pDsRed-Fank1 vectors, the RFP and green fluorescence protein fluorescence intensities were detected via fluorescence microscopy (Nikon, Tokyo, Japan). The efficiency of the Fank1 interference vector was detected by reverse transcription-PCR (RT-PCR) and Western blots.
Generation of Transgenic Mice
B6D2F1 female mice (4 to 6 weeks old) were superovulated and mated with stud B6D2F1 males. The females were killed the next morning, and the fertilized eggs were removed from the oviductal ampullae, washed and maintained in an incubator before injection. The plasmids with high interference efficiency were selected and digested by the single enzyme Hind III, purified using a gel extraction kit and then the 5489-bp fragment of pSUPER-Fank1 631 was injected into the fertilized eggs of B6D2F1 mice via microscopic injection. Two hundred and three injected two-cell embryos were transferred into the oviduct of eight pseudopregnant imprinting control region female mice.7 The tail DNA from 4-week-old mice was analyzed by PCR (Primers: Forward: 5’-GGAAGATGGCTGTGAGGG-3’ and reverse: 5’-CCAGGCTTTACACTTTATGCT-3’). The transgenic mice were mated with normal C57BL/6J mice.
Detection of Fank1 siRNA Expression in Knockdown Mice
Small RNA samples from the testes of knockdown mice were isolated using the mirVanaPTMP miRNA isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions.8 Briefly, 50–250 mg of tissue was homogenized in 10 volumes of lysis/binding buffer. A 1/10 volume of miRNA homogenate additive was added and incubated on ice for 10 min. The total RNA was extracted by adding an equal volume of acid-phenol: chloroform. Small RNA was extracted from the total RNA with a filter cartridge and 100 μl of preheated (95 °C) elution solution. The concentration of small RNA was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
The construction of small RNA cDNA (srcDNA) libraries and semiquantitative PCR were performed. The primer sequences were as follows: RTQ-UNIr primer: 5’-CGAATTCTAGAGCTCGAGGCAGG-3’; shFank1 primer: 5’-CCCGCACCTAGATGTTGTCAAA-3’; let7 primer: 5’-GAGGTAGTAGG TTGTATAGT-3’. A three-step PCR protocol (95 °C for 10 min, then 40 cycles of 95 °C for 15 s, 50 °C for 30 s, and 60 °C for 30 s) was used. After PCR, an aliquot of 2 μl of the PCR product was analyzed on a 2% (w/v) agarose gel.
Real-Time Polymerase Chain Reaction
The quantification of Fank1 expression levels was performed using a SYBR-Green I-based real-time fluorescence detection method.9 Total RNA was isolated from the testes using a kit (Qiagen, mRNA isolation kit, Crawley, West Sussex, UK) and was subsequently reverse transcribed using the Reverse Transcription PCR kit (Promega) with Oligo-dT primers. The cDNA was amplified with the SYBR Premix Ex Taq kit (TAKARA) on a Rotor-Gene 3000 (Corbett Research Co, Mortlake, Australia). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the reference gene, and samples without cDNA or without oligonucleotides were used as negative controls. The control GAPDH fragment was amplified using the following primer sequences: GAPDH-forward, 5’-AGCGAGCATCCCCCA AAGTT-3’; and GAPDH-reverse, 5’-GGGCACGAGGGCTCATCATT-3’. The Fank1 primers were as follows: Forward: 5’-GAGGACCCCAAGATGCACAG-3’ and reverse: 5’-AGGGGCTGGTGACCTTCAGT-3’. The 25 μl PCR included 5 μl 10 × diluted cDNA, 12.5 μl SYBR Premix EX TaqTM and 2 μl of primers. The reaction was run online at 95 °C for 1 min, followed by 45 cycles at 95 °C for 5 s and 60 °C for 20 s. The threshold cycle (Ct) data were determined from the default threshold settings, and all measurements were performed at least three times.
Western Blot
The testes of transgenic animals were snap-frozen in liquid nitrogen. Frozen tissues were homogenized in a glass Teflon Potter in radioimmunoprecipitation assay buffer (50 mmol l−1 Tris, 150 mmol l−1 NaCl, 1 mmol l−1 ethylenediaminetetraacetic acid and 0.2% (v/v) NP-40) containing the complete protease inhibitor, phenylmethylsulfonyl fluoride, followed by centrifugation at 12 000g at 4 °C. The protein concentration was determined from the supernatant using a bicinchoninic acid assay (Beyotime, Biotech, Shanghai, China) and analyzed using an enzyme-linked immunosorbent assay-plate reader (Sunrise RC, Tecan, Mannedorf, Switzerland). The protein extracts were diluted with 5× sodium dodecyl sulfate loading buffer (120 mmol l−1 Tris (pH 6.8), 2% (w/v) sodium dodecyl sulfate, 5% (v/v) β-mercaptoethanol, 50% (v/v) glycerol and bromophenol blue), boiled and resolved by 10% (w/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis. For testes, 150 μg protein extract was loaded. After electrophoresis, the proteins were transferred to a polyvinylidene fluoride membrane (Roche Diagnostics, Basel, Switzerland) by semidry blotting (Biorad, Hercules, CA, USA). Detection was performed using anti-FANK1 antibody (1:200), and mouse anti-ACTIN antibody (1:400) was used as the loading control (SM0441, Marker Fermentas, Lithuania). For blocking experiments, the antibody was preabsorbed with 50 μg peptide for 3 h at room temperature. For the secondary antibodies, anti-rabbit and anti-mouse immunoglobulin G coupled to horseradish peroxidase (Beyotime) were used at dilutions of 1:2000. Blots were developed with the Western Lightning Chemiluminescence Reagent Plus (Pierce, Thermo, Rockford, IL, USA) and chemiluminescence detection film.
Mating Studies
Male Fank1-knockdown mice and wildtype female mice were housed for 6 months, and the litter interval and litter sizes were recorded. Wildtype male mice were used as a comparison.
Testicular Weight
The male Fank1-knockdown mice were killed by neck dislocation and the testes were weighed. Wildtype males were used as a comparison.
Sperm isolation and Assessment of Sperm parameters
Male Fank1-knockdown mice and wildtype males were housed alone for 1 week before they were killed. The cauda epididymidis was removed from each mouse and finely diced into 500 μl of human tubal fluid (HTF) medium in a 1.5-ml microcentrifuge tube; the solution was then shaken for 15 s at 37 °C for 30 min.
The sperm concentration was measured in a hemocytometer (Conception Technologies, San Diego, CA, USA) and expressed as million per ml of suspension. From each sample, three aliquots (5 μl) were loaded into a chamber slide with a 20 μm depth, and the spermatozoa were counted in five squares of area. The sperm number was calculated as follows (dilution factor) × (sperm count in five squares) × 0.05 × 106.
Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL)
The testes from the transgenic mice were fixed in 4% (w/v) paraformaldehyde and embedded in paraffin. Next, the tissue blocks were cut into 5 μm-thick sections. The sections were dewaxed and rehydrated in serial baths of xylene and graded alcohols, hydrated by phosphate buffered saline and pretreated with 200 μg l−1 proteinase K solution for 20 min. Finally, the sections were washed three times for 5 min each with phosphate buffered saline. The sections were hybridized with reaction solution at 37 °C for 1 h according to the manufacturer's instructions for the TUNEL kit (DeadEnd™ Colorimetric TUNEL System, Promega, Madison, WI, USA). After being washed with phosphate buffered saline three times (5 min each time), the sections were incubated with streptavidin horseradish peroxidase solution for 30 min at room temperature. Apoptotic cells were visualized in a solution containing 3,3’-diaminobenzidine for 5–10 min, and the number of positive apoptotic spermatogonia per tubule section was calculated for a total of 100 tubule sections per group.
Microarray Analysis
Using TRIzol (Invitrogen, Carlsbad, CA, USA), total RNA was isolated from the testes of the positive generation of founder 2 and the control negative generation of founder 2 for quantitative microarray analyses (Gene company agent of China, Shenyang). Three independent samples were analyzed in triplicate. The generation of double-stranded cDNA, the preparation and labeling of cDNA, the hybridization to 430 2.0 Mouse Genome Arrays (Affymetrix, Santa Clara, CA, USA), washing and scanning were performed according to the standard Affymetrix protocols. The Affymetrix Expression Console software was used to quantify the hybridized arrays. Scanning and analysis were performed with Partek GS 6.5 (Partek® Genomics Suite™). Fold-changes were calculated by dividing the Fank1-knockdown mice value by the wildtype value. A fold-change of 1 or–1 indicates no change; however, a fold-change of 2 indicates product doubling and a fold change of −2 indicates the halving of transcript abundance.
Verification of Microarray Analysis Results by RT-PCR
Two overexpressed genes identified from the results of the microarray analysis (Dusp1 and Klk1b21) were confirmed by PCR. RNA was extracted from wildtype and Fank1-knockdown mice, and cDNA was generated from the total RNA with the reverse transcriptase kit (Promega, Madison, WI, USA) according to the manufacturer's protocol. PCR was performed with the cDNA as the template and the following primers: Dusp1 forward: 5’-CCTGTTGTTGGATTGTCGCTCCTTCT-3’ and reverse: 5’-GGATGCTCTTGTACTGGTAGTGACCC-3’ (the amplified fragment was 120 bp in length); Klk1b212 forward: 5’-CCGAGGATCTTCAACAGCTC-3’ and reverse: 5’-TTGTTGTAGCGGAACACAGC-3’ (the amplified fragment was 120 bp in length). The PCR protocol was as follows: 95 °C for 3 min, then 30 cycles of 95 °C for 20 s, 57 °C for 30 s, and 60 °C for 20 s. After PCR, an aliquot of 2 μl of the PCR product was analyzed on a 2% (w/v) agarose gel.
Statistical Analyses
Data are shown as the mean ± standard error of the mean (s.e.m.), and significant differences between data sets were assessed by t-test and one-way analysis of variance using Statistical Package for Social Sciences 13.5. Significance was accepted at the level of P < 0.05.
RESULTS
Identification of the Fank1 Consensus DNA-binding Sequence
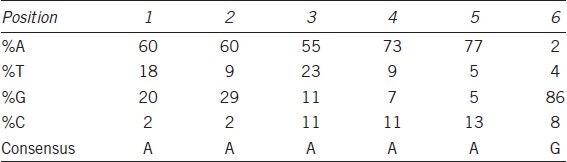
In this study, we amplified the FnIII domain (479 bp) and constructed a fusion protein with GST. The GST-FANK1 fusion proteins were detected by Western blotting with an anti-GST antibody. The molecular weight of the GST-FANK1 protein was 43 kDa (Figure 1a). This protein was applied to the CAST experiment to confirm FANK1 DNA-binding sequences. The 76 bp synthesized oligonucleotide primer contained 26 random nucleotides and bound to purified GST-FANK1. Forty-four sequencing results were obtained after five rounds of PCR (Figure 1b). The most frequently observed sequences (after comparative analysis with the LaserGene software) are shown in Table 2. The percentages of each base at each position ranged from 55 to 86%. The CAST assay also revealed a consensus sequence (5’-AAAAAG-3’).
Figure 1.

Identification of the GZF1 binding consensus sequence. (a) GST and GST-FANK1 proteins expressed in bacteria were stained with Coomassie Brilliant Blue. M indicates the protein marker (left panel). The GST-FANK1 fusion proteins were detected by Western blotting with an anti-GST antibody (right panel). (b) Alignment of individual DNA sequences recovered in the cyclic amplification of sequence target (CAST) analysis. The deduced consensus FANK1-binding sequence.
Table 2.
Consensus binding sequence of FANK1
Expression of shRNA with the pSUPER-shFank1 Vector Induces Fank1 mRNA Degradation
Two of the designed siRNA sequences of shFank (631# and 274#) were located in the ankyrin and FnIII domains of FANK1 (Figure 2a). After transfection of the 293-T cells with pDsRed-Fank1 and siFank1, the green fluorescence protein in pSUPER-shFank1 and the RFP in pDsRed-Fank1 were observed for 36 h. Fluorescence microscopy demonstrated that shFank1 631 and 247 inhibited RFP expression with efficiencies of 90 and 95%, respectively (Figure 2b). The results of RT-PCR and Western blot statistical analyses revealed that the Fank1 gene was knocked down at both the mRNA and protein levels in 293-T cells by shFank1 631 and 274, with shFank1 631 exhibiting higher activity (Figure 2c).
Figure 2.
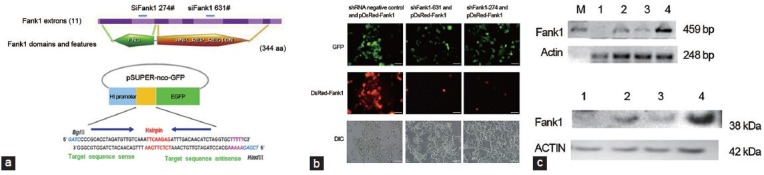
Expression of shRNA using the pSUPER-shFank1 vector induces Fank1 mRNA degradation. (a) shFank 631 and 274 are two oligonucleotides located in the ankyrin and FnIII domains of FANK1. Two oligonucleotides containing sense and antisense 20-nt sequences from the Fank1 coding region (black sequence), an 8-nt spacer sequence that provided a loop structure, and a transcription termination signal of five thymidines (purple sequence) were annealed and inserted downstream of the H1 promoter. (b) Transfection of 293 T cells with 1 mg of the pDsRed-Fank-expressing plasmid together with 2 mg of a plasmid expressing siFank. Fluorescence microscopy showed that shFank1 631 and 247 inhibited RFP expression with efficiencies of 90 and 95%, respectively, 36 h after the 293 T cells were transfected. Scale bars = 100 mm. (c) The results of reverse transcription-polymerase chain reaction (RT-PCR) and Western blots suggest that the Fank1 gene is inhibited at the mRNA and protein levels by Fank1 siRNA (P < 0.05).
Confirmation and Breeding of Fank1-Knockdown Mice Lines
The DNA of young mice was subjected to PCR. In total, 14 positive mice (founders) were observed. The numbers of the founders were as follows: male- F02, F011, F012, F021, F037, F043, F044 and F045; female- F01, F020, F024, F039, F040 and F046. The founders of F043, F044 and F046 were chimeras in which shFank1 was recombined in somatic cells but not in germ cells. F020 and F024 were not pregnant, and F040 and F046 died (Figure 3a). Therefore, seven founders were studied in subsequent experiments. We first confirmed the expression of Fank1 siRNA in the testes of the mice by PCR (Figure 3b). Then, the downregulation of Fank1 was confirmed in the testes of the transgenic mice by RT-PCR (real-time PCR) and Western blotting (Figure 3c-e).
Figure 3.
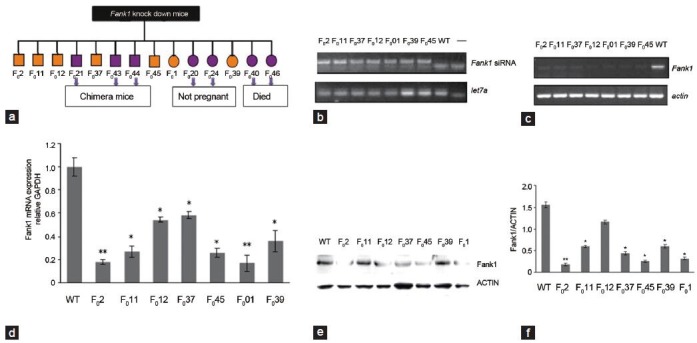
Identification and phenotype analysis of Fank1 knockdown transgenic mice. (a) In total, 14 founders of Fank1-knockdown mice, eight males and six females, were studied. F021, F043 and F044 were chimeras, F020 and F024 were not pregnant, and F040 and F046 died (marked in purple); F02, F011, F012, F037, F045, F01 and F039 were studied by phenotype analysis (marked in orange). (b) Expression of Fank1 trangenic mouse siRNAs. Fank1 siRNA expressed in the testes of seven transgenic mice, but not in the wildtype mice. let7a is the blank control. (c) Expression of Fank1 in the testes of transgenic mice and wildtype mice by RT-PCR analyses. Down-regulation of Fank1 in transgenic mice compared with the wildtype mice. Actin is the control. (d) Quantification of the Fank1 transcription level in the testes of seven knock down mice and wildtype male mice using real-time RT-PCR. Expression levels were calculated based on the threshold cycle (Ct) values and were normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA, and then the expression was determined relative to the level of Fank1. The experiments were repeated at least three times. Data were determined by one-way ANOVA with S-N-K's post-test. (*P < 0.05, **P < 0.01, compared with wildtype mice). (e) Expression of FANK1 protein in the testes of transgenic mice and wildtype mice by Western blot analyses (mean ± s.e.m.). Differences in the FANK1 protein levels between the Fank1-knockdown mice and wildtype mice were observed. Data were determined by one-way ANOVA with S-N-K's post-test (*P < 0.05, **P < 0.01, compared with wildtype mice). ANOVA, analysis of variance; s.e.m., standard error of the mean.
Phenotype Analysis of Fank1-Knockdown Transgenic Mice
We observed the testes weight, litter interval, litter size and number of spermatozoa in seven knockdown mice compared with 21 wildtype mice. The results of the statistical analysis revealed differences in litter interval, litter size and sperm number in transgenic mice compared with wildtype mice (P < 0.05), but did not show differences in the weight of the testes (P > 0.05) (Table 3). The reduction of the sperm number in Fank1-knockdown mice was associated with prolonging the litter interval and decreasing the litter size compared to the wildtype mice. Based on the above results, we examined the apoptotic cells in the testes from the transgenic mouse F02 (inhibition efficiency > 90%). TUNEL staining revealed that the average number of apoptotic cells in each group was 45.33 ± 7.5 and 121.3 ± 6.26 in the knockdown and wildtype mice, respectively (P < 0.05, n = 3), and most of the apoptotic cells were spermatogonia and spermatocytes (Figure 4a and 4b).
Table 3.
Mating study of Fank1-knockdown transgenic mice
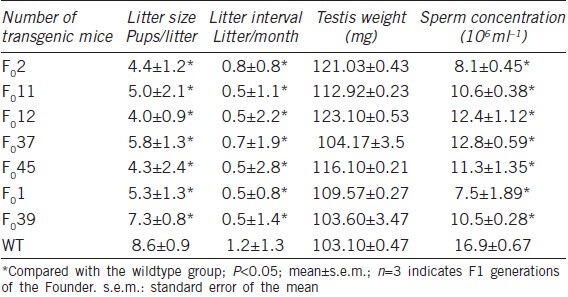
Figure 4.
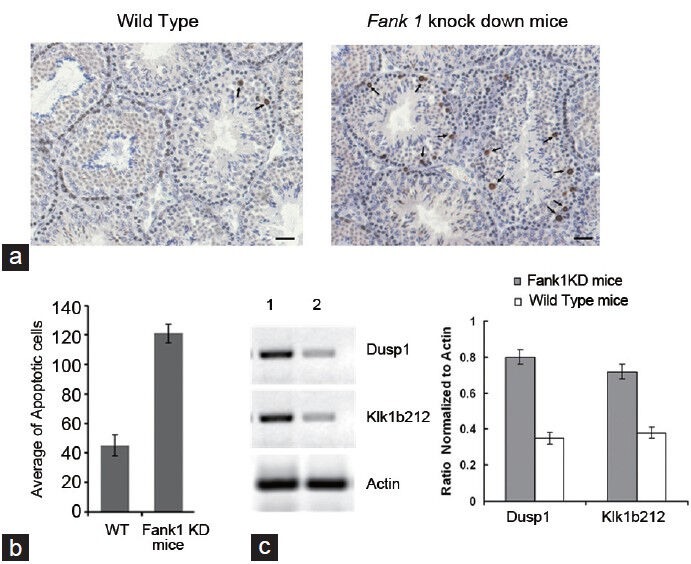
TUNEL staining and confirmation of microarray analysis. (a) and (b) TUNEL staining shows that left and right testis paraffin sections indicate wildtype and Fank1-knockdown mice, respectively. The arrow indicates apoptotic cells. Scale bars = 100 mm. TUNEL staining revealed that the average number of apoptotic cells per tubule cross section in each group was 45.3 ± 7.5 and 121.3 ± 6.3 in the knockdown mice and the wildtype mice, respectively (mean ± s.e.m., P < 0.05, n = 3, t-test), and most of the apoptotic cells were spermatogonia and spermatocytes. (c) Expression of Dusp1 and KlK1b212in Fank1-knockdown mice and wildtype mice by RT-PCR; the 1st and 2nd line indicate the Fank1-knockdown mice and wildtype mice, respectively. Dusp1 and KlKlb212 were overexpressed in Fank-knockdown mice compared to the wildtype mice (mean ± s.e.m., P < 0.05, t-test). TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
Microarray Analysis of Fank1-Knockdown Transgenic Mice
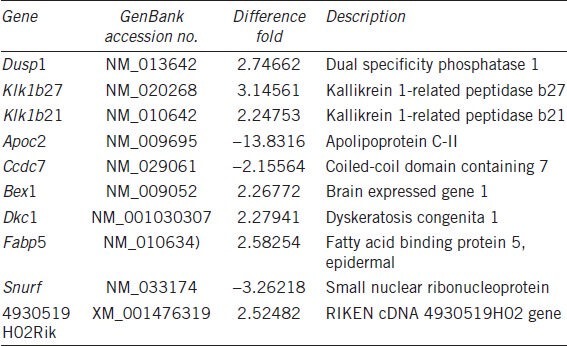
The results of the microarray analysis in the testes of Fank1-knockdown and wildtype mice showed that 17 genes were differentially expressed with a change greater than twofold. In total, 14 genes were upregulated, including Egr1, Fos and Dusp1; however, three genes were downregulated: Apoc2, Ccdc7 and Snurf. All 17 genes were analyzed by bioinformatics, and 10 genes contained the FANK1 DNA-binding sequence AAAAAG upstream of their promoter region. The results suggest that these genes are regulated by FANK1 directly or indirectly. We also found that AAAAAG was at -1616-1621 and -241-246 in the upstream regions of Dusp1 and Klk1b212, respectively. The results from our RT-PCR analysis are consistent with the results of the microarray analysis (Figure 4c).
DISCUSSION
Spermatogenesis is an elaborate cell differentiation process10 and is also a developmental process that progresses according to a genetic programme directing the sequential and coordinate expression of specific genes.11 More than 100 genes have been identified to play important roles during spermatogenesis,12,13,14,15 but the mechanisms and key genes responsible for sperm morphogenesis are poorly understood. Fank1 is exclusively expressed in the testis from the meiosis phase to the haploid phase of spermatogenesis; therefore, it may function as a testis-specific transcription factor. Fank1 is highly conserved in vertebrates, and the human and mice orthologues display an 85% amino acid sequence identity.
One method of studying gene function in vitro is to establish knockdown transgenic mice. However, this method is limited in the study of the mechanisms in spermatogenesis because knockdown technology is expensive and time consuming, and it has a low success rate. RNAi has been applied widely in gene function research since its discovery and is currently used to reduce or suppress target gene expression in a faster, cheaper and easier sequence-specific manner in which the genome information remains intact.16,17,18 Thus, in this study, we chose to obtain Fank1-knockdown mice by RNAi to study its function in the process of spermatogenesis.
In our study, we obtained 14 founders when the Fank1-knockdown mice had been established for 3 months. After breeding and selection, the fertility of the newborn F1 generation from seven founders was analyzed. We found significant differences in the litter interval, litter size and sperm number of seven founders compared to wildtype mice. The length of the litter interval and the decreasing size of the litter presumably resulted from the reduction in sperm number in Fank1-knockdown mice. The loss of Fank1 expression to different degrees was verified in the testes of seven founders using RT-PCR and Western blotting. Our result also revealed that the number of apoptotic cells was higher in the knockdown mice than in the wildtype mice (P < 0.05), according to the TUNEL and microarray analyses using the F02 founder, which possessed an interference efficiency of > 90%. Apoptosis primarily occurs in spermatogonia and spermatocytes. The number of spermatozoa depended on the proliferation and differentiation of the spermatogenic cells and was also associated with the degree of apoptosis in spermatogenic cells.19 We observed a reduction in sperm number and an increase in apoptotic spermatocytes in Fank1-knockdown mice. Fank1 expression began on the 14th postnatal day, which occurs during the pachytene stage of meiosis, which is the longest stage of meiosis, in which increasing activation of many transcription factors contributes to the activation of genes.20 However, the downregulation of Fank1 in the knockdown mice resulted in lower transcriptional activity and the decreased activation of downstream genes. Thus, we speculate that Fank1 contributes to the reduced sperm number as a result of regulating spermatogenic cell apoptosis, which is similar to the reported symptoms of several patients with oligozoospermia.21,22
During our efforts to study the function of the protein, we found that FANK1 encodes a protein containing an FnIII domain in the amino terminus and five ankyrin repeats in its carboxyl terminus. Gene Ontology Classifications in the MGI (http://www.informatics.jax.org/) suggest that FANK1 has DNA binding activity and is involved in DNA-dependent transcription. Therefore, FANK1 may function as a transcription factor to control gene expression between the late-meiotic phase and the haploid phase of spermatogenesis. We screened the FANK1 DNA-binding sequence with the CAST assay.23 The most frequently observed sequence was AAAAAG, which had the highest affinity. The microarray analysis results of the testicular tissue of Fank1-knockdown transgenic mice indicated that DUSP1, KLK1b21, KLK1b27 and 10 other genes (Table 4) contained the AAAAAG sequence in their promoter regions. This finding further supports that this sequence is the DNA-binding sequence of FANK1, and these genes are directly regulated by FANK1. We confirmed by RT-PCR that DUSP1 and KLK1b21 had similar expression levels to those observed in the microarray analysis.
Table 4.
List of genes whose upstream promoter regions contained FANK1 DNA-binding sites that were differentially expressed between wildtype and Fank1-knockdown transgenic mice
Dusp1 was identified in 1992 as an immediate early gene that was induced in response to growth factors and oxidative stress.24,25 Dusp1 has different specificity with JNK, ERK and p38 binding. In recent years, studies have shown that MKP-1 exhibits a very important negative effect on immune regulation as a result of specifically inactivating p38 and JNK.26 MKP-1 dephosphorylates the threonines and tyrosines of MAP kinase-activated structure domains and then stops the signaling pathways, which activates the transcription factors to regulate the related proteins in a cascading amplification effect.27,28 FANK1 has characteristics of a transcription factor and may be related to Dusp1. The results of the microarray analysis showed that Dusp1 may be a negative regulator because the expression of Fank1 was suppressed following Dusp1 upregulation.
Klk1b21 is a kallikrein that belongs to the subfamily of Klk. KlK1b21 is expressed in the kidney, submaxillary glands and testes of mice and is expressed exclusively in the Leydig cells of adult mice. Several studies have shown that KlK1b21 cleaves fibronectin and IGFBP-3 in the Leydig cells of the adult mouse.29,30 The Leydig cells produce 95% of androgens, which induce the synthesis and secretion of a variety of growth factors, bioactive substances and estrogens, and they have a relationship with spermatogenesis. Although the mechanism of KlK1b21 in spermatogenesis is still unclear, recent studies have shown that KlK1b21 plays an important role in the adult mouse Leydig cells.31 Our microarray analysis suggests that the overexpression of KlK1b21 in Fank1-knockdown mice causes a reduction in hormones secreted from Leydig cells, which then influence spermatogenesis in the knockdown mouse.
In summary, using RNAi, we successfully obtained Fank1- nockdown mice. The results from our study on Fank1 provide evidence that FANK1 is a vital transcription factor in spermatogenesis. Using microarray analysis, we will continue to study the genes regulated by FANK1 to potentially reveal the role and significance of Fank1 in spermiogenesis and male sterility.
AUTHOR CONTRIBUTIONS
WWD performed the molecular genetic studies, conducted the construction of pSUPER-shFank1 and pDsRed-Fank1, isolated the sperm, assessed the sperm parameters, performed the TUNEL analysis and drafted the manuscript. HLH conducted the microarray analysis and verified the microarray analysis results. WYang participated in the expression and purification of GST-FANK1 and CAST analysis. JL aided in drafting the manuscript. YY, SLZ, WW and XCL participated in the generation of transgenic mice, and ZYL participated in the real-time PCR and Western blot analysis. MYZ participated in the design of the study and performed the statistical analysis. ZHZ was involved in the project design and manuscript revising. WYan participated in the design of the study. All authors read and approved the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing interests.
ACKNOWLEDGMENTS
The authors would like to thank Dr Zheng-Li Wei and Dr Shui-Qiao Yuan at China Medical University for their technical assistance. This work was supported by The National Nature Science Foundation of China (No. 30770818) and the LiaoNing Science and Technology Project (No. 2009408001-1). Program funding was provided by the Liaoning Province Education Administration (No. 2009S109).
REFERENCES
- 1.Eddy EM, O’Brien DA. Gene expression during mammalian meiosis. Curr Top Dev Biol. 1998;37:141–200. [PubMed] [Google Scholar]
- 2.Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–61. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 3.Khaitovich P, Kelso J, Franz H, Visagie J, Giger T, et al. Functionality of intergenic transcription: an evolutionary comparison. PLoS Genetics. 2006;2:e171. doi: 10.1371/journal.pgen.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Z, Zheng H, Yan W. Fank1 is a testis-specific gene encoding a nuclear protein exclusively expressed during the transition from the meiotic to the haploid phase of spermatogenesis. Gene Expr Patterns. 2007;7:777–83. doi: 10.1016/j.modgep.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Wright WE, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11:4104–10. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Peng S, York JP, Zhang P. A transgenic approach for RNA interference-based genetic screening in mice. Proc Natl Acad Sci U S A. 2006;103:2252–6. doi: 10.1073/pnas.0511034103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ro S, Park C, Jin J, Sanders KM, Yan W. A PCR-based method for detection and quantification of small RNAs. Biochem Biophys Res Commun. 2006;351:756–63. doi: 10.1016/j.bbrc.2006.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm D, Herold S, Kuechler A, Liehr T, Laccone F. Rapid detection of subtelomeric deletion/duplication by novel real-time quantitative PCR using SYBR-green dye. Hum Mutat. 2004;23:368–78. doi: 10.1002/humu.20011. [DOI] [PubMed] [Google Scholar]
- 10.Ronfani L, Bianchi ME. Molecular mechanisms in male determination and germ cell differentiation. Cell Mol Life Sci. 2004;61:1907–25. doi: 10.1007/s00018-004-4034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grootegoed JA, Siep M, Baarends WM. Molecular and cellular mechanisms in spermatogenesis. Baillieres Best Pract Res Clin Endocrinol Metab. 2000;14:331–43. doi: 10.1053/beem.2000.0083. [DOI] [PubMed] [Google Scholar]
- 12.Zheng H, Stratton CJ, Morozumi K, Jin J, Yanagimachi R, et al. Lack of Spem1 causes aberrant cytoplasm removal, sperm deformation, and male infertility. Proc Natl Acad Sci U S A. 2007;104:6852–7. doi: 10.1073/pnas.0701669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Du YR, Qin WH, Hu YG, Huang YN, et al. RIM-BP3 is a manchette-associated protein essential for spermiogenesis. Development. 2009;136:373–82. doi: 10.1242/dev.030858. [DOI] [PubMed] [Google Scholar]
- 14.Hsu LC, Chen HY, Lin YW, Chu WC, Lin MJ, et al. DAZAP1, an hnRNP protein, is required for normal growth and spermatogenesis in mice. RNA. 2008;14:1814–22. doi: 10.1261/rna.1152808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert S, Kamino K, Bohm D, Adham I, Engel W, et al. TSPY expression is variably altered in transgenic mice with testicular feminization. Biol Reprod. 2008;79:125–33. doi: 10.1095/biolreprod.107.067025. [DOI] [PubMed] [Google Scholar]
- 16.Leung RK, Whittaker PA. RNA interference: From gene silencing to gene-specific therapeutics. Pharmacol Ther. 2005;107:222–39. doi: 10.1016/j.pharmthera.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maine EM. Studying gene function in Caenorhabditis elegans using RNA-mediated interference. Brief Funct Genomic Proteomic. 2008;7:184–94. doi: 10.1093/bfgp/eln019. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Wang GL, Li HX, Li L, Cui QW, et al. Regulation of fertilization in male rats by CatSper2 knockdown. Asian J Androl. 2012;14:301–9. doi: 10.1038/aja.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClusky LM, Barnhoorn IE, van Dyk JC, Bornman MS. Testicular apoptosis in feral Clarias gariepinus using TUNEL and cleaved caspase-3 immunohistochemistry. Ecotoxicol Environ Saf. 2008;71:41–6. doi: 10.1016/j.ecoenv.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Song W, Hu T, Zhang N, Miao S, et al. Fank1 interacts with Jab1 and regulates cell apoptosis via the AP-1 pathway. Cell Mol Life Sci. 2011;68:2129–39. doi: 10.1007/s00018-010-0559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yatsenko AN, Iwamori N, Iwamori T, Matzuk MM. The power of mouse genetics to study spermatogenesis. J Androl. 2010;31:34–44. doi: 10.2164/jandrol.109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C, Yeh S, Chen YT, Wu CC, Chuang KH, et al. Oligozoospermia with normal fertility in male mice lacking the androgen receptor in testis peritubular myoid cells. Proc Natl Acad Sci U S A. 2006;103:17718–23. doi: 10.1073/pnas.0608556103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi Y, Rajkovic A. Characterization of NOBOX DNA binding specificity and its regulation of Gdf9 and Pou5f1 promoters. J Biol Chem. 2006;281:35747–56. doi: 10.1074/jbc.M604008200. [DOI] [PubMed] [Google Scholar]
- 24.Charles CH, Abler AS, Lau LF. cDNA sequence of a growth factor-inducible immediate early gene and characterization of its encoded protein. Oncogene. 1992;7:187–90. [PubMed] [Google Scholar]
- 25.Keyse SM, Emslie EA. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature. 1992;359:644–7. doi: 10.1038/359644a0. [DOI] [PubMed] [Google Scholar]
- 26.Purwana IN, Kanasaki H, Oride A, Miyazaki K. Induction of dual specificity phosphatase 1 (DUSP1) by gonadotropin-releasing hormone (GnRH) and the role for gonadotropin subunit gene expression in mouse pituitary gonadotroph L beta T2 cells. Biol Reprod. 2010;82:352–62. doi: 10.1095/biolreprod.109.080440. [DOI] [PubMed] [Google Scholar]
- 27.Franklin CC, Kraft AS. Conditional expression of the mitogen-activated protein kinase (MAPK) phosphatase MKP-1 preferentially inhibits p38 MAPK and stress-activated protein kinase in U937 cells. J Biol Chem. 1997;272:16917–23. doi: 10.1074/jbc.272.27.16917. [DOI] [PubMed] [Google Scholar]
- 28.Hirsch DD, Stork PJ. Mitogen-activated protein kinase phosphatases inactivate stress-activated protein kinase pathways in vivo. J Biol Chem. 1997;272:4568–75. doi: 10.1074/jbc.272.7.4568. [DOI] [PubMed] [Google Scholar]
- 29.Matsui H, Takahashi T. Mouse testicular Leydig cells express Klk21, a tissue kallikrein that cleaves fibronectin and IGF-binding protein-3. Endocrinology. 2001;142:4918–29. doi: 10.1210/endo.142.11.8505. [DOI] [PubMed] [Google Scholar]
- 30.Chan CS, Harvey MB, Clements JA. Temporal and tissue-specific expression of kallikrein (Klk) genes and identification of a novel Klk messenger ribonucleic acid transcript during early development in the mouse. Bio Reprod. 1999;61:621–8. doi: 10.1095/biolreprod61.3.621. [DOI] [PubMed] [Google Scholar]
- 31.Strauss L, Kallio J, Desai N, Pakarinen P, Miettinen T, et al. Increased exposure to estrogens disturbs maturation, steroidogenesis, and cholesterol homeostasis via estrogen receptor alpha in adult mouse Leydig cells. Endocrinology. 2009;150:2865–72. doi: 10.1210/en.2008-1311. [DOI] [PubMed] [Google Scholar]


