Abstract
Although the inflammation-associated cytokine Interleukin-6 (IL-6) has been implicated in cholangiocarcinoma growth, the relationship between IL-6 and oncogenic changes is unknown. IL-6 can increase expression of DNA methyltransferase 1 (DNMT-1) and epigenetically regulate the expression of several genes, including microRNAs (miRNAs). DNMT-1 up-regulation occurs in hepatobiliary cancers and is associated with a poor prognosis. To understand the potential regulation of DNMT-1 by IL-6 dependent miRNAs, we examined the expression of a group of miRNAs which have sequence complementarity to the 3′-UTR of DNMT-1, namely miR-148a, miR-152 and miR-301. The expression of these miRNAs was decreased in cholangiocarcinoma cells. Moreover, the expression of all three miRNAs was decreased in IL-6 over-expressing malignant cholangiocytes in vitro and in tumor cell xenografts. There was a concomitant decrease in expression of the methylation-sensitive tumor suppressor genes Rassf1a, and p16INK4a. Using luciferase reporter constructs, DNMT-1 was verified as a target for miR-148a and miR-152. Precursors to miR-148a and miR-152 decreased DNMT-1 protein expression, increased Rassf1a and p16INK4a expression and reduced cell proliferation.
Conclusion
These data indicate that IL-6 can regulate the activity of DNMT-1 and expression of methylation-dependent tumor suppressor genes by modulation of miR-148a and miR-152, and provide a link between this inflammation-associated cytokines and oncogenesis in cholangiocarcinoma.
Keywords: Cholangiocarcinoma, DNMT, Interleukin 6, microRNA
Cholangiocarcinomas are primary malignancies of the biliary tract epithelia that are typically associated with chronic inflammation (1). The inflammation-associated cytokine interleukin-6 (IL-6) has been identified as contributing to the abnormal growth and survival of malignant cholangiocytes via an autocrine-paracrine mechanism (2-4). However, the precise role of IL-6 in cholangiocarcinogenesis has not been fully characterized yet. Recent studies provide evidence for the involvement of epigenetic modifications of critical genes in mediating the effects of IL-6. IL-6 can increase overall methylation activity with the suppression of key regulatory onco-suppressor genes (5).
We and others have shown that IL-6 can increase DNA methyltransferase-1 (DNMT-1), the most abundant methyltransferase in mammalian cells that play a key role in the maintenance of DNA methylation (5, 6). Although DNMT-1 is considerably more active on hemimethylated DNA as compared with unmethylated substrate in vitro, it is active also in de novo methylation similar to other DNMTs (7). Enforced expression of IL-6 in cholangiocarcinoma increases the expression of DNMT-1 and increases overall methylation activity (6, 8, 9). The modulation of methyltransferases, such as DNMT-1, provides an attractive mechanism by which IL-6 can restore and maintain methylation of critical genes. Thus, elucidating mechanisms by which IL-6 modulates DNMT-1 expression can provide insights into the role of IL-6 in the pathogenesis of cholangiocarcinoma.
We have recently shown that IL-6 contributes to tumor growth by modulation of expression of selected microRNAs (miRNAs) (6). miRNAs are important mediators of post-transcriptional regulation of mRNA expression and have been shown to modulate the expression of DNMT-3a and DNMT-3b, “de novo” methyltransferases involved in methylation of DNA during early development (10, 11). In contrast, the modulation of DNMT-1, which is involved in maintenance methylation, is unknown. Several tumor suppressor genes such as Rassf1a and p16INK4 have been shown to be modulated by promoter hypermethylation in cholangiocarcinoma (12-15). Thus, we sought to evaluate the potential role of IL-6 mediated changes in microRNA expression as a mechanism of modulation of DNMT-1 expression, and subsequently methylation-dependent regulation of oncogene or tumor suppressor gene expression in cholangiocarcinoma.
EXPERIMENTAL PROCEDURES
Cell lines and cultures
KMCH-1, Mz-ChA-1, and TFK-1 human cholangiocarcinoma cell lines and the nonmalignant human cholangiocyte H69 cell line were obtained as previously described (16). Mz-ChA-1 cells are derived from metastatic gallbladder cancer, TFK-1 cells from common bile duct cancer, and KMCH-1 from an intrahepatic mixed cholangiocellular-hepatocellular carcinoma. H69 cells are derived from nonmalignant cholangiocytes and immortalized by SV40 transfection. Mz-ChA-1 and TFK-1 cells were cultured in CMRL 1066 medium with 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% antimycotic antibiotic mix. H69 and KMCH-1 cells were cultured in Dulbecco’s modified Eagle medium (DMEM)/F-12 as previously described (16). All other cell culture media and supplements were obtained from Invitrogen (Carlsbad, CA) For methylation specific activation or inhibition studies, Mz-ChA-1 and KMCH-1 malignant cholangiocytes were stably transfected with full-length IL-6 to generate cell lines that over-expressed IL-6 (Mz-IL-6 and KM-IL-6) as previously described (3). To assess 5-aza-CdR methylation inhibitory effects, cells were grown to 75% confluency on 100 mm culture dishes and then treated with 5 μM 5-aza-CdR or diluent (acetic acid) control for 24 h at 37°C. Following treatment, cells were washed twice with cold phosphate-buffered saline (PBS) before harvesting for isolation of total RNA or protein.
Transfections
Transfections were performed by electroporation using the Nucleofector system (Amaxa Biosystems, Koln, Germany). All studies were performed in quadruplicate. Cells (1-2 × 106) were spun down at 1000 rpm for 5 minutes, and the medium was removed. Cells were then resuspended in 100 μL Nucleofector solution (Amaxa Biosystems) at room temperature followed by addition of 100 nmol/L microRNA precursor or controls (all obtained from Ambion Inc., Austin, TX). Electroporation was performed using the Nucleofector solution (Amaxa Biosystems). Transfected cells were then resuspended in regular culture medium containing 10% serum for 48-72 hours prior to study.
microRNA expression analysis
Total RNA was obtained from cell lines and tissue samples using the Totally RNA isolation kit (Ambion). The miRNA fraction was obtained using the flashPAGE Fractionator System (Ambion) as previously described (17). The size of the miRNA fractions were verified using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA). The isolated miRNA from the pooled sample were appended 3′ amine-modified tails using the mirVana miRNA Array Labeling Kit (Ambion) and then fluorescently coupled with Cy3 or Cy5 using the Post Labeling Reactive Dye kit (Amersham Bioscience, Pittsburgh, PA). miRNA arrays were generated and analyzed as previously described (6).
Real-Time PCR Assays for Mature miRNAs
Total RNA was isolated as described previously and the expression of specific mature miRNAs was confirmed using real-time PCR analysis using a TaqMan Human MicroRNA Assay kit (Applied Biosystems, Foster City, CA).
mRNA expression analysis
Microarray analysis was performed using Affymetrix U133 plus 2 chips (Affymetrix, Santa Clara, CA, USA) as previously described (6). In contrast to the former analysis, the outputs of each array were normalized by multiplying by a factor to obtain a robust average target intensity arbitrarily set at 100. Normalized values were then exported and analyzed with GeneSpring software (Silicon Genetics, Redwood City, CA) and Matlab 7 (Math Works, Inc., MA). As a complement, the data set was also analyzed by quantile normalization and a Robust MultiArray Analysis. Gene expression was expressed as a log 2 ratio of expression relative to that of α-tubulin.
Cell proliferation
Cell proliferation was assessed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI), which uses a tetrazolium compound as substrate. Following transfection, cells (10,000/well) were plated in 96-well plates (BD Biosciences, Rockville, MD) and incubated at 37°C, and cell proliferation was assessed after 24 hours.
Luciferase reporter vector
The consensus recognition sequence shared between miR-148, miR-152, miR-301 and the 3′-UTR of DNMT-1 was cloned downstream of the firefly luciferase gene as follows. Total cDNA was obtained by reverse transcription using random primers. The 3′-UTR of DNMT-1 was amplified using the following primers DNMT-1-3′UTR-F: AGGACTAGTTCTGCCCTCCC and DNMT1-3′UTR-R: GCGAAGCTTGGTTTATAGGAGAGATTT. The product was then digested with SpeI and HindIII and cloned into a pMIR-REPORT vector (Ambion, Austin, TX) to generate the DNMT1-WT reporter construct. A reporter construct with random mutations within the putative shared recognition sequence was also constructed (DNMT1-MUT). Site directed mutagenesis was performed by PCR using the primers: DNMT-1-3′UTR-sense ggcaccaggaa-tccccaacTAAATctgatgttgtg and DNMT-1-3′UTR-antisense cacaacatcagATTTAgttggggattcctggtgcc. PCR was performed for 20 cycles and using Pfu DNA polymerase as follows: 95 °C for 4 min; 95 °C for 30 sec, 57 °C for 30sec, 68 °C for 7 min. 5 μl of the reaction was treated with DpnI for 1 hour at 37 °C prior to transformation. All constructs were verified by sequencing. Cells were co-transfected with 2 μg DNMT1-WT or DNMT1-MUT construct and 2 μg pRL-TK Renilla luciferase expression construct followed by a precursor miRNA at 100 nM final concentration using TransIT-LT1 and TransIT-TKO reagents (Mirus, Madison, WI) for DNA vectors and precursor miRNA respectively. Luciferase assays were performed after 48 h using the Dual Glo Assay system (Promega, Madison, WI) and a multiwell plate luminometer (Veritas, Turner Biosystems, Sunnyvale CA).
Western Blotting
Cells grown in 100-mm culture dishes were washed twice with ice-cold PBS and then lyzed by incubation for 20 minutes in 1 mL of ice-cold cell lysis buffer (Cell Signaling Inc., Beverly, MA). For analysis of xenograft tumor tissue, the tissue was homogenized, and lysates were obtained. The protein concentration in the lysates was measured using a Bradford protein assay kit (Bio-Rad, Hercules, CA). Equivalent amounts of protein were mixed with 4× SDS-PAGE sample buffer, electrophoresed in a 4%-12% linear gradient Tris-HCl-ready gel (Bio-Rad), and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline, pH 7.4, containing 0.05% Tween 20, and were incubated with primary antibodies and IRDye700 and IRDye800-labelled secondary antibodies (Rockland, Gilbertsville, PA). The protein of interest was visualized and quantitated using the LI-COR Odyssey Infrared Imaging System (LI-COR Bioscience, Lincoln, NE).
Tumor cell xenograft studies
Eight-week-old male athymic nu/nu mice were obtained from Charles River Laboratories (Wilmington, MA) and fed food and water ad libitum. The mice were maintained in accordance with the Institutional Animal Care and Use Committee procedures and guidelines. They were housed 3 or 4 per cage, and fluorescent light was controlled to provide alternate light and dark cycles of 12 hours each. Mz-IL-6 or Mz-1 control cells (5 × 106 cells) were suspended in 0.25 mL of extracellular matrix gel, and the mixture was injected subcutaneously into the right and left flanks. Xenograft growth was monitored by serial measurements. For analysis of protein expression in vivo, the xenografts were excised once visible tumors had formed, and tissue was homogenized. An aliquot of the lysates was used for protein expression studies.
Reagents
Pre-miR miRNA precursors of pre-miR-148a, pre-miR-152, pre-miR-301 and control pre-miR-precursor were purchased from Ambion (Austin, TX). Antibodies against DNMT-1 and p16INK4a were obtained from Santa Cruz Biotechnology Inc.; Rassf1A from Abcam (Cambridge, MA); and α-tubulin from Sigma (St. Louis, MO).
RESULTS
Can IL-6 alter the expression of tumor suppressor genes regulated by DNMT1?
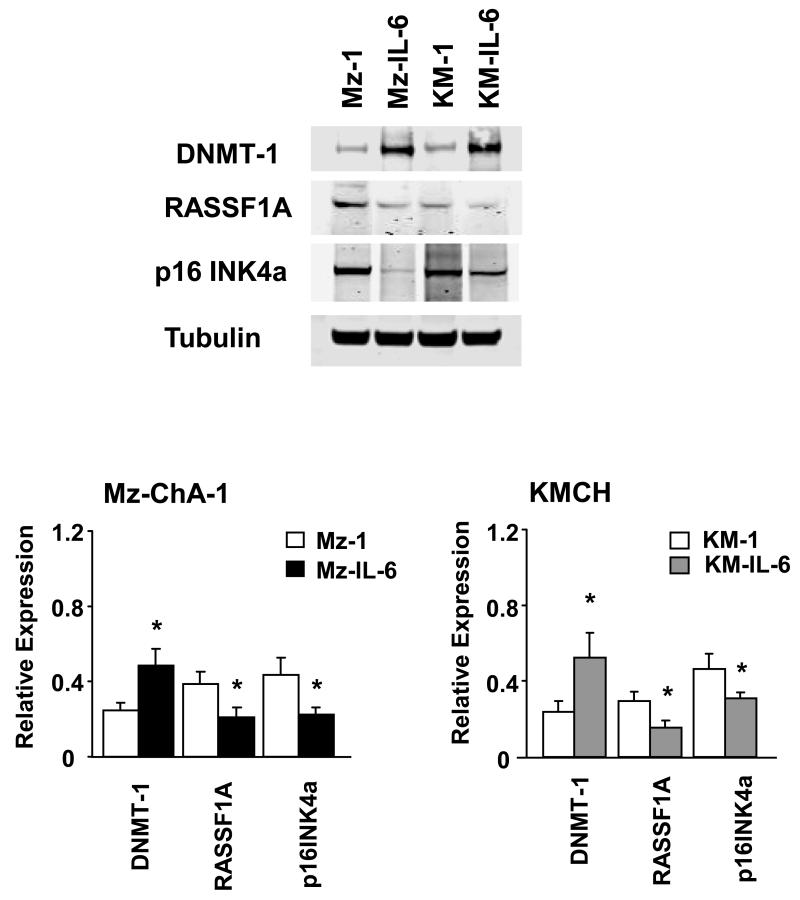
To determine whether over-expression of IL-6 can modulate DNMT-1 dependent gene expression, we generated Mz-IL-6 and KM-IL-6 cell lines by stable transfection of full length IL-6 in malignant human cholangiocyte cell lines as previously described (3). Genome-wide microarray enhanced analysis revealed an increased expression of DNMT-1 in Mz-IL-6 cells compared to control Mz-1 cells (6). The expression of DNMT-1 protein was increased to 199% ± 25% in Mz-IL-6 and to 220% ± 31 % in KM-IL-6 cells when compared to their respective controls (Figure 1). Furthermore, several tumor suppressor genes that have been shown to be frequently methylated in human cholangiocarcinoma such as Rassf1a, p16INK4a, APC and MGMT were also noted to be significantly down-regulated in IL-6 over-expressing cells (Table 1). Rassf1a is the most frequently methylated tumor-suppressor gene in human cholangiocarcinoma and its role in tumor biology is well characterized (13). p16INK4a is a tumor suppressor usually inactivated in cholangiocarcinoma (18, 19). The expressions of Rassf1a and p16INK4a was noted by immunoblot analysis to be reduced by 38 ± 10 % and 49 ± 11 % in Mz-IL-6 and by 31 ± 7 % and 33 ± 8 % in KM-IL6 cells relative to their respective controls (Figure 1). Thus IL-6 can modulate the expression of DNMT-1 as well as the expression of methylation-dependent tumor suppressor genes. The discrepancy between the mRNA and the protein levels could result from post-transcriptional changes altering protein stability and half-life. These observations suggest that regulation of methylation-dependent tumor suppressor gene expression via effects on DNMT-1 could represent an important mechanism by which IL-6 contributes to tumorigenesis.
Figure 1. IL-6 alters DNMT-1 protein expression in malignant cholangiocytes.
Cell lysates were obtained from Mz-ChA-1 or KMCH cells stably transfected to over-express IL-6 or their respective controls. Immunoblot analysis was performed to assess DNMT-1, p16INK4a, and Rassf1a protein expression. The blots were stripped and reprobed for α-tubulin as a loading control and for quantitation. Representative immunoblots are shown along with quantitative data that show the mean ± standard error from 4 separate blots. * p < 0.05 when compared with respective controls.
Can DNMT-1 be targeted by microRNAs?
Recent studies have shown that certain methyltransferases such as DNMT-3a and DNMT-3b can be directly targeted by miRNAs (10, 11). Thus, we postulated that alterations in miRNA expression could represent a mechanism by which IL-6 induces DNMT-1 expression. To explore this possibility, we analyzed the 3′-UTR of DNMT-1 as potential target sites for miRNA binding. Interestingly, a group of microRNAs, including miR-130a, mir-130b, miR-148a, mir-148b, miR-152 and miR-301, showed sequence complementarity to the same region in the 3′-UTR of DNMT-1 gene (between positions 5135-5143). Several microRNA target prediction databases were interrogated to determine if their respective algorithms would identify DNMT-1 as a target of these miRNAs. Three databases, PicTar, TargetScan and miRBase identified DNMT-1 as a predicted target for miR-148a and miR-152, whereas one database, PicTar identified DNMT-1 as a predicted target for miR-130 and miR-301.
Is the expression of these miRNAs altered in malignant cholangiocytes?
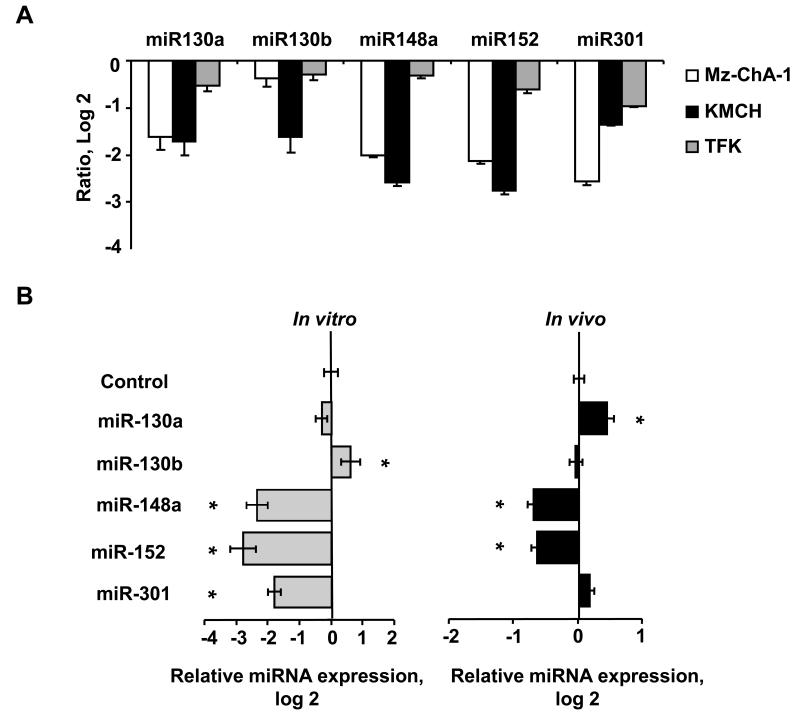
Alterations in methylation have been implicated in several malignancies, including cholangiocarcinoma. We therefore evaluated the expression of miRNAs that could potentially bind DNMT-1 in malignant and non-malignant cholangiocytes. Interestingly, the expression of all the identified miRNAs was decreased in human cholangiocarcinoma cell lines compared to non-malignant cholangiocytes (Figure 2). In Mz-ChA-1 cells, miR-148a expression was decreased to 0.25- ± 0.03-fold, and miR-152 expression was decreased to 0.23- ± 0.02-fold relative to H69 non-malignant human cholangiocytes. Similar reductions in expression were also seen in malignant KMCH and TFK cells. The reduced expression of this group of miRNAs is consistent with increased expression of DNMT-1 in cholangiocarcinoma, and suggests that this group of miRNAs may be involved in deregulation of genomic methylation in human cholangiocarcinoma. A recent study of microRNA expression in intrahepatic cholangiocarcinoma samples showed reduced expression of miR-148a and miR-152 in cholangiocytes compared to normal liver tissues (20), but these were not aberrantly expressed in malignant tissues. These may reflect differences in anatomical site of origin between these tumors and the cell lines used in our study. Notably, miR-148a expression is reduced in metastatic HCC supporting its potential as an oncosuppressor RNA gene.
Figure 2. Expression of potential DNMT-1 targeting miRNAs in cholangiocarcinoma.
miRNA was isolated and profiling performed by hybridization to miRNA-specific probes on epoxy-coated slides. Panel A: Samples from KMCH-1, Mz-ChA-1 or TFK-1 malignant cholangiocytes were labeled with Cy5 whereas samples from H69 nonmalignant cholangiocytes were labeled with Cy3. The data in the panel represent the average ± standard errors of the log2 of the relative expression (ratio of Cy5/Cy3 fluorescence intensity) for each specific miRNA. Panel B: miRNA was isolated and microarray analysis performed from IL-6-overexpressing Mz-ChA-1 cells (Mz-IL-6) and their respective control (Mz-1) cells grown in cell culture in vitro or as tumor cell xenografts in immunodeficient mice. Compared to controls, IL-6 over-expressing Mz-ChA-1 cells (Mz-IL-6) showed a reduced expression of miR-148a and miR-152 in vitro (left panel) as well as in tumor xenografts in vivo (right panel) whereas the reduction of miR-301 was only seen in vitro. Data represents the mean and standard error of the ratio Mz-IL6/Mz-1 in four determinations performed in triplicate. * p < 0.05 when compared with respective controls.
Are these miRNAs located in cancer-associated genomic regions?
Chromosomal aberrations in genomic regions encoding miRNAs could contribute to altered expression in tumors. In order to evaluate the relationship between chromosomal aberrations and miRNA expression in biliary cancers, we evaluated the frequency of chromosomal loss in the regions corresponding to miR-130a (11q12.1), miR-130b (22q11.21), miR-148a (7p15.2), miR-152 (17q21.32) and miR-301 (17q22), in intrahepatic and extrahepatic cholangiocarcinoma, using a comprehensive cytogenetic database (http://www.progenetix.de/~pgscripts/progenetix). Chromosomal losses were observed in 11% in the sites of miR-152 and miR-301 and in 22% in the site of miR-130a of extrahepatic bile ducts tumors, while no losses were detected for the location of miR-148a and miR-130b. In intrahepatic bile ducts tumors, losses in both sites of chromosome 17 were detected in 6%, while no losses were observed in the sites of miR-148a and miR-130a. The highest frequency (11.8%) of losses was observed for the site of miR-130b. Analysis of chromosomal changes in Mz-ChA-1 using a bacterial artificial chromosome array comparative genomic hybridization analysis did not show any significant changes in copy number for clones encompassing the genomic site of these miRNAs. Thus, chromosomal alterations do not account for altered expression of these microRNAs in Mz-ChA-1 cells.
Is the expression of these miRNAs altered by enforced expression of IL-6?
To determine the role of this specific group of microRNAs on IL-6 mediated DNMT-1 expression, Mz-ChA-1 human cholangiocarcinoma cells were stably transfected to overexpress IL-6 (Mz-IL-6 cells). When implanted as xenografts in athymic nude mice, the growth rate of Mz-IL-6 xenografts was increased compared with Mz-1 control cell xenografts, in conjunction with a decrease in the number of TUNEL-positive (apoptotic) cells (3). We used a miRNA microarray to assess the expression of human miRNAs in Mz-IL-6 cell lines over-expressing IL-6 and in Mz-IL-6 derived xenografts. miRNAs that were decreased with enforced IL-6 expression were identified (Figure 2). The group included miRNAs that are predicted to target DNMT-1 such as miR-148a, miR-152 and miR-301 as well as miR-370, which has been postulated as contributing to cholangiocarcinogenesis. A reduction in expression of both miR-148a and miR-152 but not miR-301 was noted in tumor cell xenografts in vivo (Figure 2). These data showing altered miRNA expression profiles in vivo suggest that some effects of IL-6 on tumor cell growth and apoptosis could be mediated by miRNA-dependent induction of methyltransferase gene expression.
Is DNMT-1 a target for miRNA silencing?
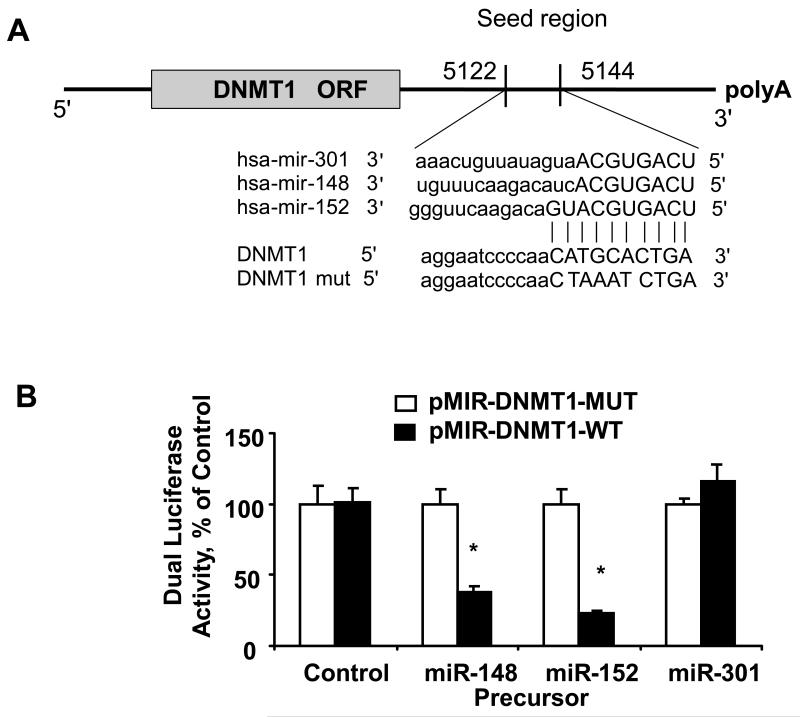
Several studies were performed to evaluate and experimentally validate DNMT-1 as a bona fide target of translational regulation by miR-148a, miR-152 and miR-301. First, we verified that the DNMT-1 3′-UTR is a target for miR-148a, miR-152 and miR-301 using luciferase reporter constructs containing the shared recognition sequence from the 3′-UTR of DNMT-1 inserted downstream of the luciferase gene (Figure 3). Transfection with precursors of miR-148a and miR-152 significantly modulated reporter activity in Mz-ChA-1 cells, whereas miR-301 precursor failed to show any effects. Next, the studies were repeated with random mutations in the shared recognition sequence, which resulted in abolition of the reporter activation by miR-148a and miR-152 precursors. Thereafter, we assessed whether ectopic expression of individual miR-148a, miR-152 and miR-301 sequences induces down-regulation of endogenous DNMT-1 protein expression. Transfection of Mz-ChA-1 and KMCH cells with either miR-148a or miR-152 decreased DNMT-1 expression after 3 days (Figure 4). Consistent with our previous observations, miR-301 did not have the same effect on DNMT-1 protein expression. Meanwhile, we did not see any significant changes of DNMT-3a and DNMT-3b in human cholangiocarcinoma cells after over-expression of these three miRNAs (Figure 4). Furthermore, transfection with a precursor to miR-370, a miRNA that is regulated in an IL-6 dependent manner but which does not have complementary sequences to the DNMT1 3′-UTR, did not significantly alter the expression of DNMT-1 with a relative expression of 0.96 ± 0.14-fold of controls. Taken together, these data confirm that DNMT-1 is a biologically relevant target of miR-148a and miR-152.
Figure 3. Targeting of DNMT-1 3′-UTR by miRNAs.
Panel A: The location of target sites of miR-148a, miR-152 and miR-301 in the DNMT-1 3′-UTR are shown. The sequence of the mutated DNMT-1 3-UTR construct with mutations to disrupt base pairing at shared putative microRNA binding sites is also displayed. Panel B: Mz-ChA-1 cells were transfected with the Renilla luciferase expression construct pRL-tk, the luciferase construct pMIR-DNMT1-WT-luc or pMIR-DNMT1-MUT-luc (in which mutations were introduced in the DNMT-1 target site) and miR-148a, miR-152, miR-301 or control precursor. After 24 h, Dual-Luciferase assays were performed. The decrease in relative firefly luciferase activity was present with miR-148a and miR-152 and the pMIR-DNMT1-WT-luc (black bars), while no effect was detected with the pMIR-DNMT1-MUT-luc construct (white bars), confirming that the 3′-UTR of DNMT-1 is a direct target of modulation by miR-148a and miR-152. The data represent the mean and standard deviations from six determinations from three independent transfections. * p < 0.05 compared to the respective controls.
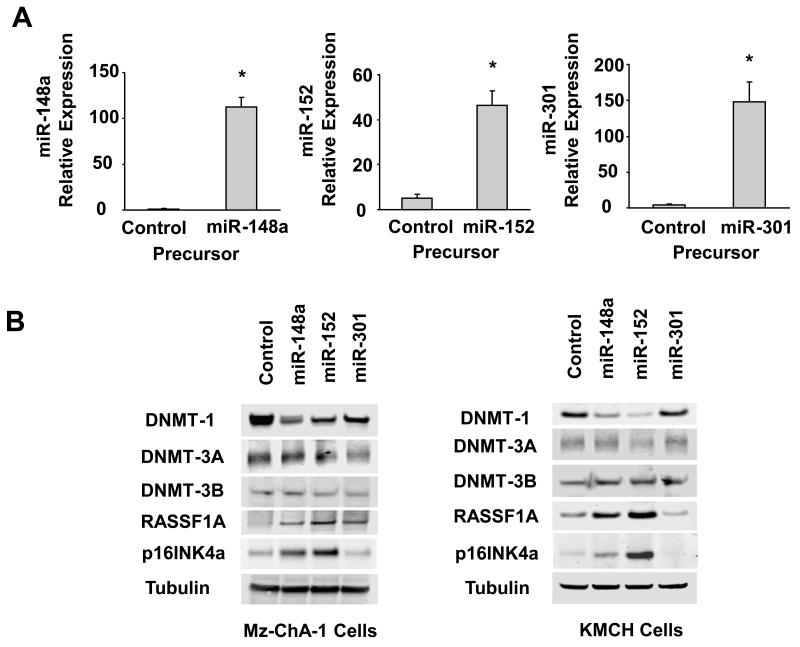
Figure 4. Increased expression of miR 148 and, miR-152 alters expression of DNMT-1 and selected tumor suppressor genes.
Panel A: Mz-ChA-1 cells were transfected with either miR-148a, miR-152, miR-301 or control precursor. After 24 h, real-time PCR assays were performed using TaqMan Human MicroRNA Assay kit and the expression of miRs was normalized to that of the small nuclear RNA U6B (RNU6B). A significant increase in microRNA expression was observed following transfection with precursor miR-148a and miR-152 compared to control miRNA precursors. The data represent the mean and standard errors from four independent transfections. * p < 0.05 relative to respective controls. Panel B: Mz-ChA-1 and KMCH cells were transfected with either miR-148a, miR-152, miR-301 or control precursor. After 72 hours, cell lysates were obtained for Western blot analysis of DNMTs, Rassf1a, and p16INK4a. The blots were stripped and re-probed for α-tubulin as a loading control.
Can miRNAs that target DNMT-1 alter tumor suppressor gene expression?
To characterize the effects of the methylation changes on tumor suppressor gene expression, we analyzed the effect of the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-aza-CdR) on the mRNA expression levels of several tumor suppressor genes implicated in cholangiocarcinogenesis. 5-aza-CdR treatment significantly decreased expression of DNMT-1 as previously shown (6). In contrast, Rassf1a and p16INK4a were significantly increased by 5-aza-CdR treatment, indicating that these genes might be regulated by DNMT-1 through promoter methylation mechanisms. Furthermore, complementary binding regions between miR-148a/miR-152 and Rassf1a/p16INK4a have not been predicted by any of the databases used, making it unlikely that these genes can be directly targeted by miR-148a or miR-152. We assessed cellular expression of Rassf1a and p16INK4a in several cholangiocarcinoma cell lines. By immunoblot analysis, Rassf1a and p16INK4a expression was decreased in all malignant cell lines compared with H69 non-malignant cholangiocytes (Figure 5). Moreover, there was a further decrease of Rassf1a and p16INK4a in IL-6 over-expressing cholangiocytes when compare to their respective controls (Figure 1). Enforced expression of miR-148a and miR-152 in Mz-ChA-1 and KMCH cells was also noted to significantly enhance Rassf1a and p16INK4a expressions concomitant with decreased DNMT-1 protein level (Figure 4).
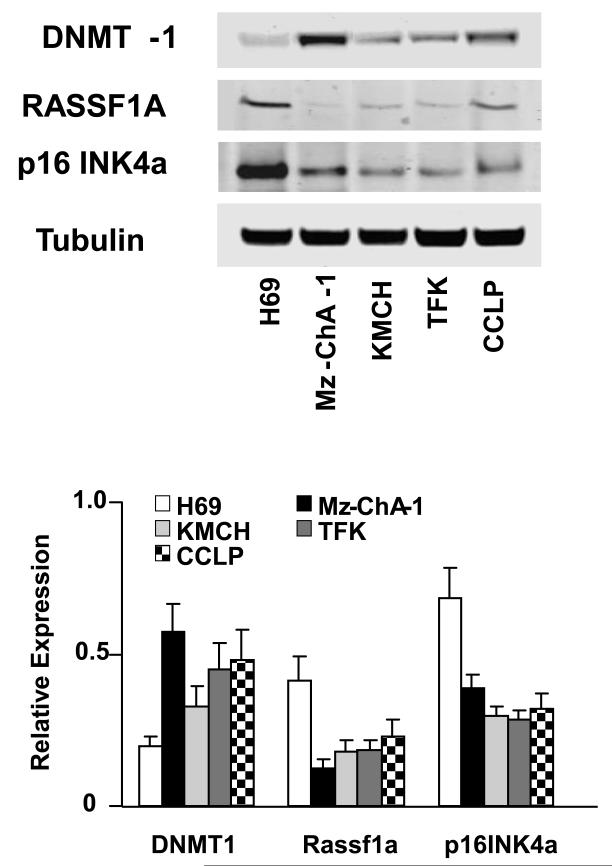
Figure 5. Expression of selected genes in normal and malignant cholangiocytes.
Cell lysates were obtained from H69 non-malignant cholangiocytes and KMCH, CC-LP, TFK and MzChA-1 cholangiocarcinoma cell lines. Immunoblot analysis was performed for DNMT-1 and for the tumor suppressor genes RASSF1a, and p16INK4a using antigen specific antibodies. The blots were stripped and reprobed for α-tubulin as a loading control and for quantitation. Representative immunoblots are shown along with quantitative data showing the mean ± standard error from 4 separate blots. For each cholangiocarcinoma cell line, DNMT expression was significantly increased and Rassf1a and p16INK4a expression significantly decreased compared with non-malignant H69 cells, with p<0.05.
Can IL-6 alter methylation dependent tumor suppressor gene expression in vivo?
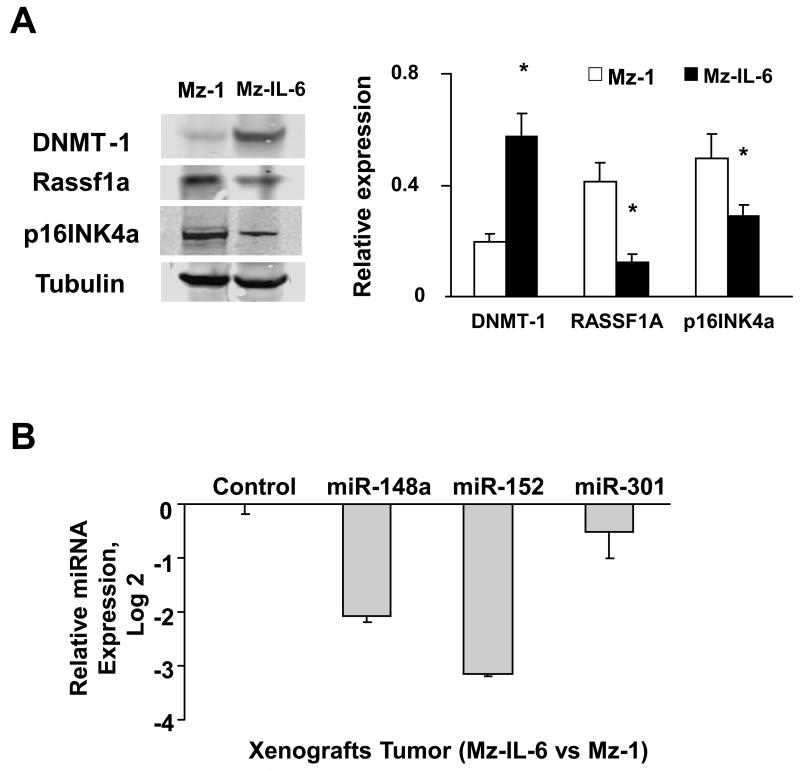
To explore the in vivo relevance of these observations, we assessed the expressions of DNMT-1, Rassf1a, p16INK4a in homogenates from xenograft tumors. The results corresponded to those observed in vitro with an increase in basal expression of DNMT-1 and decrease in Rassf1a and p16INK4a in Mz-IL-6 xenografts when compared with control cell xenografts (Figure 6). The expression of mature miR-148a and miR-152 were also confirmed to be decreased by real-time PCR in IL-6-over-expressing tumor cell xenografts in vivo when compared with controls.
Figure 6. Enforced expression of IL-6 alters expression of DNMT-1 targeting miRNA in vivo.
IL-6 over-expressing (Mz-IL-6) or control (Mz-1) cells (3×106) were injected subcutaneously into the flank of athymic BALB/c mice. Panel A: Western blot analysis was performed in tissue lysates from tumor cell xenografts using antibodies to DNMT-1, Rassf1a, p16INK4a, and α-tubulin. A representative blot image is shown on left panel whereas the densitometric analysis is shown on right panel. Panel B: miRNA was extracted from xenograft tissues and miR-148a, miR-152 and miR-301 expression was assessed by quantitative real-time PCR. Potential DNMT-1 targeting miRNA expressions were normalized to expression of RNU6B. The data represent results from three experiments performed in triplicate. *p<0.05 when compared with miRNA expression in control cell xenografts.
Can miRNAs that target DNMT-1 alter tumor cell growth?
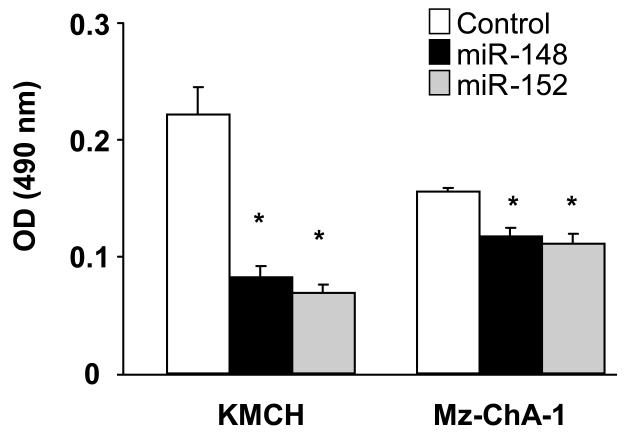
The effects of miRNAs targeting DNMT-1 on cell proliferation were further characterized. Transfection of KMCH cells with precursors of miR-148a and miR-152 decreased proliferation to 37% ± 11%, 31% ± 3% and 35% ± 4%, respectively, of controls after 24 hours (all p < 0.05). Similar effects of miR-148a and miR-152 were also seen in Mz-ChA-1 cells (Figure 7) although not to the same extent. Cell proliferation was not significantly altered (101 ± 5% of controls) by precursors to miR-17, whose expression is not altered in malignant cells. We hypothesize that aberrant expression of miR-148a and miR-152 can inhibit tumor cell growth
Figure 7. Effect of potential DNMT-1 targeting miRNAs on cell growth in cholangiocarcinoma cells.
Mz-ChA-1 and KMCH cells (5 × 104/well) were plated in 96-well plates after transfection with miRNA-specific precursors to miR-148a and miR-152 or with control precursors (100 nM). Cell proliferation was assessed after 24 hours using a viable cell assay and the proliferation index derived. The mean ± standard errors from 4 separate experiments are illustrated. *p < 0.05 when compared with controls.
DISCUSSION
The role of aberrant methylation in the pathobiology of cholangiocarcinoma is becoming increasingly recognized, but remains poorly understood. Similar to observations from many other cancers, alterations in DNA methylation can modulate the expression of specific oncogenes and tumor suppressor genes involved in cholangiocarcinoma growth (21, 22). While the focus of most studies has been on the identification of gene transcripts that are regulated by methylation, the mechanisms by which methylation itself is regulated in tumor-specific circumstances has received less attention. Environmental perturbations and enhanced expression of cytokines such as IL-6 can modulate the expression of the methyltrasferase DNMT-1 and thereby directly modulate tumorigenesis. IL-6 can alter miRNA expression, and in these studies we have shown that that these alterations can contribute to the regulation of the expression of DNMT-1 and Rassf1a / p16INK4a, emphasizing the role of aberrantly expressed non-coding RNAs as modulators of tumorigenesis in cholangiocarcinoma.
The over-expression of IL-6 in cholangiocarcinoma cells is associated with increased DNMT-1 expression, as well as with reduced expression of tumor suppressor genes such as Rassf1a and p16INK4a. Interestingly, we found that expression of miR-148a and miR-152, which were silenced in cholangiocarcinoma compared to H69 cells, were further decreased with enforced IL-6 expression both in vitro and in vivo. We further demonstrated that miR-148a and miR-152 can negatively modulate the expression of DNMT-1, by interacting with the 3′-UTR of the gene. In cells transfected with these miRNAs we observed down-regulation of endogenous DNMT-1 protein expression and decreased cell proliferation. Treatment with 5-aza-CdR, a methylation inhibitor, increased the expression of tumor suppressor genes, thus supporting the regulation by DNMT-1 through promoter methylation mechanisms.
Hypermethylation at promoter CpG islands and inactivation of multiple tumor suppressor genes are common in cholangiocarcinoma, and contribute to tumor growth (13). Indeed, the number of methylated CpG island loci in extra-hepatic cholangiocarcinoma specimens increases significantly with the stage of the disease and with the presence of node metastasis (23). DNMT-1 has a role in the establishment and regulation of tissue-specific patterns of methylated cytosine residues. DNMT-1 contributes more to maintenance of methylation than to de novo methylase activity and deregulated expression of DNMT-1 may play a causal role in cellular transformation (24).
Aberrant DNMT-1 expression has been detected in several tumors, including liver tumors (25), supporting a role for DNMT-1 in tumorigenesis. Reduced expression of specific miRNAs such as miR-148 and miR-152 can directly modulate the expression of DNMT-1 in cholangiocytes. In human cholangiocarcinoma, loss of expression of these miRNAs by genetic or environmental factors such as IL-6 over-expression can contribute to enhanced DNMT-1 level with subsequent silencing of expression of critical tumor suppressor genes through altered promoter methylation.
Reduced expression of miR-301 in malignant cholangiocytes in vitro suggests a potential role of this miRNA in tumorigenesis, albeit not one that involves modulation of DNMT-1 expression. To gain insight into potential functional effects of miR-301, we performed a gene annotation enrichment analysis of predicted target genes using the Database for Annotation, Visualization and Integrated Discovery (26). KEGG pathway mapping of the results showed enrichment of genes involved in the Wnt, Notch and TGF-β signaling pathways. These observations warrant further study.
Rassf1a has been shown to be the most frequently (65%) hypermethylated tumor suppressor gene in cholangiocarcinoma compared to normal cholangiocytes and is silenced both in extrahepatic and intrahepatic bile ducts tumors (13, 23). Rassf1a can mediate the apoptotic effects of Ras and act as a negative Ras effector, inhibiting cell growth and inducing cell death (27). Inactivation of Rassf1a can thus enhance cellular proliferative capacity. The genomic location of Rassf1a on 3p21.3 is a region which is frequently subjected to loss of heterozygosity (14, 28, 29). Within the remaining allele mutations are rare and promoter methylation has been recognized as a primary mechanism for gene inactivation. Inactivation of the p16INK4a gene is also a frequent event in human cholangiocarcinoma. The most common somatic alteration is promoter methylation of the p16INK4a gene (18). Although the specific mechanisms by which gene methylation are regulated during tumorigenesis are not well understood, we show herein that IL-6 can modulate the expression of certain genes such as Rassf1a and p16INK4a expressions by a miRNA-dependent up-regulation of DNMT-1. These observations thus provide a direct mechanism by which IL-6 could contribute to tumorigenesis.
Our studies indicate the presence of IL-6 mediated mechanisms of regulation of methylation dependent gene expression that may act in combination with other identified non -IL-6 dependent mechanism. Both de novo and maintenance methyltransferases share a common regulating mechanism via miR-148, at the same time preserving individual regulating mechanisms, for example miR-152 controls expression of DNMT-1 while miR-29 regulates DNMT-3 (10). Additionally, other unique miRNA dependent pathways may be active, and further study of potential links between enzymes involved in gene methylation or demethylation and aberrantly expressed miRNA in cancer may be fruitful.
Supplementary Material
Acknowledgment
We acknowledge with gratitude the expert assistance of Fanyin Meng, Erica Swenson, Lyudmyla Khrapenko and Melinda Freed with these studies.
Financial support: Supported by National Institutes of Health grant DK06370
Abbreviations
- IL-6
Interleukin-6
- DNMT-1
DNA methyltransferase 1
- miR, miRNA
microRNA
- p16INK4a
p16 inhibitor of CDK4
- Rassf1a
Ras association domain family member 1
- DMEM
Dulbecco’s modified Eagle medium
- MEM
minimum essential medium
- FBS
fetal bovine serum
- 5-Aza-CdR
5-Aza-2′-deoxycytidine
- PBS
phosphate-buffered saline
- APC
adenomatous polyposis coli
- MGMT
O-6-methylguanine-DNA methyltransferase
- UTR
Untranslated region
REFERENCES
- 1.Patel T. Cholangiocarcinoma. Nat Clin Pract Gastroenterol Hepatol. 2006 Jan;3(1):33–42. doi: 10.1038/ncpgasthep0389. [DOI] [PubMed] [Google Scholar]
- 2.Park J, Tadlock L, Gores GJ, Patel T. Inhibition of interleukin 6-mediated mitogen-activated protein kinase activation attenuates growth of a cholangiocarcinoma cell line. Hepatology. 1999 Nov;30(5):1128–1133. doi: 10.1002/hep.510300522. [DOI] [PubMed] [Google Scholar]
- 3.Meng F, Yamagiwa Y, Ueno Y, Patel T. Over-expression of interleukin-6 enhances cell survival and transformed cell growth in human malignant cholangiocytes. J Hepatol. 2006 Jun;44(6):1055–1065. doi: 10.1016/j.jhep.2005.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blechacz B, Gores GJ. Cholangiocarcinoma: Advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008 Mar 3;48(1):308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodge DR, Xiao W, Clausen PA, Heidecker G, Szyf M, Farrar WL. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem. 2001 Oct 26;276(43):39508–39511. doi: 10.1074/jbc.C100343200. [DOI] [PubMed] [Google Scholar]
- 6.Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008 Jan 10;27(3):378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 7.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000 Oct;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 8.Wehbe H, Henson R, Meng F, Mize-Berge J, Patel T. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res. 2006 Nov 1;66(21):10517–10524. doi: 10.1158/0008-5472.CAN-06-2130. [DOI] [PubMed] [Google Scholar]
- 9.Hodge DR, Peng B, Cherry JC, Hurt EM, Fox SD, Kelley JA, et al. Interleukin 6 supports the maintenance of p53 tumor suppressor gene promoter methylation. Cancer Res. 2005 Jun 1;65(11):4673–4682. doi: 10.1158/0008-5472.CAN-04-3589. [DOI] [PubMed] [Google Scholar]
- 10.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duursma AM, Kedde M, Schrier M, le SC, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008 May;14(5):872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Kim WH, Jung HY, Yang MH, Kang GH. Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Am J Pathol. 2002 Sep;161(3):1015–1022. doi: 10.1016/S0002-9440(10)64262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang B, House MG, Guo M, Herman JG, Clark DP. Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol. 2005 Mar;18(3):412–420. doi: 10.1038/modpathol.3800287. [DOI] [PubMed] [Google Scholar]
- 14.Chen YJ, Tang QB, Zou SQ. Inactivation of RASSF1A, the tumor suppressor gene at 3p21.3 in extrahepatic cholangiocarcinoma. World J Gastroenterol. 2005 Mar 7;11(9):1333–1338. doi: 10.3748/wjg.v11.i9.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foja S, Goldberg M, Schagdarsurengin U, Dammann R, Tannapfel A, Ballhausen WG. Promoter methylation and loss of coding exons of the fragile histidine triad (FHIT) gene in intrahepatic cholangiocarcinomas. Liver Int. 2005 Dec;25(6):1202–1208. doi: 10.1111/j.1478-3231.2005.01174.x. [DOI] [PubMed] [Google Scholar]
- 16.Marienfeld C, Tadlock L, Yamagiwa Y, Patel T. Inhibition of cholangiocarcinoma growth by tannic acid. Hepatology. 2003 May;37(5):1097–1104. doi: 10.1053/jhep.2003.50192. [DOI] [PubMed] [Google Scholar]
- 17.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006 Jun;130(7):2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 18.Tannapfel A, Benicke M, Katalinic A, Uhlmann D, Kockerling F, Hauss J, et al. Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut. 2000 Nov;47(5):721–727. doi: 10.1136/gut.47.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniai M, Higuchi H, Burgart LJ, Gores GJ. p16INK4a promoter mutations are frequent in primary sclerosing cholangitis (PSC) and PSC-associated cholangiocarcinoma. Gastroenterology. 2002 Oct;123(4):1090–1098. doi: 10.1053/gast.2002.36021. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009 Feb;50(2):358–369. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Esteller M. Epigenetics in cancer. N Engl J Med. 2008 Mar 13;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 22.Sandhu DS, Shire AM, Roberts LR. Epigenetic DNA hypermethylation in cholangiocarcinoma: potential roles in pathogenesis, diagnosis and identification of treatment targets. Liver Int. 2008 Jan;28(1):12–27. doi: 10.1111/j.1478-3231.2007.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim BH, Cho NY, Choi M, Lee S, Jang JJ, Kang GH. Methylation profiles of multiple CpG island loci in extrahepatic cholangiocarcinoma versus those of intrahepatic cholangiocarcinomas. Arch Pathol Lab Med. 2007 Jun;131(6):923–930. doi: 10.5858/2007-131-923-MPOMCI. [DOI] [PubMed] [Google Scholar]
- 24.Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992 Jul;11(7):2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh BK, Kim H, Park HJ, Shim YH, Choi J, Park C, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med. 2007 Jul;20(1):65–73. [PubMed] [Google Scholar]
- 26.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 27.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000 Jul;25(3):315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 28.Tischoff I, Markwarth A, Witzigmann H, Uhlmann D, Hauss J, Mirmohammadsadegh A, et al. Allele loss and epigenetic inactivation of 3p21.3 in malignant liver tumors. Int J Cancer. 2005 Jul 10;115(5):684–689. doi: 10.1002/ijc.20944. [DOI] [PubMed] [Google Scholar]
- 29.Wong N, Li L, Tsang K, Lai PB, To KF, Johnson PJ. Frequent loss of chromosome 3p and hypermethylation of RASSF1A in cholangiocarcinoma. J Hepatol. 2002 Nov;37(5):633–639. doi: 10.1016/s0168-8278(02)00269-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.