Summary
Bacteria that cause disease rely on their ability to counteract and overcome host defenses. Here we present a genome-scale study of Mycobacterium tuberculosis (Mtb) that uncovers the bacterial determinants of surviving host immunity, sets of genes we term “counteractomes.” Through this, we find that CD4 T cells attempt to starve Mtb of tryptophan through a mechanism that limits Chlamydia and Leishmania infections. In those cases, tryptophan starvation works well, since those pathogens are natural tryptophan auxotrophs. Mtb, however, can synthesize tryptophan, and thus starvation fails as an Mtb-killing mechanism. We then describe a small molecule inhibitor of Mtb tryptophan synthesis, which turns Mtb into a tryptophan auxotroph and restores the efficacy of a failed host defense. Together, our findings demonstrate that the Mtb determinants for surviving host immunity—Mtb’s immune counteractomes—serve as probes of host immunity, uncovering immune-mediated stresses that can be leveraged for therapeutic discovery.
Introduction
Mycobacterium tuberculosis (Mtb), the etiologic agent of tuberculosis (TB), remains one of the world’s major bacterial pathogens. After a centuries-long decline, the last few decades have seen a resurgence in TB, with an estimated 2 billion people infected and about 1.7 million deaths per year (WHO, 2011). The success of Mtb as a pathogen lies in its adaptation to the human host and its ability to counteract the many arms of antibacterial immunity (Ernst, 2012). Its ability to survive host defenses is directly responsible for the large reservoir of infected people, and its ability to subvert bactericidal mechanisms allows it to replicate in vivo and cause disease (Ehrt and Schnappinger, 2009). Elucidating these mechanisms will help us understand the complex host-pathogen interface, and targeting these mechanisms is an underutilized therapeutic strategy that can help patients’ immune systems kill Mtb.
In the multifaceted immune response against Mtb, CD4 T cells make up one of the most biologically and epidemiologically important compartments. Humans and mice generally cannot clear Mtb during infection, but both are able to limit bacterial growth, and in the case of immunocompetent humans, prevent disease (Ernst, 2012; Flynn, 2006). This response is dependent on CD4 T cells. The CD4-deficient MHC Class II knockout mice, and other mice that lack CD4 T cells, cannot stop mycobacterial growth and rapidly succumb to disease and death (Caruso et al., 1999; Cosgrove et al., 1991; Grusby et al., 1991; Mogues et al., 2001; Scanga et al., 2000). In human disease, progressive CD4 T cell loss due to HIV infection also increases the risk for TB disease and death (Macpherson et al., 2011; McDermid et al., 2013; Pawlowski et al., 2012; Selwyn et al., 1989).
However, crucial as they are for TB immunity, CD4 T cells ultimately fail to sterilize infection. The surviving bacteria remain latent, with the potential to cause disease in the future (Ernst, 2012; Flynn, 2006). The nature of the environment imposed by CD4 T cells, enough to limit growth but not kill Mtb, is not well studied. Reports have shown that the Th1 subset is especially effective in limiting Mtb growth, and cytokines such as IFN-γ and TNF-α are needed in some, but not all, models of CD4 T cell mediated defenses (Bold et al., 2011; Cooper et al., 1993; Flynn et al., 1995; Flynn et al., 1993; Gallegos et al., 2011; Nandi and Behar, 2011; Russell, 2007; Scanga et al., 2000). But the exact nature of CD4-mediated stress—the repertoire of anti-pathogen effectors induced by CD4 T cells—is poorly understood. Knowing how CD4 T cells attempt to kill Mtb and how Mtb survives could identify Mtb vulnerabilities and aid drug discovery efforts.
Here we profile the mycobacterial genetic requirements for surviving the CD4 response, the CD4 “counteractome”. We compare this with relevant in vitro survival signatures such as acid, oxidative stress and nutrient starvation, creating counteractomes for physical stresses encountered in the host. Using these, we focus on a particular bacterial pathway, tryptophan (Trp) synthesis, and find the host mechanism for inducing amino acid starvation in intracellular mycobacteria. We go on to find a small molecule that targets Trp synthesis, determine the structural and biochemical basis of its activity and show that it acts together with the host to kill Mtb both in vitro and during a model infection. Together, our findings demonstrate the utility of profiling the pathogen response to host-mediated stresses. We characterize the stresses induced by CD4 T cells, and leverage one stress, Trp starvation, to synergize with a small molecule to more effectively treat tuberculosis.
Results
Genes required for survival during infection: virulence factors
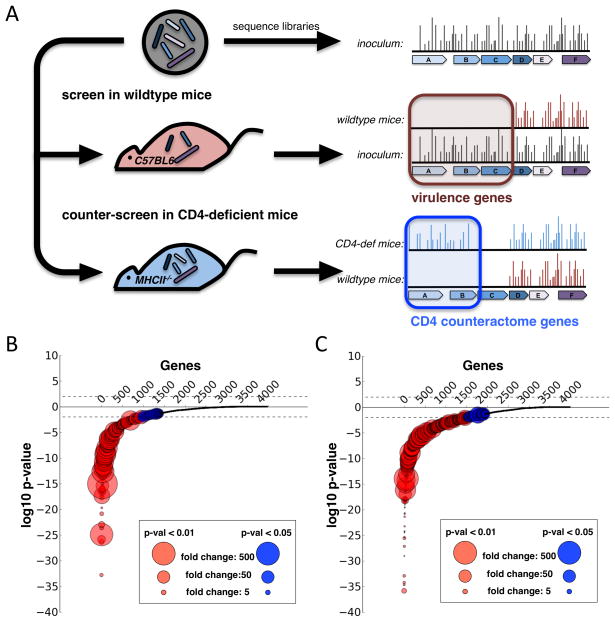
In order to define the set of Mtb genes required for surviving the CD4 T cell response, we infected both wild type and MHC Class II knockout (MHCII−/−) mice with a library of Mtb transposon mutants (Zhang et al., 2012). We injected 106 bacteria intravenously and plated surviving bacteria from infected spleens at 10 days and 45 days after infection (Figure 1A). To identify mutant Mtb in the surviving pools, we deep sequenced transposon junctions to map the insertion site of each mutant. For each time point, we made two comparisons. First, we compared the surviving pool of mutants from wild type mice (wt output library) to the inoculating pool (input library), defining genes with a statistically validated decrease in output library as required for growth during infection, commonly known as virulence factors (Figure 1A). Second, we compared the output library from wild type mice to the output library from MHCII−/− mice (Figure 1A). In effect, we screened for Mtb mutants with in vivo growth defects that were rescued in the absence of CD4 T cells. We reasoned that genes required for surviving the CD4 T cell response would be required for growth in wild type mice, but not required for growth in MHCII−/− mice, which lack CD4 T cells (Figure 1A). Since these genes are responsible for counteracting the effects of CD4 T cells, we refer to this set of genes as Mtb’s CD4 counteractome.
Figure 1.
A. MHC Class II KO and wild type mice were infected with 106 bacilli from our transposon library. At 10 days and 45 days after infection, 4 mice in each group were sacrificed and spleen homogenates were plated to recover surviving bacteria. To discover genes required for surviving CD4 T Cell immunity, we searched for genes that were required for growth in a wild type mouse but not required in the MHC Class II mice. For both the d10 (B.) and d45 (C.) timepoints, we calculated for each gene the insertion count differences between the input library and the recovered libraries from wild type mice, and a p-value expressing the significance of this differences. We then log-transformed the p-values and gave each gene with a loss of insertions a negative log-p-value and each gene with a gain of insertions a positive log-p-value. After ordering the genes based on their p-values, we plotted each gene as a dot with log-p-values on the y-axis and the size of the circles representing the fold-change of insertion count differences.
We found 576 genes that were required for growth during infection (Figure 1B and 1C, Figure S1A). These genes had a statistically significant (FDR q-value < 0.01) 10-fold or more decrease in insertion counts across the gene. On day 10, 405 genes were required for growth in vivo, and on day 45, 317 genes were required. A total of 146 genes were required at both time points (Figure 1B and 1C, Table S1). Genes required late but not early could represent mutants that are able to establish infection but unable to sustain long-term in vivo growth. Genes required early but not late could represent mutants that grow slowly in mice. These mutants would be underrepresented at day 10, but catch up by day 45 post infection. In fact, genes which were required at day 10 but not at day 45 were enriched for loss-of-insertions (Fig S1B). The average in vitro:in vivo ratio for these genes was 4.3, compared to the non-required average of 1.3 (p-value < 0.001) (Figure S1C). While this set significantly overlapped with a previously defined set of genes required for growth in mice (Gene Set Enrichment Analysis P-val < 0.01), it also includes more than 400 newly discovered genes required for in vivo growth (Figure S2A) (Sassetti and Rubin, 2003).
Genes required for surviving CD4-mediated stress: the CD4 counteractome
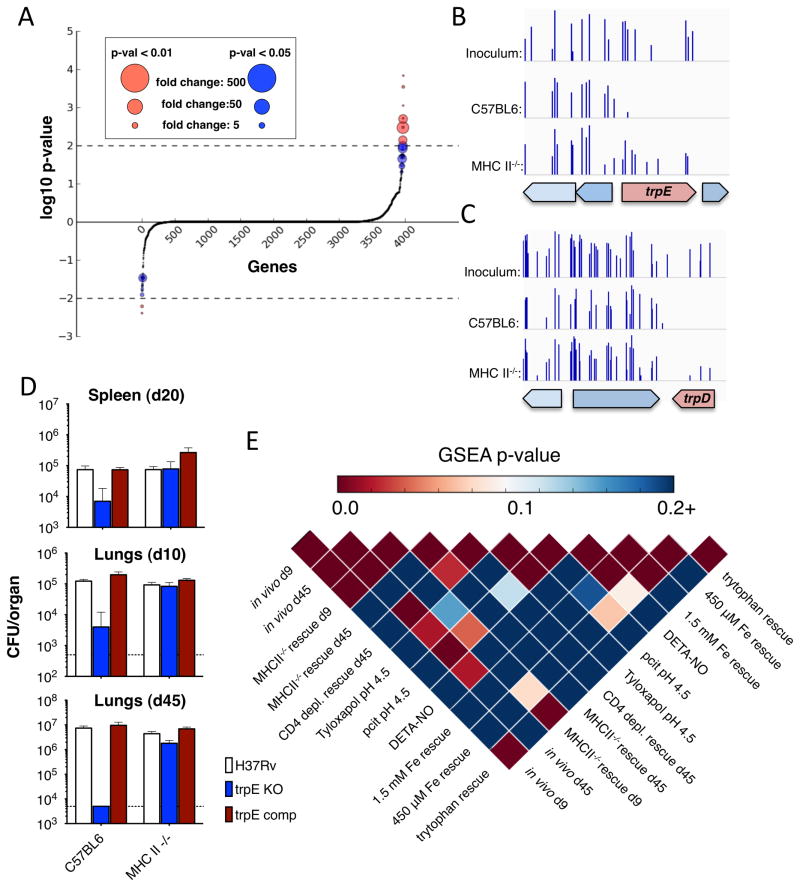
To screen for the CD4 T cell counteractome, the set of mycobacterial genes required for counteracting CD4 mediated stress, we searched for genes wherein mutations caused a growth defect in wild type mice but not in CD4-deficient mice. By comparing the wild type output libraries to the MHCII−/− output libraries, we found that 58 genes had a statistically validated increase in insertions in MHCII−/− mice compared to wild type mice. These genes had at least a 5-fold increase in insertions and a Mann-Whitney U p-value of less than 0.05 (Figure 2A and Table S2). Two biochemical pathways, gluconeogenesis and tryptophan biosynthesis, were enriched in this list, suggesting that CD4 T cells are responsible for inducing a shift in Mtb metabolic demands (Table S2).
Figure 2.
A. To search for genes required for surviving the CD4 T cell response, we identified genes that had a statistically validated increase in insertion counts (above the axis). Transposon insertion counts in the regions containing trpE (B) and trpD (C). Histograms represent the number of times insertions were found at each potential insertion site. Both trpE and trpD sustained insertions in our library, but whereas we were unable to retrieve insertions in these genes from wild type mice, we were able to recover trpE and trpD mutants from MHC Class II KO mice. D. We infected wild type, trpE KO, and complemented strain Mtb into wild type and MHC Class II KO mice. Growth of the three strains was determined, confirming the results of our transposon screen. E. By comparing gene requirement signatures, we profiled the similarity of CD4-mediated stress to in vitro models of potential immune-mediated stresses. Each box represents a pairwise comparison between two gene sets, where the larger gene set was ordered by p-value and ratio and the smaller gene set was used by the GSEA pre-ranked tool to search for enrichment of second set genes in the first
The CD4 counteractome could be required for the bacterium’s physiologic changes in response to CD4-driven environmental changes. Thus, the counteractome is a signature that cues us into the nature of CD4-mediated stress. To identify these cues, we selected our Mtb transposon library in a series of in vitro stress conditions that model stresses thought to be present during in the immune competent host and compared the counteractome signatures. Using the same transposon mapping technique on our selected library, we tested carbon starvation, amino acid starvation (and isolated tryptophan starvation), iron depletion, acid stress and nitrosative stress (Table S3A, B, C and D). We created conditionally required gene sets as well as ranked gene lists for each condition, which allowed us to compare gene sets using the running-sum enrichment analysis in the Gene Set Enrichment Analysis (GSEA) tool (Subramanian et al., 2005). We found that both nitrosative stress and acid stress had significantly similar profiles to genes required for in vivo growth (Figure 2E, pairwise comparisons of “Tyloxapol pH 4.5,” “pcit pH 4.5,” “DETA-NO” and “in vivo d45”). This is consistent with the reactive nitrogen species and low pH of Mtb’s in vivo niche (Ehrt and Schnappinger, 2009; Vandal et al., 2008). Interestingly, we also found that tryptophan starvation was enriched in the CD4 counteractome (Figure 2E, intersection of “tryptophan rescue” and “MHCII−/− d9 rescue”). Two genes in the tryptophan biosynthesis pathway, trpD and trpE, had no insertions in the wild type output library but multiple insertions in the MHCII−/− output library (Figure 2B and 2C). To confirm this finding, we infected wild type and MHCII−/− mice with strains of Mtb auxotrophic for Trp. The growth of the auxotrophs was significantly limited in wild type mice but restored in MHCII−/− mice (Figure 2D). This suggested that CD4 T cells induced the requirement for bacterial tryptophan biosynthesis suggesting that interfering with tryptophan metabolism might offer an appealing therapeutic strategy.
Tryptophan auxotrophy is bactericidal
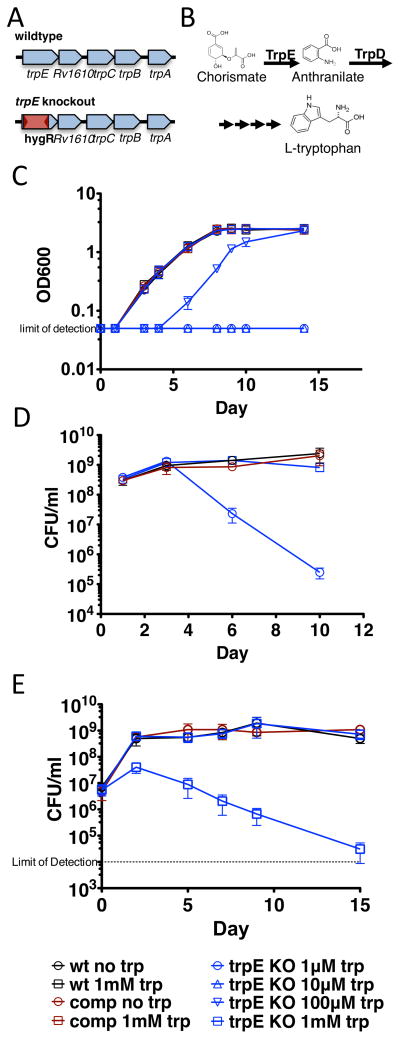
To study the effects of tryptophan auxotrophy and starvation during Mtb infection, we deleted the trpE gene in Mtb and replaced it with a hygromycin resistance gene (Figure 3A). TrpE, or anthranilate synthase, converts chorismate to anthranilate in the first committed step of tryptophan biosynthesis. The next enzyme in the pathway, TrpD, converts anthranilate to N-(5′-phosphoribosyl)-anthranilate. An Mtb strain carrying a deletion of the trpD gene has been shown to be a tryptphan auxotroph (Figure 3B) (Parish, 2003; Smith et al., 2001). The trpE deletion strain grew normally in liquid media only when supplemented with 1 mM tryptophan (Figure 3C). Interestingly, lower amounts of tryptophan delayed entry into the logarithmic phase of growth, but the growth dynamics from that point onwards was similar to wild type. Genetic complementation with the trpE gene restored normal growth in media lacking tryptophan (Figure 3C).
Figure 3.
A. We made a tryptophan auxotroph by replacing trpE with a hygromycin resistance cassette. B. Tryptophan biosynthesis starts with the conversion of chorismate to anthranilate by TrpE, followed by ribosylation by TrpD. C. Tryptophan auxotrophs, complemented strains, and wild type Mtb were grown in the presence and absence of tryptophan in 7H9 media. No tryptophan was required for wild type and complement growth, but 1 mM tryptophan was required to restore normal growth of the tryptophan auxotroph. Auxotrophs grown to mid-log (D.) and stationary (E.) phase were washed, starved of tryptophan and plated for CFU. Starvation of the auxotroph resulted in rapid mycobacterial death.
Auxotrophy in Mtb is not always bactericidal (Parish, 2003). Our results suggested that CD4 T cells help starve Mtb of exogenous tryptophan, so we sought to test the bactericidal potential of blocking endogenous tryptophan biosynthesis in the face of this exogenous starvation. To do so, we cultured the auxotroph in tryptophan to both mid-log and stationary phase, and then continued the culture either in the presence or absence of tryptophan and plated to measure survival. The auxotroph was rapidly killed when starved of tryptophan, suggesting that tryptophan biosynthesis is an attractive target for a bactericidal drug (Figure 3D and 3E). Interestingly, the trpE deletion strains dies far more rapidly than a trpD deletion (Parish, 2003). The trpE deletion strain has about a 100,000-fold loss of viability at two weeks, a level which is reportedly not achieved after 13 weeks of starvation of the trpD deletion strain. This may be due to accumulation of intermediary metabolites or the presence of an as yet undescribed alternative tryptophan synthesis pathway.
Tryptophan auxotrophs are hypersusceptible to macrophages activated by CD4 T cells
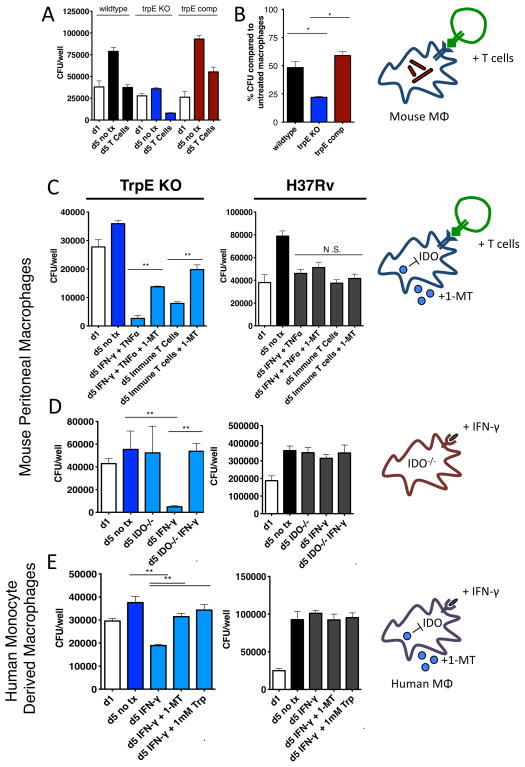
During a typical lung infection, Mtb first enters alveolar macrophages, which then form an inflammatory structure known as the granuloma (Russell, 2007). Upon adaptive immune activation, CD4 T cells enter the granuloma and stimulate the infected macrophages (Ernst, 2012). Our screen results suggested that one of these stimulatory mechanisms imposes a need for bacterial tryptophan biosynthesis, and we sought to delineate the mechanism by which CD4 T cells exert this need. To do so, we infected thioglycolate-elicited mouse peritoneal macrophages with wild type Mtb, the trpE deletion strain and the complemented strain. Over a five-day infection, the wild type and complemented strains increased in colony forming units (CFU) by about 5-fold, while the trpE deletion strain had no measureable growth, demonstrating that tryptophan biosynthesis is required for growth even in unstimulated macrophages (Figure 4A).
Figure 4.
A. Wild type, tryptophan auxotroph, and complemented strains were used to infect macrophages. One day after infection, macrophages were either stimulated with CD4 T cells in co-culture or remained unstimulated. The tryptophan auxotroph grew poorly in unstimulated macrophages and were hypersuscpetible to the effects of CD4 T cells (B). C. Macrophages were stimulated by either immune CD4 T cells (CD4 T cells isolated from spleens of Mtb-infected mice) or IFN-γ and TNF-α. To test whether the hypersusceptibility of the auxotroph was dependent on IDO, we also inhibited IDO with its small molecule inhibitor, 1-MT, or used macrophages isolated from IDO KO mice (D). E. Human monocyte derived macrophages were infected with wild type and tryptophan auxotroph bacteria. After stimulation with IFN-γ, we showed that the tryptophan auxotroph was also hypersusceptible to the effects of human IFN-γ
Our mouse findings predicted that the trpE deletion strain would be particularly sensitive to CD4-mediated stress. Indeed, the trpE deletion strain was significantly hypersusceptible to the effects of CD4 T cells in co-culture with macrophages (Figure 4A and 4B). By adding CD4 T cells harvested from the spleens of Mtb-infected mice, macrophages were able to kill the auxotroph more effectively. Compared to unstimulated macrophages, CD4 T cell-stimulated macrophages decreased growth of wild type Mtb by about 50%, whereas CD4 co-culture decreased trpE deletion strain growth by about 80% (Figure 4B). Additionally, the trpE deletion strain was also hypersusceptible to IFN-γ and TNF-α, cytokines secreted upon the arrival of CD4 T cells to an Mtb lesion (Figure S3). These data confirmed our findings in mice, and showed that CD4 T cells and the cytokines they secrete demand the need for mycobacterial tryptophan biosynthesis during infection. Since human macrophages differ from mouse macrophages in their mycobacterial killing strategies, we tested the growth of our Mtb strains in monocyte-derived macrophages from human donors. As with previous reports, we observed that IFN-γ alone does not measurably inhibit wildtype Mtb growth (Fabri et al., 2011). However, while the trpE deletion strain grew slightly over the 5-day infection, its growth was inhibited by IFN-γ (Figure 4E).
IFN-γ induction of IDO necessitates mycobacterial tryptophan biosynthesis
To show that the auxotroph’s hypersusceptibility to IFN-γ was tryptophan dependent, we added tryptophan to the media in IFN-γ-treated macrophages, and showed that the addition of tryptophan reversed the IFN-γ hypersusceptibility (Figure S4). As expected, tryptophan did not change the bacterial growth inhibitory effect of IFN-γ in wild type Mtb, showing that tryptophan supplementation does not have a general growth effect on Mtb (Figure 4E). Interestingly, tryptophan supplementation could not restore growth of the auxotroph in unstimulated macrophages to wild type Mtb levels. It is possible that the levels of tryptophan needed to restore wild type growth (1 mM in liquid broth) cannot be reached intracellularly, while the amount of tryptophan required to protect from IFN-γ mediated killing could.
Many intracellular pathogens are natural tryptophan auxotrophs whose intracellular growth is also inhibited by IFN-γ. Thus, we attempted to determine if similar processes affected tryptophan availability in Mtb infection. One of the transcriptionally induced genes in response to IFN-γ is a tryptophan-catabolizing enzyme, indoleamine-2,3-dioxygenase (IDO) (Alberati-Giani et al., 1997). IDO utilizes tryptophan as a synthetic precursor for kynurenines, immune signaling molecules that help control inflammation (Zelante et al., 2009a). In this synthetic process, it also greatly decreases the intracellular tryptophan pool. IDO is thus required for IFN-γ-mediated growth inhibition of Chlamydia and other tryptophan auxotrophic intracellular pathogens (Beatty et al., 1994; Fujigaki, 2002; Ibana et al., 2011; Zelante et al., 2009b).
We tested the role of IDO in the trpE deletion strain’s hypersusceptibility to CD4 T cell and IFN-γ by either a) inhibiting IDO in both human and mouse macrophages with a specific chemical inhibitor, 1-methyl tryptophan (1-MT) (Figure 4C and 4E) or b) using mouse macrophages derived from IDO knockout mice (Figure 4D) (Baban et al., 2004; Belladonna et al., 2006). In both cases, the hypersusceptibility was reversed. The inhibitor acts on the macrophage rather than the pathogen as it had no effect on bacteria grown in a defined medium (Figure S5). These data support that CD4 T cells, likely acting through IFN-γ, stimulate intracellular tryptophan depletion, forcing Mtb to synthesize its own tryptophan.
Halogenated anthranilate analogs disrupt tryptophan biosynthesis to kill Mtb in vitro
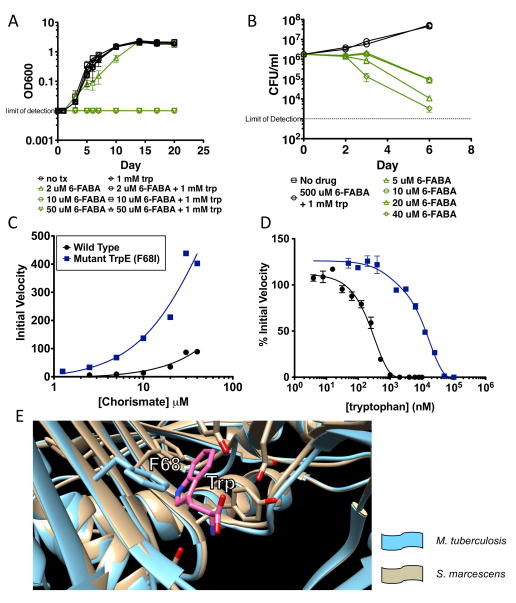
Since tryptophan is required for intracellular growth in the face of CD4-mediated immunity, compounds that inhibit the bacterial tryptophan synthesis pathway should synergize with host immunity. We focused on anthranilate analogues, compounds that have been shown to inhibit the synthesis of quorum-sensing molecules in Pseudomonas aeruginosa that, like Trp synthesis, also have an anthranilate intermediate (Lesic et al., 2007). We tested a panel of anthranilate analogs for Mtb growth inhibition in the presence and absence of tryptophan. Two fluorinated anthranilates, 2-amino-5-fluorobenzoic acid (5-FABA) and 2-amino-6-fluorobenzoic acid (6-FABA) had an MIC of 5 μM in liquid broth in the absence of tryptophan (Figure 5A). The addition of tryptophan blocked toxicity (Figure 5A). 6-FABA was less active against Mycobacterium smegmatis (Msm), with an MIC of 65 μM but this was, again, blocked by added tryptophan (Figure S6). As predicted from the experiments with auxotrophs, 6-FABA was also bactericidal in liquid broth (Figure 5B) with an ~100-fold and ~10,000-fold decrease in CFU compared to the starting inoculum or the untreated control on day 6 respectively.
Figure 5.
A. 6-FABA inhibits the growth of Mtb in 7H9. B. To test the bactericidal potential of 6-FABA, wild type Mtb was treated with 6-FABA in 7H9, and cultures were plated for CFU at various time points. C. Wild type (blue) and mutant (black) TrpE (F68I) were isolated and enzymatic activity was assessed by measuring chorismate concentration. D. The inhibitory affect of tryptophan on both wild type (blue) and mutant (black) TrpE was measured as % initial velocity of the no-inhibitor control reaction. E. Mtb TrpE structure (blue) was modeled based on the homologous enzyme from S. macrcescens (beige), showing F68 in the allosteric binding pocket of tryptophan.
C. Wild type Mtb was used to infect macrophages. On day 1 after infection, macrophages were treated with 6-FABA, and on day 5, cells were lysed and bacteria plated, showing the 6-FABA was bactericidal in macrophages. D. To test the synergy between 6-FABA and IFN-γ, we added 6-FABA and IFN-γ at concentrations that inhibited approximately half of bacterial growth. E. Together, their effect was greater than the estimated additive effect of the two molecules in the absence of synergy, showing that they indeed work synergistically to kill Mtb in macrophages.
To determine the likely target of these compounds we selected for mutants on solid media containing 150 μM 5-FABA or 300 μM 6-FABA. We plated 109 bacteria treated with 0.25% ethyl methanesulfonate on 150 μM 5-FABA, which resulted in ~30 resistant colonies. We then sequenced the genome of a resistant clone, and found the following polymorphisms: TrpE:F68I, Rv2585c:A154V, Pks13:P568A, 4359303:A>G (downstream of RD1 deletion). The trpE mutation was surprising, as the enzyme converts chorismate into anthranilate, and we had hypothesized that TrpD, which utilizes anthranilate, would be the target of these anthranilate analogs. This led to two competing hypotheses for the mechanism of resistance. The FABA molecules could target TrpE by inhibiting the enzyme at its allosteric site, and the F68I mutation might confer resistance to this targeting. Alternatively, the F68I mutant could be a hypermorph, and insensitivity to allosteric inhibition by tryptophan might confer resistance through increased tryptophan synthesis.
Testing the possibility that the FABAs are inhibitors of TrpE activity was technically challenging. We cloned and purified both wild type and the F68I mutant form of TrpE and assayed their activity by measuring the fluorescent product, anthranilate (Baker and Crawford, 1966; Bauerle et al., 1987). Both anthranilate derivative FABA compounds had fluorescent spectra that overlapped with anthranilate. Therefore, we were unable to assess the possibility of the FABA compounds being inhibitors of TrpE. However, we were able to test the enzymatic properties of both mutant and wildtype TrpE and address the possibility of a hypermorphic allele. Indeed, we found that the mutant enzyme had a 3-fold increase in in vitro activity compared to the wild type enzyme (Figure 5C). Furthermore, the mutant enzyme was ~50 times less sensitive to allosteric inhibition by tryptophan in vitro (Figure 5D). When we modeled the Mtb TrpE structure using the Serratia marcescens homologue, we found that F68 resides in the allosteric binding pocket of tryptophan (Figure 5E). Altogether, this suggests that the F68I mutant is a hypermorphic allele that is also less sensitive to allosteric inhibition by tryptophan. The likely increase in tryptophan production, then, confers resistance to the FABA compounds in the same way that added exogenous tryptophan does (Figure 3C, D, and E).
6-FABA synergizes with IFN-γ to kill Mtb in macrophages
If these compounds act through the tryptophan biosynthetic pathway, they should phenocopy trpE deletion strains during macrophage infection. To test this we infected macrophages with Mtb and, after one day, added 6-FABA and measured bacterial growth by plating for CFU on day 5. At concentrations as low as 10 μM, 6-FABA significantly limited growth by over 10-fold (Figure 6A). The number of CFU at d5 was lower than at d1, demonstrating that 6-FABA had bactericidal activity in macrophages. To ensure that activity was not due to the death of macrophages, we tested cytotoxicity and found that 6-FABA did not affect cell viability (Figure S7).
Figure 6.
A. Wild type Mtb was used to infect macrophages. On day 1 after infection, macrophages were treated with 6-FABA, and on day 5, cells were lysed and bacteria plated, showing the 6-FABA was bactericidal in macrophages. B. To test the synergy between 6-FABA and IFN-γ, we added 6-FABA and IFN-γ at concentrations that inhibited approximately half of bacterial growth. C. Together, their effect was greater than the estimated additive effect of the two molecules in the absence of synergy, showing that they indeed work synergistically to kill Mtb in macrophages. Mice were infected with 102 aerosolized Mtb bacilli. After 8 days of infection, mice were treated with INH (25 mg/kg), 6-FABA (200 mg/kg) or with the ester derivative of 6-FABA (200 mg/kg). At 2 weeks (D and E) and 4 weeks (F and G) after infection, lungs and spleens were homogenized and plated for CFU. *: P-val < 0.05. **: P-val < 0.01. Insets: linear scale CFU.
Since the tryptophan auxotroph was hypersusceptible to IFN-γ, we hypothesized that 6-FABA’s block of tryptophan biosynthesis would work in synergy with IFN-γ to kill Mtb in macrophages. To test this, we dosed both IFN-γ (10U/ml) and 6-FABA (0.2 μM) to a level where each individually had about a 2-fold inhibitory effect on bacterial growth (Figure 6B). Without synergy, we predicted that the combined effect of 6-FABA and IFN-γ would be about 4-fold. Instead, the effect was ~40-fold in mouse and ~9-fold in human macrophages, demonstrating clear synergy (Figure 6C and Figure S8).
6-FABA inhibits Mtb growth during infection
While many compounds are active in vitro, few have adequate bioavailability to be used in animal infections without considerable modification. Fortunately, however, we found that both 6-FABA and an ester derivative (that is rapidly cleaved to the free acid in the circulation) are absorbed orally and achieve high serum concentrations, though with relatively short half-lives (figure S9). Moreover, doses of up to 250 mg/kg/day did not result in clinical illness or weight loss during a five-day tolerability trial. This allowed us to test the activity of these compounds in a mouse model of TB.
We infected mice with 102 aerosolized Mtb bacilli, and allowed infection to establish in the lungs for 8 days. We treated mice six times a week with either INH (25 mg/kg/day), 6-FABA (200 mg/kg/day) or the ester derivative (200 mg/kg/day), and planned to measure bacterial growth at 2 weeks and 4 weeks after initiating treatment. Unfortunately, prolonged treatment with 6-FABA resulted in 50% death of animals within 4 weeks. Thus, we could only evaluate 2-week efficacy with this compound. Notably, the ester form of 6-FABA was not toxic, even though it releases the same amount of 6-FABA in mouse serum. This suggested to us that toxicity of 6-FABA was due to an as-of-yet undetermined off-target effect that might be influenced by an interaction with anesthesia used for gavaging Mtb-infected animals (anesthesia was not used for tolerability trial). Importantly, the non-toxic ester derivative allowed us to assess the in vivo efficacy of pharmaceutical tryptophan biosynthesis targeting over a longer time period. At 2 weeks after infection, growth in mouse spleens was decreased by greater than 10-fold in both 6-FABA and the ester-derivative treated mice (Figure 6E). 6-FABA also decreased bacterial growth in the lungs, but to a lesser extent (Figure 6D). At 4 weeks after infection, the ester continued to significantly decrease mycobacterial growth in both spleens and lungs (Figure 6F and 6G). The effect of these molecules on decreasing bacterial growth in vivo demonstrates that tryptophan biosynthesis is a viable target for drug development. Combined with the fact that CD4 T cells are required for optimal killing of tryptophan auxotrophs (Figure 2D), we have shown that this therapeutic strategy leverages CD4 T cell activity to kill Mtb in vivo.
Discussion
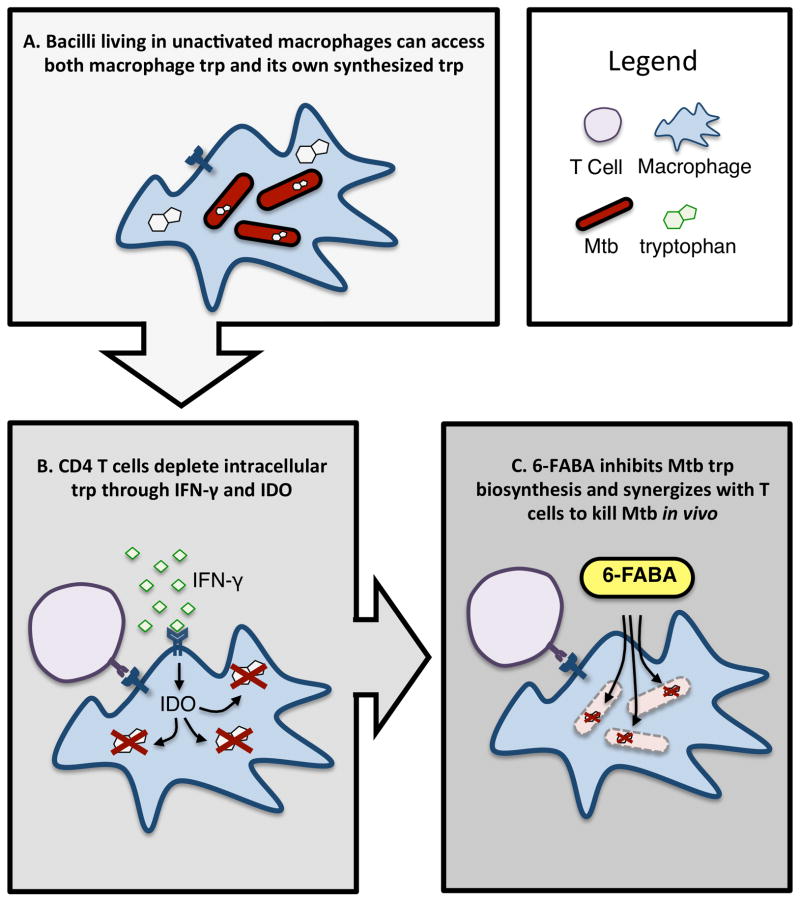
CD4 T cells are paramount in the host defense against TB, but are insufficient to clear the bacteria from a diseased patient. We found that one of the mechanisms by which bacteria survive host CD4-generated stress is through the production of tryptophan, thus avoiding starvation and death. Loss or inhibition of the tryptophan biosynthetic pathway renders Mtb hypersusceptible to IFN-γ-mediated killing within macrophages, both in vitro and during infection (Figure 7).
Figure 7.
A. Upon infection, unstimulated macrophages have a lot amount of tryptophan, which together with tryptophan synthesized by the bacterium is able to support Mtb growth. B. CD4 T cells, through IFN-γ and the induction of IDO, decreases the amount of intracellular tryptophan available to Mtb, demanding the need for mycobacterial tryptophan biosynthesis for bacterial survival. C. Treatment with 6-FABA chemically induces Mtb tryptophan auxotrophy. Together with immune-mediated tryptophan starvation, this results in mycobacterial death.
This observation highlights one of the major mechanisms of host protection against invading organisms—depriving them of key nutrients. Infection triggers host responses that sequester important compounds such as amino acids and iron (Hood and Skaar, 2012; Skaar, 2010; Zhang and Rubin, 2013). While these non-specific responses are likely important for the control of poorly-adapted invaders, successful pathogens have developed strategies to counteract these defenses, including biosynthesis of key compounds and efficient scavenging of others. Mtb in particular relies upon cholesterol and fatty acid catabolism, gluconeogenesis and the glyoxylate shunt to survive the host environment (Lee et al., 2013; Marrero et al., 2010; McKinney et al., 2000; Munoz-Elias and McKinney, 2005; Muñoz-Elías and McKinney, 2006; Pandey and Sassetti, 2008).
Tryptophan starvation, through IDO-mediated depletion of cytoplasmic pools of this amino acid, appears to be a mechanism that has the added benefit of leading to kynurenine production and prevention of immunopathology (Desvignes and Ernst, 2009). This clearly benefits the host in blocking the growth of cytoplasmic pathogens. However, it is less obvious that it should be a successful strategy for vacuolar organisms that do not have ready access to the cytoplasm. In the case of mycobacteria, susceptibility of tryptophan auxotrophs to IDO-mediated killing means that bacteria must be able to access this amino acid. This could occur through specific host or bacterial transport mechanisms (that have not been defined) or direct access to the cytoplasm through either partial or complete loss of vacuolar integrity, a model consistent with recent observations of Mtb escape into the cytoplasm (Hagedorn et al., 2009; McDonough et al., 1993; van der Wel et al., 2007; Welin et al., 2011). The apparent ability of Mtb to scavenge for tryptophan points towards a capability to uptake other amino acids. A host of other Mtb amino acid auxotrophs are attenuated for growth in mice; it is possible that this is due to immune-driven starvation mechanisms (Hondalus et al., 2000; Pavelka et al., 2003; Price et al., 2011; Tattoli et al., 2012; Zhang and Rubin, 2013).
While scavenging Trp in MHCII−/− mice allows the Trp deletion strains to survive, it is important to note that these mutants are eventually attenuated even in MHCII−/− mice. Similarly, we showed that the TrpE deletion strain is attenuated in macrophages even before stimulation by CD4 T cells or IFN-γ and TNF-α. So while our data clearly demonstrate the importance of CD4 T cells in demanding bacterial tryptophan synthesis, it appears that there may also be CD4-independent mechanisms of tryptophan starvation. Importantly, this suggests that targeting tryptophan biosynthesis would retain some effectiveness even in patients, such as HIV-positive individuals, who lack a normal CD4 compartment.
Here we used an improved analytic approach to identify conditionally essential genes using a transposon library. Each gene has multiple potential insertion sites, and most insertion count comparisons sum counts across each gene to compare between conditions. However, in reasonably saturated libraries, treating each potential insertion site as an independent assessment of gene requirement drastically increased the statistical power of such comparisons. Most transposon insertion sequencing techniques currently use insertion count totals summed across all sites in a gene (Barquist et al., 2013; Goodman et al., 2009; Griffin et al., 2011; Zomer et al., 2012). We found that this drastically decreases the power of statistical hypothesis testing and increases the likelihood that a single differential site could make the entire gene look differentially required. We developed a more discriminating method for assessing conditional requirement of genes in genome-wide bacterial transposon screens. Using an easily scalable non-parametric test to comprehensively compare the distribution of insertion counts (rather than the sums of insertion counts) across every gene, we were able to assess genetic requirement across a host of different conditions.
In contrast to similar microarray-based screens, deep sequencing allowed us to directly map insertion sites for each surviving mutant, and thus improved significantly on the sensitivity and specificity of insertion count comparisons. We found 576 genes that were required for growth in vivo, most of which are newly described as required. Furthermore, by searching for mutants rescued for growth in MHCII−/− mice, we described 58 genes that were likely required for surviving CD4-mediated host defenses.
One power of this approach is that we can easily compare gene requirements in multiple conditions, essentially using complex profiles to identify critical environmental determinants. We showed that the in vivo environment as a whole, as well as the specific CD4-generated component of that environment, could be understood by comparing multiple gene-requirement profiles. GSEA analysis comparing in vivo profiles to in vitro profiles confirmed that Mtb’s infectious niche is characterized by acidic and oxidative stress, and that CD4 T cells impose tryptophan starvation. Gene-requirement signatures are an effective way to understand both mycobacterial physiology as well as the environments created by components of host immunity. Our study is limited to the determination of the CD4 counteractome, but other counteractomes—those specific to IFN-γ, iNOS, CD8 T cells, or any immune system component that can be perturbed in a mouse—will further our understanding of the tug-of-war between host and pathogen. In fact, given that many CD4-mediated mechanisms are IFN-γ dependent, we would predict that bacterial strains rescued in the MHCII−/− mice, like the trpE deletion strain, would also be rescued in IFN-γ knockout mice. Thus, on a genomics scale, the IFN-γ counteractome should look similar to the CD4 counteractome. Further profiling can test such hypotheses and uncover new mechanistic differences between the many arms of host immunity.
Tryptophan biosynthesis is an attractive target for antibiotic development. Small molecule inhibitors of tryptophan and other aromatic amino acid biosynthesis exist for other organisms, suggesting that certain enzymes in these pathways might be “druggable,” or suitable for chemical inhibition (Coggins et al., 2003). Furthermore, tryptophan is an essential amino acid for humans (John and Bell, 1976; WHO, 2007) Since hosts get all of their needed tryptophan from their diets, any cross-species activity of anti-tryptophan synthesis drugs should be non-toxic (though off-target toxicity remains a possibility).
Halogenated anthranilates were first described as inhibitors of both quorum sensing and growth in P. aeruginosa infection (Lesic et al., 2007). We found that two of these compounds, 6-FABA and 5-FABA had bactericidal activity against Mtb at low concentrations, though only in the absence of tryptophan. Two lines of evidence suggest that tryptophan biosynthesis pathway is the physiologic target for both of these compounds. First, normal bacterial growth is restored during FABA treatment by the addition of tryptophan. Second, resistant mutants harbor a hypermorphic enzyme that is resistant to allosteric inhibition by tryptophan. Together, these provide chemical and genetic confirmation that tryptophan can rescue the effect of FABA, but it does not prove that FABA inhibits tryptophan production. Because TrpD utilizes anthranilate as a substrate, we speculated that 6-FABA might be a competitive inhibitor of TrpD. However, it is also possible that 6-FABA could be a substrate of TrpD, resulting in the production of fluorinated intermediates that inhibit tryptophan biosynthesis or even fluorinated tryptophan that incorporates into and poisons protein synthesis or function. Our data clearly demonstrate that the bactericidal activity of 6-FABA is due to its negative effect on tryptophan biosynthesis, and further studies to elucidate the precise mechanism of toxicity should focus on this tryptophan effect.
The vast majority of antibiotics are effective against organisms grown in vitro. However, one of the most effective antituberculous drugs, pyrazinamide, is poorly active under most in vitro growth conditions(Tarshis and Weed, 1953; Zhang and Mitchison, 2003). Instead, it relies on the host environment for efficacy. Similar compounds, which target processes critical in the host even if not in vitro could effectively synergize with the host response to infection and have substantial efficacy. Probing CD4-mediated immunity helped us identify a specific facet of the host-derived environment that could be utilized for bacterial killing. While we do not yet have a compound that is highly effective during infection, our results provide genetic and chemical validation of the tryptophan biosynthesis pathway as a novel target for highly active antibiotics.
Experimental Procedures
Library generation
Transposon libraries were created as described previously (Zhang et al., 2012), but plated on 7H10 plates with glycerol, OADC, Tween80, Cas-amino acids and tryptophan. 500,000 mutants were scraped, frozen and saved for future use.
Mouse infections and harvests
Wild type mice (C57BL/6) were obtained from Jackson Laboratory. MHC Class II knockout mice (Abb H2-Ab1) were obtained from Taconic Farms. These mice have a disruption in the H2-Ab gene, do not express any MHC Class II molecules, and lack CD4 T cells. Mice were infected with 106 bacteria via tail vein injection. At 10 days and 45 days post-infection, spleens were harvested and plated for bacteria. For each mouse, 106 surviving colonies were scraped and DNA was extracted for analysis. The protocols, personnel and animal use was approved and monitored by the Institutional Animal care and use committee. The animal facilities are AAALAC accredited.
In vitro transposon library selections
For acid stress, the library was suspended in liquid media at a starting concentration of 108 CFU/ml and selected in 7H10 with tyloxapol (in place of Tween80) buffered to pH 6.5 or pH 4.5, and in phosphate-citrate buffer at pH 4.5. Bacteria was plated after 6 days and scraped for DNA prep. For nitrosative stress, the library was suspended in 5 mM DETA-NO for 3 days. Tryptophan starvation and amino acid starvation was measured using libraries that were created either on normal 7H10, or on 7H10 supplemented with 1 mM tryptophan or 1% Cas-amino acids, respectively. Iron supplemented libraries were plated on 7H10 with 450 μM and 1.5 mM iron.
Statistical Analysis
For each insertion count comparison, the control libraries were combined using a script that normalized insertion counts to the sequencing run’s total read counts. This combined control library was then used to compare to each experimental library. For each gene, we treated the insertion counts at sites within the middle 90% of the gene as non-parametric distributions, and assumed the null hypothesis that the distributions would be the same between conditions. We used a Mann-Whitney U test for hypothesis testing. A p-value was thus calculated for each replicate, and a composite p-value was generated by using a Bonferroni correction and Fisher’s method. We then generated Benjamini-Hochberg false discovery rates using the composite p-values. Ratios were calculated by averaging the read counts per gene for all replicates and comparing to the combined control library, after normalizing for total read counts.
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis was performed using the pre-ranked tool in GSEA. Genes were stratified by p-value (< 0.01 or 0.05 and > 0.01 or 0.05), and then ranked by ratio within the strata. For each conditional gene-requirement experiment, a ranked list and a conditional essential “calls” gene set was created. Each ranked list was then assessed against each gene set, and Family-wise Error Rate was used to generate the p-values for significant enrichment.
Construction of trpE deletion and complement strains
The hygromycin resistance gene was amplified with flanks containing 500 bp regions upstream and downstream (and slightly overlapping with) of trpE. This construct was electroporated into a strain of Mtb containing the plasmid pNIT(kan)::RecET-SacB, which contains the machinery for mycobacterial recombineering. Recombineering-competence was achieved by induction of the RecET complex by the addition of 1 mM isovaleronitrile (IVN, Sigma Aldrich) to a culture at OD600 0.8. IVN addition induced expression of the recombineering machinery on pNit(kan)::RecET-SacB. After 8h, 10 mL of 2M glycine was added, and the culture was grown overnight to yield recombineering-competent Mtb. Transformations were plated on 7H10 agar with 1 mM tryptophan. Positive clones were plated on 7H10 agar containing 10% sucrose to counterselect against the recombinase plasmid, and scored for growth on kanamycin-containing agar to confirm the loss of pNit::RecET. Deletion of the endogenous locus was confirmed by PCR and by phenotypic tests for auxotrophy. Finally, to complement, the trpE-rv1610 two-gene operon was amplified and cloned, along with an artificial promoter, using multisite gateway into pDE43-MCK, which integrates into the L5 site.
Macrophage infections
Peritoneal macrophages were stimulated with Thioglycollate Medium (3%) by intraperitoneal infection. Three – five days after injecting, the peritoneum was exposed and macrophages were harvested by adding 10 mL of RPMI media with penicillin and streptomycin into the peritoneal cavity and retrieving the fluid. Cells were washed and incubated with CD11b microbeads (Miltenyi) for 15 minutes on ice. Magnetic separation of CD11b+ cells was completed, and cells were plated for one day before use. Human monocyte-derived macrophages were made from donated buffy coats. Buffy coats were diluted and layered on Ficoll for gradient centrifugation. The middle cellular layer was harvested and washed multiple times, and then incubated with CD14 microbeads (Miltenyi) on ice for 15 minutes for magnetic isolation. Cells were then grown in GM-CSF (10 ng/ml) for 5 days before use. Cells were infected with Mtb at an MOI of 10:1 for 2 hours at 37° C. They were then washed 4 times with RPMI media and grown overnight. On day 1 post-infection, most conditions were added. To lyse cells, Triton-X 100 (0.1%) in PBS was added to each well. Serial dilutions of the lysis were made and plated. For CD4 T cell co-culture, we infected mice with 100 aerosolized CFU of Mtb. Spleens from infected mice were harvested no earlier than 6 weeks post infection, and CD4 T cells were magnetically isolated using a T cell negative selection kit (Miltenyi) and concurrent negative selection of CD8 T cells.
Kinetic Analysis of Wild Type and Mutant Anthranilate Synthase
A master mix containing 100mM NH4Cl, 10mM MgCl2, 0.1mM EDTA, 1μM of wildtype or mutant TrpE, 20μM chorismate, 20mM of Tris pH-9 was made for each reaction to a final volume of 199μl. Either tryptophan or 5-flurotryptophan was also added in 1μl amounts at concentrations ranging from 0.025uM to 24 uM. The readings for fluorescence emission (anthranilate) within the range of 480–512nM were taken using polar star omega plate reader.
Alamar Blue Assays
In a 96-well plate, bacterial cultures were started at an OD600 of 0.003. After five days, resazurin reagent was added and cells were kept in a shaking 37° C incubator for another 1–2 days. Plates were read and the MIC was determined as the first concentration at which the color changes.
Mouse pharmacokinetic study and sample preparation
A single dose of 5-FABA, 6-FABA and the corresponding esters was administered intraperitoneally or orally at 25mg/kg to female CD-1 mice, 4 to 6 weeks of age. Blood samples were collected in heparinized tubes at 6 to 8 serial timepoints between 0 and 24h post-dosing. Plasma was recovered following centrifugation for 10min at 1,000g. Plasma levels of 5-FABA, 6-FABA and the corresponding esters were quantified by high pressure liquid chromatography coupled to tandem mass spectrometry (LC/MS/MS) following protein precipitation with a 9:1 acetonitrile-to-plasma ratio, with diclofenac as internal standard. Standards, quality control samples and blanks in commercial mouse plasma were used.
LCMS conditions
The analysis was performed on a Sciex Applied Biosystems API4000 triple-quadrupole mass spectrometer coupled to an Agilent 1260 HPLC system. Sample analysis was accepted if the low level quality control samples were within ±20% of nominal concentration and ±15% for mid and high level quality control samples.
Acetonitrile with 0.1% formic acid was used as extractant. Gradient elution conditions with a Fluophase 2.1 × 100mm 5u column were used. The mobile phase A was 0.1% formic acid in deionized water and mobile phase B was 0.1% formic acid in acetonitrile. The flow rate was 0.6mL/min.
Supplementary Material
Highlights.
We define the Mtb pathways used to survive the CD4 T cell response.
CD4 T cells attempt to starve Mtb of tryptophan, an essential amino acid.
We block Mtb trp synthesis using an inhibitor, allowing CD4 T cells to kill Mtb.
Acknowledgments
We thank Michael DeJesus, Chris Sassetti and Justin Pritchard for their ongoing collaboration and thoughtful conversations about genomic profiling in Mtb. We thank Xiaohua Li and Spandana Valluru for technical support. Noman Sidddiqi and Larry Pipkin manage and run our BL3 facility; we thank them for their support. We thank the constant support and feedback from our tremendous scientific community, including Marcia Goldberg, Laurence Rahme, Sarah Fortune and Barry Bloom.
Funding: The project described was supported by the National Institutes of Health (T32 GM007753 to YJZ, R01 AI 098637 to SMB, P01 AI095208 to JCS and EJR), the Herchel Smith Graduate Fellowship of Harvard and Cambridge University (to YJZ) and the Wolfe-Welch Chair (to JCS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions: YJZ, EJR, VD, SMB, and JCS conceived and designed the experiments. YJZ, ACR, BMS, MCR, SG, DEW and AT performed the experiments. YJZ, CH and TRI built and utilized the tools for genomic analysis of the transposon screen. YJZ and EJR prepared the manuscript, and TRI, JCS, VD, ACR and SMB edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberati-Giani D, Malherbe P, Ricciardi-Castagnoli P, Kohler C, Denis-Donini S, Cesura A. Differential Regulation of Indoleamine 2,3-Dioxygenase Expression by Nitric Oxide and Inflammatory Mediators IFN-gamma-Activated Murine Macrophages and Microglial Cells. Journal of immunology (Baltimore, Md : 1950) 1997;159:419–426. [PubMed] [Google Scholar]
- Baban B, Chandler P, McCool D, Marshall B, Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase expression is restricted to fetal trophoblast giant cells during murine gestation and is maternal genome specific. Journal of reproductive immunology. 2004;61:67–77. doi: 10.1016/j.jri.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Baker TI, Crawford IP. Anthranilate synthetase: partical purification and some kinetic studies on the enzyme from Eschericia coli. Journal of Biological Chemistry. 1966;241:5577–5584. [PubMed] [Google Scholar]
- Barquist L, Langridge GC, Turner DJ, Phan MD, Turner AK, Bateman A, Parkhill J, Wain J, Gardner PP. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic acids research. 2013 doi: 10.1093/nar/gkt148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerle R, Hess J, French S. Anthranilate synthase-anthranilate phosphoribosyl transferase complex and subunits of Salmonella typhimurium. Methods Enzymol. 1987;142:366–386. doi: 10.1016/s0076-6879(87)42049-1. [DOI] [PubMed] [Google Scholar]
- Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infection and immunity. 1994;62:3705–3711. doi: 10.1128/iai.62.9.3705-3711.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belladonna ML, Grohmann U, Guidetti P, Volpi C, Bianchi R, Fioretti MC, Schwarcz R, Fallarino F, Puccetti P. Kynurenine Pathway Enzymes in Dendritic Cells Initiate Tolerogenesis in the Absence of Functional IDO. Journal of immunology (Baltimore, Md : 1950) 2006;177:130–137. doi: 10.4049/jimmunol.177.1.130. [DOI] [PubMed] [Google Scholar]
- Bold TA, Banaei N, Wolf AJ, Ernst JD. Suboptimal Activation of Antigen-Specific CD4 Effector Cells Enables Presistence of M. tuberculosis In Vivo. PLoS Pathogens. 2011;7:e1002063. doi: 10.1371/journal.ppat.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso AM, Serbina N, Klein E, Treibold K, Bloom BR, Flynn JL. Mice Deficient in CD4 T Cells Have Only Transiently Diminished Levels of IFN-{gamma}, Yet Succumb to Tuberculosis. Journal of immunology (Baltimore, Md : 1950) 1999;162:5407–5416. [PubMed] [Google Scholar]
- Coggins JR, Abell C, Evans LB, Frederickson M, Robinson DA, Roszak AW, Lapthorn Ap. Experiences with the shikimate-pathway enzymes as targets for rational drug design. Biochemical Society Transactions. 2003:31. doi: 10.1042/bst0310548. [DOI] [PubMed] [Google Scholar]
- Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated Tuberculosis in Interferon-gamma Gene-disrupted Mice. Journal of Experimental Medicine. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, Mathis D. Mice Lacking MHC Class II Molecules. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- Desvignes L, Ernst JD. Interferon-gamma-responsive nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity. 2009;31:974–985. doi: 10.1016/j.immuni.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell Microbiol. 2009;11:1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JD. The immunological life cycle of tuberculosis. Nature reviews Immunology. 2012;12:581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, Realegeno S, Lee HM, Krutzik SR, Schenk M, Sieling PA, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci Transl Med. 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JL. Lessons from experimental Mycobacterium tuberculosis infections. Microbes Infect. 2006;8:1179–1188. doi: 10.1016/j.micinf.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer R, Mak TW, Bloom BR. Tumor Necrosis Factor-alpha Is Required in the Protective Immune Response Against Mycobacterium tuberculosis in Mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- Flynn JL, JC, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An Essential Role for Interferon gamma in Resistance to Mycobacterium tuberculosis Infection. Journal of Expermental Medicine. 1993:178. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujigaki S. L-Tryptophan-L-Kynurenine Pathway Metabolism Accelerated by Toxoplasmagondii Infection Is Abolished in Gamma Interferon-Gene-Deficient Mice: Cross-Regulation between Inducible Nitric Oxide Synthase and Indoleamine-2,3-Dioxygenase. Infection and immunity. 2002;70:779–786. doi: 10.1128/iai.70.2.779-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos AM, van Heijst JWJ, Samstein M, Xiaodi S, Pamer EG, Glickman MS. A Gamma Interferon Independent Mechanism of CD4 T Cell Mediated Control of M. tuberculosis Infection in vivo. PLoS Pathogens. 2011;7:e1002052. doi: 10.1371/journal.ppat.1002052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell host & microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusby MJ, Johnson RS, Papaioannou VE, Glimcher LH. Depletion of CD4+ T Cells in Major Histocompatibility Complex Class II-Deficient Mice. Science. 1991;253:1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by Tubercular Mycobacteria Is Spread by Nonlytic Ejection from Their Amoeba Hosts. Science. 2009:323. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondalus MK, Bardarov S, Russell R, Chan J, Jacobs WR, Bloom BR. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infection and immunity. 2000;68:2888–2898. doi: 10.1128/iai.68.5.2888-2898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nature reviews Microbiology. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibana JA, Belland RJ, Zea AH, Schust DJ, Nagamatsu T, AbdelRahman YM, Tate DJ, Beatty WL, Aiyar AA, Quayle AJ. Inhibition of indoleamine 2,3-dioxygenase activity by levo-1-methyl tryptophan blocks gamma interferon-induced Chlamydia trachomatis persistence in human epithelial cells. Infect Immun. 2011;79:4425–4437. doi: 10.1128/IAI.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John A, Bell JM. Amino Acid Requirements of the Growing Mouse. The Journal of Nutrition. 1976;106:1361–1367. doi: 10.1093/jn/106.9.1361. [DOI] [PubMed] [Google Scholar]
- Lee W, VanderVen BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesic B, Lepine F, Deziel E, Zhang J, Zhang Q, Padfield K, Castonguay MH, Milot S, Stachel S, Tzika AA, et al. Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 2007;3:1229–1239. doi: 10.1371/journal.ppat.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson P, Dimairo M, Bandason T, Zezai A, Munyati SS, Butterworth AE, Mungofa S, Rusakaniko S, Fielding K, Mason PR, et al. Risk factors for mortality in smear-negative tuberculosis suspects: a cohort study in Harare, Zimbabwe. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2011;15:1390–1396. doi: 10.5588/ijtld.11.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A. 2010;107:9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermid JM, Hennig BJ, van der Sande M, Hill AV, Whittle HC, Jaye A, Prentice AM. Host iron redistribution as a risk factor for incident tuberculosis in HIV infection: an 11-year retrospective cohort study. BMC infectious diseases. 2013;13:48. doi: 10.1186/1471-2334-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough K, Kress Y, Bloom BR. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infection and immunity. 1993;61:2763–2773. doi: 10.1128/iai.61.7.2763-2773.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- Mogues T, Goodrich ME, Ryan L, Lacourse R, North RJ. The Relative Importance of T Cell Subsets in Immunity and Immunopathology of Airborne Mycobacterium tuberculosis Infection in Mice. Journal of Experimental Medicine. 2001;193:271–280. doi: 10.1084/jem.193.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Elías EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nature medicine. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Elías EJ, McKinney JD. Carbon metabolism of intracellular bacteria. Cellular microbiology. 2006;8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- Nandi B, Behar SM. Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. J Exp Med. 2011;208:2251–2262. doi: 10.1084/jem.20110919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish T. Starvation survival response of Mycobacterium tuberculosis. Journal of Bacteriology. 2003;185:6702–6706. doi: 10.1128/JB.185.22.6702-6706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka MS, Chen B, Kelley CL, Collins FM, Jacobs WR., Jr Vaccine efficacy of a lysine auxotroph of Mycobacterium tuberculosis. Infection and immunity. 2003;71:4190–4192. doi: 10.1128/IAI.71.7.4190-4192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski A, Jansson M, Skold M, Rottenberg ME, Kallenius G. Tuberculosis and HIV co-infection. PLoS Pathog. 2012;8:e1002464. doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CTD, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science (New York, NY) 2011;334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- Russell DG. Who puts the tubercle in tuberculosis? Nature reviews Microbiology. 2007;5:39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanga CA, Mohan VP, Yu K, Joseph H, Tanaka KE, Chan J, Flynn JL. Depletion of CD4+ T Cells Causes Reactivation of Murine Persistent Tuberculosis Despite Continued Expression of Interferon-gamma and Nitric Oxide Synthase 2. Journal of Experimental Medicine. 2000;192:347–358. doi: 10.1084/jem.192.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn PA, Hartel D, Lewis VA, Schoenbaum EE, Vermund SH, Klein RS, Walker AT, Friedland GH. A Prospective Study of the Risk of Tuberculosis Among Intravenous Drug Users with Human Immunodeficiency Virus Infection. The New England Journal of Medicine. 1989;320:545–550. doi: 10.1056/NEJM198903023200901. [DOI] [PubMed] [Google Scholar]
- Skaar EP. The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts. PLoS Pathogens. 2010 doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Parish T, Stoker NG, Bancroft GJ. Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infection and immunity. 2001;69:1142–1150. doi: 10.1128/IAI.69.2.1142-1150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarshis M, Weed W. Lack of significant in vitro sensitivity of Mycobacterium tuberculosis to pyrazinamide on three different solid media. Am Rev Tuberc. 1953;67:391–395. doi: 10.1164/art.1953.67.3.391. [DOI] [PubMed] [Google Scholar]
- Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LAM, Yang C, Emili A, Philpott DJ, Girardin SE. Amino Acid Starvation Induced by Invasive Bacterial Pathogens Triggers an Innate Host Defense Program. Cell Host and Microbe. 2012;11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Vandal OH, Pierini LM, Schnappinger D, Nathan CF, Ehrt S. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nature medicine. 2008;14:849–854. doi: 10.1038/nmXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welin A, Eklund D, Stendahl O, Lerm M. Human macrophages infected with a high burden of ESAT-6-expressing M. tuberculosis undergo caspase-1- and cathepsin B-independent necrosis. PloS one. 2011;6:e20302. doi: 10.1371/journal.pone.0020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Protein and Amino Acid Requirements in Human Nutrition. 2007 [PubMed] [Google Scholar]
- WHO. Global Tuberculosis Control 2011. 2011 ( http://www.who.int/tb/publications/global_report/2011/en/)
- Zelante T, Fallarino F, Bistoni F, Puccetti P, Romani L. Indoleamine 2,3-dioxygenase in infection: the paradox of an evasive strategy that benefits the host. Microbes Infect. 2009a;11:133–141. doi: 10.1016/j.micinf.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Zelante T, Fallarino F, Bistoni F, Puccetti P, Romani L. Indoleamine 2,3-dioxygenase in infection: the paradox of an evasive strategy that benefits the host. Microbes and infection/Institut Pasteur. 2009b;11:133–141. doi: 10.1016/j.micinf.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: a review. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2003;7:6–21. [PubMed] [Google Scholar]
- Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Rubin EJ. Feast or famine: the host-pathogen battle over amino acids. Cell Microbiol. 2013;15:1079–1087. doi: 10.1111/cmi.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, Burghout P, Bootsma HJ, Hermans PW, van Hijum SA. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PloS one. 2012;7:e43012. doi: 10.1371/journal.pone.0043012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.