Abstract
Anisotropic nanoparticles can provide considerable opportunities for assembly of nanomaterials with unique structures and properties. However, most reported anisotropic nanoparticles are either difficult to prepare, have a low yield, or difficult to functionalize. Here we report a facile one-step solution-based method to prepare anisotropic DNA-functionalized gold nanoparticles (a-DNA-AuNP) with 96% yield and with high DNA density (120 ± 20 strands on the gold hemisphere surface). The method is based on the competition between a thiolated hydrophilic DNA and a thiolated hydrophobic phospholipid and has been applied to prepare a-DNA-AuNP with different sizes of nanoparticles and a variety of DNA sequences. In addition, DNA strands on the a-DNA-AuNP can be exchanged with other DNA strands with a different sequence. The anisotropic nature of the a-DNA-AuNPs allows regioselective hetero- and homo-nuclear assembly with high monodispersity, as well as regioselective functionalization of two different DNA strands for more diverse applications.
DNA-functionalized gold nanoparticles (DNA-AuNPs) are among the most useful building blocks for nanoscale assembly. They have been used to form wide variety of structures from discrete clusters1 to one dimensional chains,2 two dimensional assemblies consisting of regular and periodic sheets of AuNP,3 and to three dimensional superlattices.4 The programmable nature of the DNA strands allows tuning and engineering of a variety of superlattices not achievable by conventional methods and introduces functionality to the particles.5 Most DNA-AuNPs used in assembly, however, are isotropic ones with DNA molecules distributed evenly around the AuNP surface. While a number of nanoscale assembly has been reported using these isotropic nanoparticles, it is desirable to explore the use of anisotropic nanoparticles in the assembly, as the unique directionality in interaction provided by anisotropically shaped or functionalized particles such as Janus particles or patchy particles, can result in complex structures with novel properties which are not achievable by using isotropic nanoparticles.6 As a result, Janus or anisotropic particles are attracting huge amount of interest, as these particles, which contain spatially separated functionalities, are capable of directing the orientation of particle interactions.7 These particles have been demonstrated to form novel lattices, discrete and chiral assemblies with unique ensemble properties.8 They are potential building blocks for smart artificial plasmonic structures that exhibit chiral optical properties in the visible spectra interval.9 Chiral metamaterials have promising applications in optics such as circular polarizers,10 negative refractive index materials,11 and chiral plasmonic rulers.12 Additionally, these materials interact asymmetrically with chiral molecules permitting chiral-selective sensing.13 Particle assemblies with chiral properties are typically achieved from careful design that results in special geometric property which are challenging to synthesize but achievable via anisotropic particles.1c, 8d, 14 In addition, regioselective assembled systems with nanorods and nanospheres were also shown to serve as biosensor with very low LOD and intracellular SERS labels.6c, 15 Despite being promising building blocks for self-assembly, DNA-conjugated anisotropic particles are challenging to synthesize,15 with only few efficient methods.16 Most synthesis methods require careful surface-based fabrication, long incubation time, or have low yields that require additional step of purification.8a, b, 17 Furthermore, the prepared anisotropic nanoparticles are often difficult to functionalize selectively.18 Here, we report a facile method to synthesize DNA-functionalized anisotropic nanoparticles with high yields, and subsequently functionalized with DNA for chemoselective and regioselective assembly of a variety of AuNPs. We further demonstrate that this particle could be used to regioselectively functionalize two different DNA strands on the same particle.
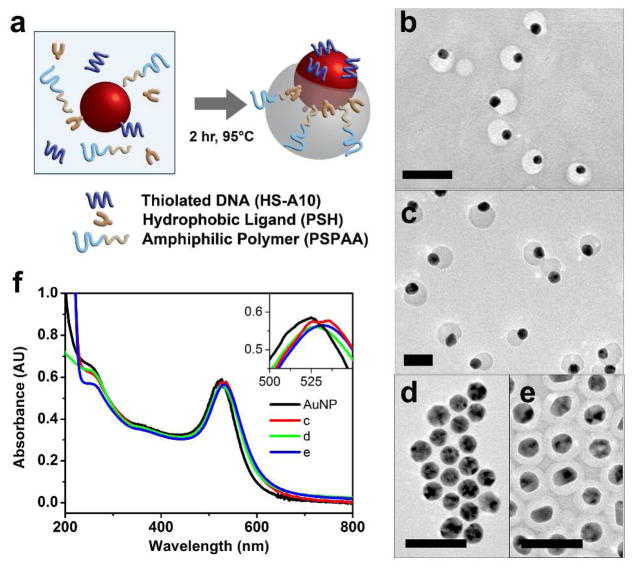
The anisotropic AuNP (a-AuNP) was formed based on competition between hydrophobic and hydrophilic ligands on gold nanoparticle surface to produce anisotropic attachment of polymers, as reported by Chen et al..18–19 This method allows facile synthesis of a-AuNP in high yield. However, the a-AuNP have not been applied for anisotropic functionalization due to the low efficiency for post-modification nor applied for selective assembly due to the limited functional groups that can be used for synthesis of these anisotropic AuNP. Attempts to attach DNA to this anisotropic gold covered with polymers by directly mixing thiolated DNA with the particles have not been successful, probably due to the high density of the small thiolated ligands already present on the gold surface and low thiolated ligand exchange rate, as observed previously.18 To overcome this limitation, we used a 10-mer oligo of adenine with thiol at the 5′ end (HS-A10) as the hydrophilic ligand and a thiolated phospholipid (PSH) as the hydrophobic ligand (shown in Figure 1a). Both ligands were incubated with either 15 nm or 20 nm AuNP and an amphiphilic polymer polystyrene-b-poly(acrylic acid) (PSPAA) in DMF/H2O at 95 °C for two hours and the mixtures were then cooled to room temperature. Transmission electron microscope (TEM) images of the samples synthesized with 15 nm (Figure 1b, large-area-view in SI Figure S1) or 20 nm AuNPs (Figure 1c and Figure S2) indicates that a-DNA-AuNPs were formed uniformly throughout, suggesting that this method can be applied for synthesis of a-DNA-AuNPs of different sizes. To quantify the synthesis yield, we counted a total of 633 particles from the TEM micrographs and found ~96% of 20 nm a-DNA-AuNPs, with the remaining 4% either having no polymer attachment, full polymer encapsulation or encapsulated dimer of particles.
Figure 1.
(a) Schematic for the synthesis of a-DNA-AuNP where HS-A10, PSH, PSPAA and AuNP are incubated at 95°C for two hours in DMF/H2O. TEM micrographs of (b) 15 nm a-DNA-AuNP, (c) 20 nm a-AuNP, (d) completely unencapsulated AuNP when no PSH was added, (e) fully encapsulated AuNP when no SH-A10 was added. (f) UV-vis absorption spectra of citrate-capped AuNP, the a-DNA-AuNP sample in (b), and fully encapsulated AuNP (d), inset shows the shift of plasmon peak from 525 nm to 535 nm. Scale bar = 50 nm.
Since DNA molecules are negatively charged, the ratio of DNA to PSH can be tuned to balance the competition between the two ligands. As shown in Figure S3, a PSH to DNA ratio of 2 gave a larger polymer but still uniform coverage on the AuNP, while a PSH to DNA ratio of 1 resulted in a mixture of unencapsulated particles and partial encapsulated particles (Figure S4). A ligand ratio of 1:2.8 and 1:1.5 was found to be optimal for 15 nm and 20 nm a-DNA-AuNPs, respectively. Control experiments performed using only DNA or only PSH as the ligand resulted in unencapsulated AuNP (Figure 1d) and the AuNP fully encapsulated with PSPAA (Figure 1e), respectively. These results support the roles of DNA as hydrophilic ligand and PSH as hydrophobic ligand. The areas on the AuNP not encapsulated by polymer are covered by DNA.
To investigate whether this method can be generally applied to DNA strands with different sequences, thiolated A10, A30 and R20 (full sequence available in SI) were used to repeat the above procedures and all of them produced anisotropic structures, as long as proper PSH to DNA ligand ratios of 1.5, 2.2 and 5.6 are maintained respectively (Figure S2, S5 and S6). These results suggest that DNA sequences only have minimal effects on the formation of anisotropic structure. Instead, the ligand ratio is critical, especially to produce uniform anisotropic encapsulation of AuNP.
Localized surface plasmon resonance (LSPR) spectra of AuNPs are strongly correlated to the structure of the AuNP and its monodispersity. From the UV-vis absorbance spectra of the a-DNA-AuNPs, we observed a clear peak at 535 nm, which is slightly red-shifted from the absorbance peak of citrate capped AuNPs (Figure 1f). The peak width was retained and there were no additional peak or shoulder peak suggesting that the AuNPs are well dispersed and individually modified with polymer after synthesis. This result is consistent with the TEM micrographs in Figure 1c. The slight red-shift of the plasmon resonance of the synthesized a-DNA-AuNP is expected due to the functionalization of the AuNP with a ligand shell.
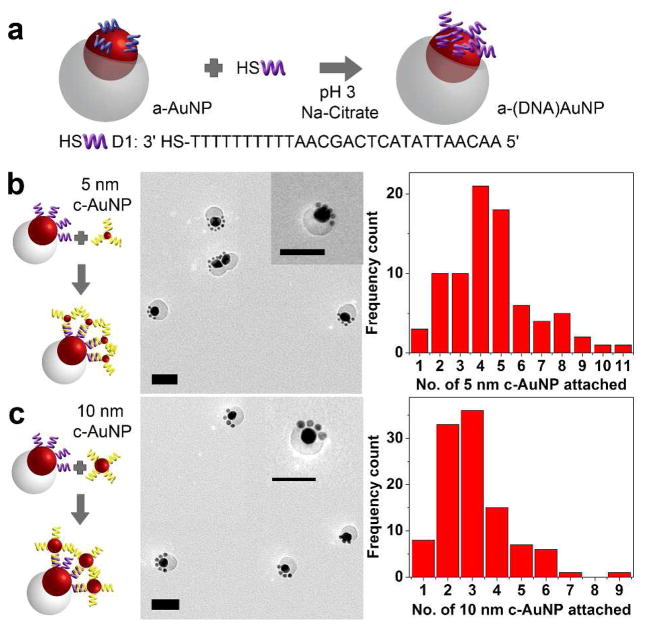
Upon demonstrating the one-pot synthesis of these a-DNA-AuNPs, we next investigated selective ligand exchange on the a-DNA-AuNPs to introduce any desired functionality for further applications of these particles. The functionalization should be regioselective so that only specific locations of the anisotropic particle are functionalized. The ligand exchange on gold surfaces, which occurs via an associative mechanism, is known to be very inefficient at room temperatures, depending on several factors such as the steric bulk and length of the incoming and existing ligands.20 To achieve this goal, we adopted a quick functionalization method reported by Zhang et al.21 where thiolated DNA (D1) was incubated with a-DNA-AuNP in 50 mM of pH 3 Na-Citrate solution to give a-(D1)-AuNP (Figure 2a). The UV-vis absorption spectra of a-DNA-AuNP before and after functionalization with D1 were nearly identical indicating that the functionalization process did not disrupt the integrity of the a-DNA-AuNP particle or cause it to aggregate (Figure S7). The DNA attachment was confirmed by functionalizing the a-DNA-AuNP with fluorescein (FAM) labeled DNA at 5′ end. From the fluorescence measurement, we found that ~ 120 ± 20 strands of DNA were functionalized per particle and 100 ± 20 complementary DNA strands can be attached per particle.(Figure S8 and Table S1) Control experiments using non-thiolated DNA strands indicated a negligible amount of DNA (0.3 strands attached per particle, data in Table S1) was detectable, suggesting that the attachment of DNA was via the thiol-gold interaction and there were little nonspecific binding of DNA to the polymer layer. In comparison to the maximum loading of 180 ± 20 DNA strands that were isotropically functionalized on a spherical 20 nm AuNP,22 the DNA density on one a-DNA-AuNP is comparable considering that only half of the particle surface was available for functionalization. Therefore, the high-density DNA functionalization suggests that the surface of AuNP is suitable to be functionalized with any thiolated ligand, similar to the versatility of the citrate-capped AuNP. Functionalization of high-density DNA on the a-DNA-AuNP is also advantageous as such feature has been shown to increase AuNP stability and allow cooperative effects in DNA directed assembly.23
Figure 2.
(a) Schematic for the functionalization of a-AuNP with D1 resulting in a-(D1)-AuNP. Schematic of selective assembly of a-(D1)-AuNP (b) with 5 nm c-AuNPs and (c) 10 nm c-AuNPs with its corresponding TEM micrographs and histogram showing particle analysis. Scale bars are 50 nm.
Progressing from the fluorescence results that indicates successful DNA (D1) attachment on the a-DNA-AuNP, we probed the regioselectivity of the functionalization of D1 on the a-(D1)-AuNP. AuNPs of 5 nm functionalized with complementary sequence to D1 (c-AuNP) were used to probe the hybridization ability and position of the DNA. Excess 5 nm c-AuNPs were removed via centrifugation. TEM micrographs of the mixture showed that the 5 nm c-AuNPs only attached on the exposed AuNP region and no particles where found on the polymer shell (Figure 2b and Figure S9), indicating that the D1 DNA on a-(D1)-AuNP not only retained its hybridization ability but also regioselectivity as demonstrated. When 5 nm AuNPs with non-complementary strands were used, most of the 5 nm AuNPs were washed away leaving no significant attachment of 5 nm AuNPs on a-(D1)-AuNP (Figure S10). These results clearly indicate that a-(D1)-AuNP interacts and assembles with other particles via specific hybridization of DNA and non-specific interaction was minimal.
Since the interaction is sequence-specific, we then expand the scope of asymmetric assembly with the use of AuNP of various sizes but functionalized with the same cDNA. As an example, the same a-DNA-AuNP was conjugated to either 5 nm or 10 nm AuNP and a cat-paw like structure was formed (see Figure 2b and 2c, respectively). For the assembly with 5 nm c-AuNPs, we found that there were on average 5 ± 2 particles regioselectively attached on each anisotropic particle from a count of total 81 particles tabulated from TEM micrographs (see histogram in Figure 2b). For 10 nm c-AuNPs, the average particle number on one a-DNA-AuNP was lower, at 3 ± 1 from a total of 107 particles (see histogram in Figure 2c and large area view in Figure S11). The standard deviation was also slightly lower for the 10 nm c-AuNP as compared to the 5 nm c-AuNP, as the sizes of the particle is more comparable to the a-DNA-AuNP. With regioselectivity of a-(D1)-AuNP, the interaction between particles is well controlled to have specific and directional cluster assemblies.
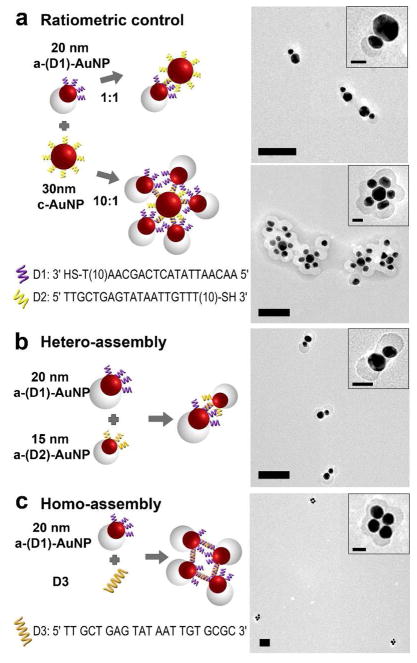
Having demonstrated regioselectivity of the a-DNA-AuNP, we further explored its application in controlling formation of more complex structures. As shown in Figures 3a and S12, using 30 nm AuNP as an example, when the concentration of a-DNA-AuNP is the same as that of c-AuNP, an interaction of one a-(D1)-AuNP with one 30 nm c-AuNP was achieved. When an excess of a-(D1)-AuNP to 30 nm c-AuNP (10:1) was used, satellite assemblies were formed with multiple a-(D1)-AuNPs surrounding the 30 nm c-AuNP (Figure 3a, and Figure S13). Previous reports of AuNP assembly using isotropically-labeled DNA-AuNPs often resulted in AuNPs aggregates.5i, 24 While herein the protection offered by the organic polymers on the a-DNA-AuNP made it possible to prepare monodispersed satellite assemblies.
Figure 3.
(a) a-(D1)-AuNP – 30 nm c-AuNP assemblies at a ratio of 1:1 (upper) and 10:1 (lower). (b) Hetero-assembly of 20 nm and 13 nm a-(D1)-AuNP. (c) Homo – assembly of a-(D1)-AuNP using a linker strand D3. Scale bar = 100 nm; (insets) 25 nm.
To demonstrate even better control over assemblies between two particles, instead of using one anisotropic particle and one isotropically functionalized particle, we investigated the use of only anisotropic particle for the assembly. We synthesized 20 nm a-(D1)-AuNP and 13 nm a-(D2)-AuNP functionalized respectively with D1 and D2. Mixture of the two particles at a ratio of 1 resulted in hetero-assembly of dimers consisting of a 20 nm and a 13 nm a-DNA-AuNP (Figure 3b and Figure S14), observed as snowman-like assemblies. From the TEM micrographs, we observed 85% of the assemblies formed dimers with the correct combination from a total of 47 assemblies. The remaining 15% of the assemblies were either dimers with the wrong orientation, combination or trimers and tetramers.
In addition to hetero-assembly, homo-assembly of a-(D1)-AuNP with the use of a linker strand, D3, was also investigated. The linker strand consists of the complementary sequence to D1, a single base spacer and CGCG bases at the 3′ end which can self-hybridize.25 Since the direction and area of interaction is controlled in the a-(D1)-AuNP, we expect the formation of clusters of particles in the presence of D3. Figure 3c and Figure S15 shows Au nanoclusters consisting of 3, 4 or 5 AuNPs. However, without such directional control, aggregations of the particles will occur.5a
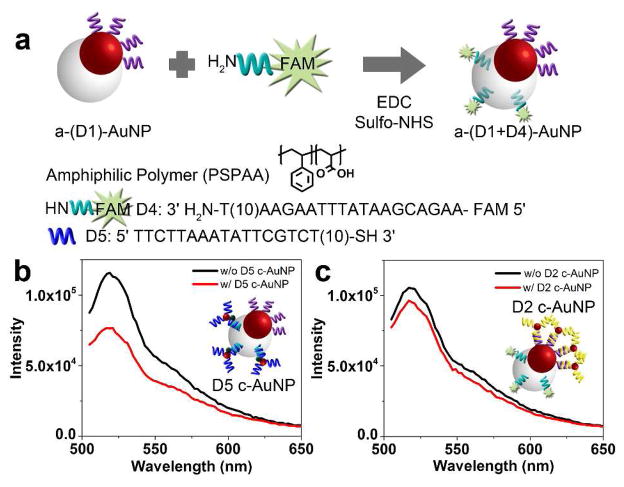
In addition to regioselective functionalization of DNA onto the asymmetric hybrid particles and its use in selective and directional assemblies, a major advantage of the a-DNA-AuNPs is that they can provide two different functionalities with spatial separation preventing crosstalk or interference. Since this particle is an anisotropic gold-polymer hybrid particle, we can further introduce a second functionality through the polymer. The polymer is an amphiphilic polymer (polystyrene-b-polyacrylic acid). The carboxylic acid chain on the polyacrylic acid tail allows functionalization via 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) coupling chemistry with amine-based substrates. To test the localization of functionalities, we attached amine functionalized DNA with a fluorophore (FAM) label (D4) on the polymer side of a-(D1)-AuNP giving a-(D1+D4)-AuNP (Figure 4a). The yield of DNA coupling was calculated to be ~ 30 DNA strands per particle. We then conjugated the a-(D1+D4)-AuNP to two different c-AuNPs, one functionalized with D2 and the other with D5. If the D1 and D4 functionalities are segregated, hybridization with D5 c-AuNP will selectively quench the fluorescence from D4, but D2 c-AuNP will not quench the fluorescence from D4. Our fluorescence results in Figure 4b and c strongly support that the D1 and D4 are functionalized respectively on the Au and polymer surface respectively.
Figure 4.
(a) Scheme showing functionalization of D4 on the polymer side of the particle via EDC coupling. Fluorescence spectra of a-(D1+D4)-AuNP before and after incubation with (b) D5 c-AuNP and (c) D2 c-AuNP.
In Figure 4b, the quenching of the fluorescence signal was observed to be incomplete. Since the a-(D1+D4)-AuNP were washed via centrifugation at least 5 times and the fluorescence signal of the supernatant solution was found to be insignificant and similar to that of the buffer solution, we conclude that the fluorescent signal is not due to free FAM-DNA. The same centrifugations should also be able to remove free polymer micelles, which are small (10 nm for amphiphilic DNA block polymers DNA-b-PPO (MW 13 kDa) and 15 nm for amphiphilic PSPAA (MW 19 kDa).26 Since the quenching efficiency of AuNP is known to be highly distance dependent,27 the incomplete quenching of the fluorescence signal is a result of relatively long distance between the fluorophore and the AuNPs.
In summary, we have demonstrated a facile method to prepare anisotropic DNA-functionalized AuNPs with very high yield. The method can be applied to prepare anisotropic DNA-functionalized gold nanoparticles with different sizes and a variety of DNA sequences in high density. In addition, the DNA on the a-DNA-AuNP can be readily exchanged with another DNA of difference sequences, allowing more sequence-specific control of nanoparticle assembly. The role of DNA in this ligand competition method serves as a unique bulky ligand which allows efficient post-functionalization on the gold surface. Taking advantage of this property, we have shown that the combination of a-DNA-AuNP and c-AuNP results in cat-paw and satellite flower assemblies. Furthermore, a-DNA-AuNP functionalized with different DNA strands were used to demonstrate hetero- and homo-assemblies, all with high regioselectivity and monodispersity. More importantly, the anisotropic nature of our a-DNA-AuNP allows us to regioselectively functionalize two different DNA strands and to demonstrate the localized quenching reactions. These asymmetric colloidal particles which are easily synthesized in high yields and versatile to be functionalized on both sides with desired DNA sequence or components which may interfere with each other can have great potential in biological applications for precise and specific targeting and response.28 In addition, with dual functionality and directional hybridization, these anisotropic particles are promising candidate for creating optically active nano-assemblies not achievable by isotropic particles.29
Supplementary Material
Acknowledgments
This work has been supported by the US National Science Foundation (CMMI 0749028). L. H. Tan was funded at UIUC from NIH National Cancer Institute Alliance for Nanotechnology in Cancer ‘Midwest Cancer Nanotechnology Training Center’ Grant R25 CA154015A. Transmission Electron Microscopy was carried out in part in the Frederick Seitz Materials Research Laboratory Central Facilities, University of Illinois.
Footnotes
Notes
The authors declare no competing financial interests.
Experimental procedures and additional TEM images can be found in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]; (b) Sheikholeslami S, Jun Y-w, Jain PK, Alivisatos AP. Nano Lett. 2010;10:2655–2660. doi: 10.1021/nl101380f. [DOI] [PubMed] [Google Scholar]; (c) Yan W, Xu L, Xu C, Ma W, Kuang H, Wang L, Kotov NA. J Am Chem Soc. 2012;134:15114–15121. doi: 10.1021/ja3066336. [DOI] [PubMed] [Google Scholar]
- 2.(a) Lee JH, Wernette DP, Yigit MV, Liu J, Wang Z, Lu Y. Angew Chem Int Ed. 2007;46:9006–9010. doi: 10.1002/anie.200702569. [DOI] [PubMed] [Google Scholar]; (b) Deng Z, Tian Y, Lee SH, Ribbe AE, Mao C. Angew Chem Int Ed. 2005;44:3582–3585. doi: 10.1002/anie.200463096. [DOI] [PubMed] [Google Scholar]; (c) Ding B, Deng Z, Yan H, Cabrini S, Zuckermann RN, Bokor J. J Am Chem Soc. 2010;132:3248–3249. doi: 10.1021/ja9101198. [DOI] [PubMed] [Google Scholar]; (d) Zhao Y, Xu L, Liz-Marzán LM, Kuang H, Ma W, Asenjo-García A, García de Abajo FJ, Kotov NA, Wang L, Xu C. J Phys Chem Lett. 2013;4:641–647. doi: 10.1021/jz400045s. [DOI] [PubMed] [Google Scholar]
- 3.(a) Aldaye FA, Sleiman HF. Angew Chem Int Ed. 2006;45:2204–2209. doi: 10.1002/anie.200502481. [DOI] [PubMed] [Google Scholar]; (b) Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sharma J, Chhabra R, Cheng A, Brownell J, Liu Y, Yan H. Science. 2009;323:112–116. doi: 10.1126/science.1165831. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Tan SJ, Campolongo MJ, Luo D, Cheng W. Nat Nanotechnol. 2011;6:268–276. doi: 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- 4.(a) Nykypanchuk D, Maye MM, van der Lelie D, Gang O. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]; (b) Maye MM, Kumara MT, Nykypanchuk D, Sherman WB, Gang O. Nat Nano. 2010;5:116–120. doi: 10.1038/nnano.2009.378. [DOI] [PubMed] [Google Scholar]; (c) Macfarlane RJ, Lee B, Jones MR, Harris N, Schatz GC, Mirkin CA. Science. 2011;334:204–208. doi: 10.1126/science.1210493. [DOI] [PubMed] [Google Scholar]; (d) Xu L, Ma W, Wang L, Xu C, Kuang H, Kotov NA. Chem Soc Rev. 2013;42:3114–3126. doi: 10.1039/c3cs35460a. [DOI] [PubMed] [Google Scholar]
- 5.(a) Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]; (b) Katz E, Willner I. Angew Chem Int Ed. 2004;43:6042–6108. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]; (c) Lau PS, Li YF. Curr Org Chem. 2011;15:557–575. [Google Scholar]; (d) Tikhomirov G, Hoogland S, Lee PE, Fischer A, Sargent EH, Kelley SO. Nat Nano. 2011;6:485–490. doi: 10.1038/nnano.2011.100. [DOI] [PubMed] [Google Scholar]; (e) Du J, Jiang L, Shao Q, Liu X, Marks RS, Ma J, Chen X. Small. 2013;9:1467–1481. doi: 10.1002/smll.201200811. [DOI] [PubMed] [Google Scholar]; (f) Kumar A, Hwang JH, Kumar S, Nam JM. Chem Commun. 2013;49:2597–2609. doi: 10.1039/c2cc37536b. [DOI] [PubMed] [Google Scholar]; (g) Barrow SJ, Funston AM, Wei X, Mulvaney P. Nano Today. 2013;8:138–167. [Google Scholar]; (h) Xing H, Wong NY, Xiang Y, Lu Y. Curr Opin Chem Biol. 2012;16:429–435. doi: 10.1016/j.cbpa.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Wang Z, Lu Y. J Mater Chem. 2009;19:1788–1798. doi: 10.1039/B813939C. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Lu Y, Liu J. Acc Chem Res. 2007;40:315–323. doi: 10.1021/ar600053g. [DOI] [PubMed] [Google Scholar]
- 6.(a) Aldaye FA, Sleiman HF. J Am Chem Soc. 2007;129:4130–4131. doi: 10.1021/ja070017i. [DOI] [PubMed] [Google Scholar]; (b) Hu SH, Gao X. J Am Chem Soc. 2010;132:7234–7237. doi: 10.1021/ja102489q. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Xu L, Kuang H, Xu C, Ma W, Wang L, Kotov NA. J Am Chem Soc. 2011;134:1699–1709. doi: 10.1021/ja2088713. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Yan J, Bloom M, Bae SC, Luijten E, Granick S. Nature. 2012;491:578–581. doi: 10.1038/nature11619. [DOI] [PubMed] [Google Scholar]
- 7.(a) Wang F, Cheng S, Bao Z, Wang J. Angew Chem Int Ed. 2013;52:10344–10348. doi: 10.1002/anie.201304364. [DOI] [PubMed] [Google Scholar]; (b) Suzuki K, Hosokawa K, Maeda M. J Am Chem Soc. 2009;131:7518–7519. doi: 10.1021/ja9011386. [DOI] [PubMed] [Google Scholar]; (c) Du J, O’Reilly RK. Chem Soc Rev. 2011;40:2402–2416. doi: 10.1039/c0cs00216j. [DOI] [PubMed] [Google Scholar]; (d) Zhang S, Li Z, Samarajeewa S, Sun G, Yang C, Wooley KL. J Am Chem Soc. 2011;133:11046–11049. doi: 10.1021/ja203133h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wang Y, Wang Y, Breed DR, Manoharan VN, Feng L, Hollingsworth AD, Weck M, Pine DJ. Nature. 2012;491:51–55. doi: 10.1038/nature11564. [DOI] [PubMed] [Google Scholar]
- 8.(a) Xu X, Rosi NL, Wang Y, Huo F, Mirkin CA. J Am Chem Soc. 2006;128:9286–9287. doi: 10.1021/ja061980b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sardar R, Heap TB, Shumaker-Parry JS. J Am Chem Soc. 2007;129:5356–5357. doi: 10.1021/ja070933w. [DOI] [PubMed] [Google Scholar]; (c) Claridge SA, Liang HW, Basu SR, Fréchet JMJ, Alivisatos AP. Nano Lett. 2008;8:1202–1206. doi: 10.1021/nl0802032. [DOI] [PubMed] [Google Scholar]; (d) Mastroianni AJ, Claridge SA, Alivisatos AP. J Am Chem Soc. 2009;131:8455–8459. doi: 10.1021/ja808570g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Maye MM, Nykypanchuk D, Cuisinier M, van der Lelie D, Gang O. Nat Mater. 2009;8:388–391. doi: 10.1038/nmat2421. [DOI] [PubMed] [Google Scholar]; (f) Yoon JH, Lim J, Yoon S. ACS Nano. 2012;6:7199–7208. doi: 10.1021/nn302264f. [DOI] [PubMed] [Google Scholar]
- 9.Fan Z, Govorov AO. Nano Lett. 2010;10:2580–2587. doi: 10.1021/nl101231b. [DOI] [PubMed] [Google Scholar]
- 10.Gansel JK, Thiel M, Rill MS, Decker M, Bade K, Saile V, von Freymann G, Linden S, Wegener M. Science. 2009;325:1513–1515. doi: 10.1126/science.1177031. [DOI] [PubMed] [Google Scholar]
- 11.(a) Zhang S, Park YS, Li J, Lu X, Zhang W, Zhang X. Phys Rev Lett. 2009;102:023901. doi: 10.1103/PhysRevLett.102.023901. [DOI] [PubMed] [Google Scholar]; (b) Lilly GD, Agarwal A, Srivastava S, Kotov NA. Small. 2011;7:2004–2009. doi: 10.1002/smll.201100536. [DOI] [PubMed] [Google Scholar]
- 12.Hentschel M, Schäferling M, Weiss T, Liu N, Giessen H. Nano Lett. 2012;12:2542–2547. doi: 10.1021/nl300769x. [DOI] [PubMed] [Google Scholar]
- 13.(a) Abdulrahman NA, Fan Z, Tonooka T, Kelly SM, Gadegaard N, Hendry E, Govorov AO, Kadodwala M. Nano Lett. 2012;12:977–983. doi: 10.1021/nl204055r. [DOI] [PubMed] [Google Scholar]; (b) Hendry E, Carpy T, Johnston J, Popland M, Mikhaylovskiy RV, Lapthorn AJ, Kelly SM, Barron LD, Gadegaard N, Kadodwala M. Nat Nano. 2010;5:783–787. doi: 10.1038/nnano.2010.209. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Bian A, Agarwal A, Liu L, Shen H, Wang L, Xu C, Kotov NA. Nano Lett. 2009;9:2153–2159. doi: 10.1021/nl900726s. [DOI] [PubMed] [Google Scholar]
- 15.(a) Perro A, Reculusa S, Ravaine S, Bourgeat-Lami E, Duguet E. J Mater Chem. 2005;15:3745–3760. [Google Scholar]; (b) Walther A, Muller AHE. Soft Matter. 2008;4:663–668. doi: 10.1039/b718131k. [DOI] [PubMed] [Google Scholar]; (c) Jiang S, Chen Q, Tripathy M, Luijten E, Schweizer KS, Granick S. Adv Mater. 2010;22:1060–1071. doi: 10.1002/adma.200904094. [DOI] [PubMed] [Google Scholar]; (d) Lee JH, Kim GH, Nam JM. J Am Chem Soc. 2012;134:5456–5459. doi: 10.1021/ja2121525. [DOI] [PubMed] [Google Scholar]
- 16.(a) Ohnuma A, Cho EC, Camargo PHC, Au L, Ohtani B, Xia Y. J Am Chem Soc. 2009;131:1352–1353. doi: 10.1021/ja8079934. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chen T, Chen G, Xing S, Wu T, Chen H. Chem Mater. 2010;22:3826–3828. [Google Scholar]; (c) Li Z, Cheng E, Huang W, Zhang T, Yang Z, Liu D, Tang Z. J Am Chem Soc. 2011;133:15284–15287. doi: 10.1021/ja205712a. [DOI] [PubMed] [Google Scholar]; (d) Gröschel AH, Walther A, Löbling TI, Schmelz J, Hanisch A, Schmalz H, Müller AHE. J Am Chem Soc. 2012;134:13850–13860. doi: 10.1021/ja305903u. [DOI] [PubMed] [Google Scholar]; (e) He J, Perez MT, Zhang P, Liu Y, Babu T, Gong J, Nie Z. J Am Chem Soc. 2012;134:3639–3642. doi: 10.1021/ja210844h. [DOI] [PubMed] [Google Scholar]
- 17.(a) Loweth CJ, Caldwell WB, Peng X, Alivisatos AP, Schultz PG. Angew Chem Int Ed. 1999;38:1808–1812. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1808::AID-ANIE1808>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]; (b) Worden JG, Shaffer AW, Huo Q. Chem Commun (Cambridge, U K) 2004:518–519. doi: 10.1039/b312819a. [DOI] [PubMed] [Google Scholar]; (c) Sung KM, Mosley DW, Peelle BR, Zhang S, Jacobson JM. J Am Chem Soc. 2004;126:5064–5065. doi: 10.1021/ja049578p. [DOI] [PubMed] [Google Scholar]; (d) Xing H, Wang Z, Xu Z, Wong NY, Xiang Y, Liu GL, Lu Y. ACS Nano. 2011;6:802–809. doi: 10.1021/nn2042797. [DOI] [PubMed] [Google Scholar]
- 18.Chen T, Yang M, Wang X, Tan LH, Chen H. J Am Chem Soc. 2008;130:11858–11859. doi: 10.1021/ja8040288. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Chen L, Feng Y, Chen H. Acc Chem Res. 2013;46:1636–1646. doi: 10.1021/ar400020j. [DOI] [PubMed] [Google Scholar]
- 20.(a) Hostetler MJ, Templeton AC, Murray RW. Langmuir. 1999;15:3782–3789. [Google Scholar]; (b) Guo R, Song Y, Wang G, Murray RW. J Am Chem Soc. 2005;127:2752–2757. doi: 10.1021/ja044638c. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Servos MR, Liu J. J Am Chem Soc. 2012;134:7266–7269. doi: 10.1021/ja3014055. [DOI] [PubMed] [Google Scholar]
- 22.Hill HD, Millstone JE, Banholzer MJ, Mirkin CA. ACS Nano. 2009;3:418–424. doi: 10.1021/nn800726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Lu Y. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 25.Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 26.Alemdaroglu FE, Alemdaroglu NC, Langguth P, Herrmann A. Adv Mater. 2008;20:899–902. [Google Scholar]
- 27.Reineck P, Gómez D, Ng SH, Karg M, Bell T, Mulvaney P, Bach U. ACS Nano. 2013;7:6636–6648. doi: 10.1021/nn401775e. [DOI] [PubMed] [Google Scholar]
- 28.(a) Verma A, Uzun O, Hu Y, Hu Y, Han HS, Watson N, Chen S, Irvine DJ, Stellacci F. Nat Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wu LY, Ross BM, Hong S, Lee LP. Small. 2010;6:503–507. doi: 10.1002/smll.200901604. [DOI] [PubMed] [Google Scholar]; (c) Walther A, Müller AHE. Chem Rev. 2013;113:5194–5261. doi: 10.1021/cr300089t. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, Whitmer JK, Jiang S, Bae SC, Luijten E, Granick S. Science. 2011;331:199–202. doi: 10.1126/science.1197451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.