Abstract
While the standard therapy of AML has been relatively constant over the past two decades, this may be changing with enhanced technologies allowing for the classification of acute myeloid leukemia (AML) into molecularly distinct subsets. Some specific subsets of AML have an excellent prognosis in response to standard therapy while the poor prognosis of AML associated with specific sets of mutations or chromosomal anomalies require the development of new therapies. Elucidation of the molecular pathogenesis of AML has led to the development and of therapies that affect signaling, apoptosis, protein and intermediate metabolism, the surface of the leukemia cell, leukemia cell/stromal interaction and epigenetic regulation of gene expression.
Background
Despite decades of clinical research, induction therapy of Acute Myeloid Leukemia (AML) has remained virtually unchanged for 30 years. Between 20% to 40% of patients fail to achieve remission with induction chemotherapy, and 50%-70% of patients who achieve a complete remission (CR) relapse within 3 years. Over the past decades our insight into the pathogenesis and prognosis of AML has evolved substantially. In the 1980s markers of poor prognosis included age, elevated WBC count and an antecedent hematologic disorder. In late 1980s and 1990s recurrent chromosomal anomalies were formally incorporated into the World Health Criteria for the diagnosis of AML (1). Nearly 15% of patients have favorable karyotypic abnormalities comprised of t(15;17) and the core binding factor (CBF) leukemias which include t(8;21) or inversion 16, with 5-year survival rates of 65%. Another 15% have poor karyotypic features including deletion of chromosome 7, 5q or a complex karyotype comprising 3 of more chromosomal abnormalities with 5-year survival rates of 10-15%. However cytogenetic prognostication is limited by heterogeneity in the intermediate risk group, comprising 50% of patients with AML. This group encompasses patients characterized by +8, -Y, +6, del(12p), or cytogenetically normal AML (CN-AML) This cytogenetic prognostication was validated in large clinical trials (2, 3). A more comprehensive classification system proposed by the ELN incorporates cytogenetic and molecular abnormalities resulting in four prognostic sub-groups - favorable; intermediate I; intermediate II ; and adverse (4).
Advances in the Prognostic Classification of AML
Advances in molecular technologies have led to ever-finer classification of AML. Cytogenetic studies of the 1970s and 1980 were followed by the identification of fusion genes in the 1990s and elucidation of their mode of action in blocking myeloid differentiation and stimulating proliferation. In the 1990s and 2000s point mutations in key signaling molecules and transcription factors were identified. More recently microarray technologies allowed for the classification of AML by virtue of gene expression patterns, patterns of gene segment gain or loss DNA methylation patterns and microRNA expression. Over the past 3-4 years high throughput sequencing of genomes has led to yet further sub-classification of AML (5). Hence morphologically identical disease may encompass dozens of different molecular genetic subsets. This represents a challenge in designing therapy for the individual patient.
Gene Mutations
To date, assays for NPM1, CEBPA, FLT3, and cKIT mutations have entered clinical practice, affecting risk stratification and guiding therapy. Whole genome sequencing of AML identified new recurrent mutations in AML including DNMT3A (DNA methyl transferase 3A) and IDH1 and IDH2 (isocitrate dehydrogenase 1 and 2). Subsequently, mutations in DNMT3A, TET2, and ASXL1 have emerged as important adverse prognosticators in subsets of AML patients independent of FLT3 (6). Molecular profiling will guide dosing in induction chemotherapy as certain mutations (MLL and NPM1) respond better to higher doses of Daunorubicin while others such as FLT3 do not benefit. Furthermore, genotyping of AML patients will be used to guide post-induction strategies by avoiding high risk procedures such as transplant in the favorable prognostic sub-group. (See Table 1 for a compendium of recurrent AML mutations).
| Gene Mutation | Prevalence in CN-AML | Function | Prognostic Impact | Management strategy |
|---|---|---|---|---|
| NPM-1 (Nucleophosmin-1) | 50% | Nuclear shuttle protein that is aberrantly located in cytoplasm in leukemic blasts | Favorable in absence of Flt3-ITD mutation, benefit in older population | Standard induction chemotherapy followed by 3-4 cycles of high-dose cytarabine. Benefit for ATRA + chemotherapy in patients with the NPM-1 mutation. |
| Flt-3 ITD (Fms-like tyrosine kinase internal tandem domain mutation) | 20% | Class III receptor tyrosine kinase. Constitutively activated causing proliferation, survival and differentiation | Significantly worse than unmutated FLT-3. A higher ratio of mutant:wild type alleles predicts worse outcome | OS with allogeneic stem cell transplant equivalent to patients with wild-type FLT-3. New FLT-3 inhibitors currently in clinical trials. |
| CEBP-α (CCAAT/enhancer binding protein alpha) | 15-19% | Transcription factor critical to granulocyte maturation | With biallelic mutations, benefit in OS and RFS equivalent to the favorable sub-group of AML | Standard induction therapy followed by repetitive cycles of high-dose cytarabine is first-line treatment option; patients may not benefit from allogeneic HSCT in first CR |
| c-kit | Only relevant in core binding factor leukemias, mutated or over-expressed in 15-20% of CBF-AML | The SCF receptor tyrosine kinase. Mutations results in constitutive activation | In patients with CBF-AML, greater probability of relapse following CR | Low-dose Ara-C (LDAC) with Imatinib in elderly with c-kit over-expression non-inferior to standard chemotherapy. NCT00850382 is currently looking at Dasatinib in combination with 7+3 in CBF-AML. |
| MLL | 7-10% | Partial tandem duplication results in wild type allelic silencing by promoter DNA methylation and histone modifications | Shorter remission duration | Improved outcomes with stem cell transplant. Preclinical data suggest a therapeutic role of HDAC inhibitors and hypomethylating agents. |
| WT1 | 10% | transcriptional regulator | Inferior prognosis. Frequent coincident FLT3mutations | No impact on treatment at this time |
| RUNX | 10% | Transcription factor | Chemotherapy resistant disease. Worse EFS, RFS and OS | Patients do better with allogeneic stem cell transplants |
| RAS | 12-27% | Constitutively activating mutations in N-Ras and K-Ras, members of GTP-ase family | No prognostic significance | Patients benefit from high dose cytarabine consolidation. Consider MEK inhibitors on clinical trials in patients with Ras mutations |
| TET2 | 23% | Redced levels of 5-OH methylcytosine which is needed for DNA demethylation | Lower CR rate and shorter DFS among favorable-risk CN-AML in CALGB study. In AMLSG study, no prognostic impact. | None known |
| DNMT3A | 20% | Reduced enzymatic activity of DNA methytransferase results in decreased DNA methylation | Independently predict shorter overall survival (OS) | Benefit from high dose anthracycline induction chemotherapy (90mg/m2 daunorubicin) |
| IDH1, IDH2 | 33% | Gain of function mutations allowing them to convert α-ketoglutarate to 2-hydroxy-glutarate, which is a competitive inhibitorof histone demthylases | Prognostic impact depends on molecular context. IDH mutations co-existent with NPM-1 are associated with a good prognosis. | None known |
Single Gene Expression
Misexpression of specific genes has been associated with AML prognosis. For example, BAALC (brain and acute leukemia cytoplasmic) gene expression is frequently associated with other adverse molecular prognostic features, including FLT3-ITD, lack of NPM1 expression, and high ERG (ETS-related gene) transcription factor expression (7). Among the favorable-prognosis FLT3-ITD–negative/NPM1-positive patient subset, high ERG expressers had a ~4-fold higher risk of adverse outcome (8). High expression of the meningioma1 gene (MN1) also confers an unfavorable outcome in CN-AML (9).
Methylation profiles
Methylation profiling of newly diagnosed AML revealed distinct subgroups. Some methylation profiles segregated according to known cytogenetic and molecular abnormalities, additional clusters showed unique epigenetic signatures. Moreover, a 15-gene methylation classifier was defined and validated as an independent risk factor for survival in a multivariate analysis (10). The presence of a strong hypermethylation signature in some gene clusters is associated with a poor prognosis and this sub-group may benefit from hypomethylating agents.
Gene-Expression Profiling
Early gene expression profiling (GEP) of AML identified molecular subgroups with distinct gene-expression signatures (11). The clinical utility of GEP was established with the Microarray Innovations in Leukemia (MILE) multiple laboratory study, to assess the clinical accuracy of gene expression profiles of 16 acute and chronic leukemia subclasses in 3,334 patients (12). Profiling of CD34+, leukemic stem cells revealed that elevated expression of ANKRD28, GNA15 and UGP2 was correlated with poorer overall survival (OS) in CN-AML (13). Moreover, GEP of AML cells exposed to cytarabine allowed identification of critical mediators of AML cell survival such as the cell-cycle checkpoint protein WEE1 (14).
MicroRNA analysis
Expression profiles of micro-RNA in AML correlate with cytogenetic and molecular profile. While genome-wide microarray profiling is relatively cumbersome for risk assessment in individual patients, changes in a single microRNA (ie, miR-181a) were shown to independently predict for remission, disease free survival (DFS) and OS in CN-AML (15). Subsequently the functional relevance of microRNAs has been demonstrated in leukemogenesis, with some microRNAs acting as oncogenes, and others as tumor suppressors. For example, miR-29b modulates DNA methylation by targeting DNMT3a and DNMT3b and elevated levels can be utilized as a predictor of response to decitabine, a DNMT inhibitor (16).
Alterations in Metabolic Pathways
Neomorphic mutations in the metabolic enzymes IDH1 and IDH2 in AML result in the conversion of α-ketoglutarate to 2-hydroxyglutarate (2HG), an oncogenic metabolite that inhibits the action of the methylcytosine oxidase TET2 and results in DNA hypermethylation. 2HG-producing IDH mutants can prevent the histone demethylation that is required for lineage-specific progenitor cells to differentiate cells (17). These mutations were identified in 30% of patients with CN-AML and confer an adverse prognosis in younger (< 60 years), molecular low risk patients (18). Loss of function mutations in the α-ketoglutarate-dependent enzyme TET2 are mutually exclusive with IDH mutations, and are similarly associated with a worse prognosis among favorable-risk patients (19). The growth of myeloid cells harboring IDH1/2 mutations can be blocked by a specific inhibitor of the mutant enzyme representing a future clinical strategy (20).
On the Horizon
First Generation FLT3 inhibitors- lestaurtinib (CEP-701), midostaurin (PKC412), and sunitinib
The FLT3-ITD mutation leads to constitutive activation of the FMS-like tyrosine kinase 3 (FLT3) and proliferation of leukemic blasts. The first generation of FLT3 inhibitors had limited specificity and potency. A multicenter phase III trial that randomized patients with relapsed AML to induction chemotherapy alone or followed by lestaurtinib showed no survival benefit (21), attributed to the small proportion of patients that achieved FLT3 inhibition in vivo. Based on encouraging phase I data (22) a phase III study of daunorubicin/Ara-C with or without midostaurin for newly diagnosed FLT3+ AML was completed with results forthcoming.
Second Generation FLT3 inhibitors
AC220 demonstrates greater potency and selectivity in biochemical and cellular assays compared to first generation inhibitors (23). A phase I study of AC220 in relapsed/refractory AML reported 24% transient clinical responses, and 4/45 patients achieved a CR. The majority of the responders harbored FLT3 mutations (24). A phase II trial of AC220 in relapsed/refractory patients with mutant FLT3 AML showed reductions in marrow blasts in 45% of, patients and 1/3 of these patients were successfully bridged to hematopoietic stem cell transplantation (HSCT) (25).
Future of FLT3 inhibition
A retrospective study showed that the FLT3-ITD mutation resulted in a higher relapse rate after HSCT (26). As a result, a phase I trial is underway to find a safe dose of sorafenib for maintenance therapy after HSCT (NCT01398501). A phase I trial is also studying the combination of AC220 plus daunorubicin/Ara-C induction followed by consolidation high dose cytarabine plus AC220 (NCT01390337).
MEK inhibitors
Aberrant signaling through growth factor and cytokine receptors, Ras mutations, and Raf overexpression all converge on the MEK/ERK cascade. The oral MEK inhibitor AZD6244 (27) showed only minor responses in AML perhaps due to concomitant activation of the phosphoinositol 3-kinase (PI3K/AKT/mToR) pathway. Leukemia cells can be more effectively targeted by combination using combined MEK and PI3K/mToR inhibitors (28) and clinical trials of these combinations should be encouraged.
PI3K/mTORC inhibitors
Constitutive activation of the PI3K/AKT/mToR pathway occurs in 60-80% of cases of AML and is associated with shorter disease-free and overall survival (29). In vitro, the PI3K inhibitor LY294002 induced apoptosis of AML cells in a dose dependent manner (30). mTOR, a key kinase which activates metabolic pathways and cell growth exists in functionally distinct TORC1 and TORC2 complexes. Rapamycin analogues targeting mTORC1 were used in AML (31) with no evidence of synergy when combined with chemotherapy. This may be due to the formation of mTORC2 and Rapamycin-insensitive (RI)-mTORC1 complexes. Dual TORC1/2 inhibitors such as OSI-027 elicited far more potent suppression of leukemic cells (32). Dual inhibition of PI3K and mTORC1/2 with BEZ-235 also suppresses growth of AML cells (33).
Epigenetic modulation
HDAC Inhibitors (HDACI) can potentially activate genes abnormally suppressed in cancer cells, hence reversing the malignant phenotype. These agents also alter chromatin structure and may lead to increased genomic fragility and DNA damage. Single-agent HDACI therapy only yields 10-20% response rates (RR). Vorinostat, the first approved HDACI, had modest single agent activity, but combined with 5-azacitidine yielded a RR of 30% (34). The combination of vorinostat, with idarubicin and ara-C (AI) has synergistic activity with optimal effect when vorinostat precedes ara-C. In a phase II trial, the RR of 85% was superior to standard responses to AI alone (35)-notably there were responses in all patients with FLT3-ITD mutations. Median survival was 82 weeks, with a trend towards better survival in the Flt3-ITD patients. A phase I trial (NCT00875745) is examining the combination of Vorinostat and Sorafenib in AML and high-risk MDS. Encouraging results have also been reported using HDAC inhibitor MGCD0103 (36) with 5-azacytidine. Entinostat synergizes with GM-CSF to promote growth of mature myeloid cells and improves marrow function, minimizing the need for platelet transfusions (37). This strategy may be applied to patients with low disease burden.
Histone methyl-transferase activity
The MLL translocated leukemias result in recruitment of DOT1L, a histone 3 lysine 79 (H3K79) methyltransferase activity, to activate critical target genes (38). EPZ0l, a small molecule DOT1L inhibitor blocked tumor growth in a mouse model of MLL-fusion gene mediated leukemia (39). Due to its short half-life, further medicinal chemistry will be required to develop a clinical reagent.
Targeting histone-protein interactions
BRD4, a protein that reads the histone code, is critical for the growth of acute myeloid leukemia cells (40). JQ1, a recently developed small molecule inhibitor that blocks BRD4 from binding to acetylated histones (41). Treatment of AML cells with JQ1 suppressed the expression of the c-myc protooncogene and resulted in marked cell death (42). Another bromodomain inhibitor GSK1210151A was highly effective against human MLL-fusion cell lines and mouse models of MLL-fusion leukemia (43).
Aberrant DNMT activity
Through empirical clinical research, the DNA demethylating agents 5-azacitidine and decitabine have come into clinical use. The use of 5-azacytidine predated knowledge of the DNMT3A mutations or documentation of aberrant patterns of gene methylation in AML. These agents yield a RR of 20-3 % in MDS and AML (44). Clinical factors associated with 5-azacitidine response included untreated disease and WBC count <10×106/dl (45). While the administration of these agents leads to large scale loss of DNA methylation in vivo, it is uncertain whether this leads to normalization of aberrant patterns of gene regulation or leads to responses through induction of DNA damage. In AML, aberrant DNMT activity may play a role in epigenetic silencing of genes involved in hematopoiesis. DNMT3A mutation status was evaluated in 46 older patients with untreated AML who received decitabine and patients with low DNMT3A activity appeared to benefit (46). Further genome-wide studies of DNA methylation patterns in AML will be required determine if the response to DNMTi can be linked to the presence of DNMT3A or other mutations that affect DNA methylation such as TET2, IDH1 or IDH2.
Targeting Protein Metabolism
Hsp90 inhibitors
Heat shock protein 90 (Hsp90) is a molecular chaperone for many oncogenic client proteins such as receptor tyrosine kinases. In a phase I trial, the Hsp90 inhibitor alvespimycin, increased apoptosis of marrow blasts and induced complete responses in 3/17 patients (47). Hsp90 inhibition can be potentiated through increased acetylation by histone deacetylase inhibitors (48). Mutated forms of FLT3 are more dependent on chaperone molecules than the wild-type molecules. 17-AAG, an Hsp inhibitor shows additive efficacy with the FLT3 inhibitor PKC412 in preclinical models (49).
IMIDS-Lenalidomide
Patients with del(5q) MDS display a unique sensitivity to lenalidomide where the drug exerts karyotype-specific clonal suppression. Lenalidomide may upregulate tumor suppressor genes activated by azacitidine (50); in addition, lenalidomide can upregulate the p21 gene through activation of lysine demethylases (51). A phase I trial used sequential azacitidine and lenalidomide in elderly, untreated AML patients, resulting in a 44% CR rates with a median response of 6.2 months. In a phase 2 study, elderly patients received high-dose (HD) lenalidomide at 50 mg daily for up to two cycles followed by maintenance. The CR rate was 30%, with a median duration of 10 months (52). The elusive target of this class of drugs was recently shown to be Cereblon (53). When bound by thalidomide, cereblon inhibits the oncogenic Cul4A E3 ligase. Whether this is how lenalidomide affects AML remains to be determined.
Targeting Apoptosis
Elevated expression of anti-apoptotic molecules is associated with chemotherapy resistance in AML. Oblimersen sodium, a BCL-2 antisense oligonucleotide, was evaluated in combination with daunorubicinin/AraC in a phase I trial but a phase III trial was halted as it did not result in improved OS. ABT-737, another small molecule BCL-2 inhibitor slowed tumor growth in xenograft models of AML and potentiated a number of chemotherapeutic agents (54). AEG35156, an X-linked inhibitor of apoptosis (XIAP) antisense oligonucleotide, when combined with idarubicin/cytarabine reinduction resulted in a CR/CRp rate of 91% (10/11) in refractory patients and 9 of these 11 patients could then be transplanted (55).
Leukemia/Stromal Interactions
The interaction of AML blasts with the marrow microenvironment through the CXCR4/CXCL1 axis appears to be an important mediator of resistance to chemotherapy. In a murine model the CXCR4 antagonist plerixafor released of leukemic cells from protective marrow niches, enhancing the efficacy of chemotherapy (56). In a phase I/II study, 52 patients with relapsed/refractory AML were treated with plerixafor plus mitoxantrone, etoposide and cytarabine, leading to a 46% CR rate (57) in association with a two-fold mobilization in leukemic blasts into the peripheral circulation. The utility of this strategy requires confirmation in a randomized trial.
Targeting the Cell Surface- Gemtuzumab Ozogamicin
Gemtuzumab ozogamicin (GO), an anti-CD33 immunoconjugate showed remission rates of about 25% in relapsed/refractory AML resulting in FDA approval of GO nearly a decade ago. GO was subsequently removed from the market after a large SWOG trial failed to showed that GO+chemotherapy yielded no survival benefit and a high early death rate (58). GO may yet have a role in subsets of AML. In young patients with favorable cytogenetics, GO increased survival when combined with induction chemotherapy (59). Two phase III trials of elderly AML using a fractionated dose of GO in combination with Daunorubicin/AraC in elderly AML showed significant improvement in RFS and OS (60) (61). Whether this drug will be re-introduced into clinical practice remains to be determined.
Conclusion
Molecular profiling of AML is now impacting treatment decisions in AML. Genetic analysis of samples from E1900 demonstrated that the more intensive 90 mg/m2 dose of daunorubicin in induction chemotherapy was associated with improved survival in patients with DNMT3A, NPM1 mutations or MLL translocations. The favorable impact of IDH1/NPM1 mutations in CN-AML, if confirmed, might obviate the need for aggressive consolidation and stem cell transplant in these patients. Conversely, patients with FLT3 mutations have a poor prognosis and may need anti-kinase therapy added to typical regimens. Other agents such as MEK/ERK and AKT axis inhibitors are well tolerated, display modest efficacy, but may yet find a place in AML treatment in combination with chemotherapy. Hypomethylating agents have been empirically used in hypoproliferative AML with moderate success, in some cases prolonging survival in the absence of achieving a CR. Whether specific anomalies in DNA methylation patterns of mutations in the DNA methylation machinery predicts success of these agents remains to be determined. HDAC inhibitors may have a role in combination with either chemotherapy or hypomethyalting agents.
It is important to emphasize the need for biologic insight-directed use of novel agents. The high expression of CD33 on the leukemic blasts of patients with NPM1 mutations suggests that trials using GO in this population may have yielded better results. There remains interest in developing targeting monoclonal antibodies against antigens unique to the leukemia stem cells and vaccine trials against antigens such as WT1 continue.
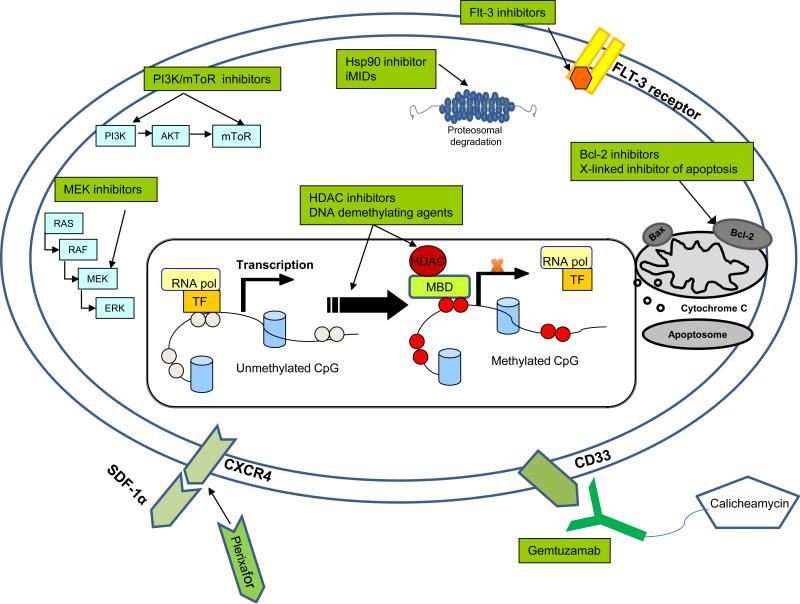
Advances in next-generation sequencing technologies should soon lead to implementation of comprehensive genetic profiling in the clinical care of AML patients. As the list of genes mutated and pathways deregulated in AML grows, more targets for therapy will be investigated (see Figure 1). AML will be subclassifed into ever-smaller subsets some of which may not be amenable to specific targeted therapy. Challenges ahead include dissecting the hierarchical significance of multiple mutations identified in these patients and finding therapies robust enough to cross over many of the genetic subsets.
Figure 1. Therapeutic Targets in AML.
Therapy may be directed against signals generated by growth factor receptors (IL3-R, FLT3) by direct kinase inhibition, by inhibition of downstream signaling (MEK), (PI3K/AKT/mTOR) or by interference with the HSP90 chaperone protein. Additional targets on the horizon include migration/stromal interactions (SDF-1/CXCR4), anti-apoptotic molecules (BCL2), or aberrant DNA methylation and histone methylation. The leukemia cell surface may be targeted by anti-CD33 antibodies, other monoclonal antibodies or tumor vaccines.
Acknowledgments
Supported by NCI T32 CA079447 (IK), A Leukemia and Lymphoma Society Specialized Center of Research Excellence (JDL) and a K12 award (JKA) through 8UL1TR000150.
Footnotes
Conflict of Interest: JDL- Research Support from Epizyme, Inc. JKA- Advisory Boards for Cellgene, Astellas, Teva
References
- 1.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 2.Farag SS, Archer KJ, Mrozek K, Ruppert AS, Carroll AJ, Vardiman JW, et al. Pretreatment cytogenetics add to other prognostic factors predicting complete remission and long-term outcome in patients 60 years of age or older with acute myeloid leukemia: results from Cancer and Leukemia Group B 8461. Blood. 2006;108:63–73. doi: 10.1182/blood-2005-11-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100:4325–36. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 4.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 115:453–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 5.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzeler KH, Dufour A, Benthaus T, Hummel M, Sauerland MC, Heinecke A, et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27:5031–8. doi: 10.1200/JCO.2008.20.5328. [DOI] [PubMed] [Google Scholar]
- 8.Marcucci G, Maharry K, Whitman SP, Vukosavljevic T, Paschka P, Langer C, et al. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B Study. J Clin Oncol. 2007;25:3337–43. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]
- 9.Schwind S, Marcucci G, Kohlschmidt J, Radmacher MD, Mrozek K, Maharry K, et al. Low expression of MN1 associates with better treatment response in older patients with de novo cytogenetically normal acute myeloid leukemia. Blood. 2011;118:4188–98. doi: 10.1182/blood-2011-06-357764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–28. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 12.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–37. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Jonge HJ, Woolthuis CM, Vos AZ, Mulder A, van den Berg E, Kluin PM, et al. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia. 2011;25:1825–33. doi: 10.1038/leu.2011.172. [DOI] [PubMed] [Google Scholar]
- 14.Porter CC, Kim J, Fosmire S, Gearheart CM, van Linden A, Baturin D, et al. Integrated genomic analyses identify WEE1 as a critical mediator of cell fate and a novel therapeutic target in acute myeloid leukemia. Leukemia. 2012 doi: 10.1038/leu.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwind S, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:5257–64. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blum W, Garzon R, Klisovic RB, Schwind S, Walker A, Geyer S, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci U S A. 2010;107:7473–8. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012 doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–43. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 19.Metzeler KH, Maharry K, Radmacher MD, Mrozek K, Margeson D, Becker H, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2011;29:1373–81. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yen K, Wang F, Choe S, Schalm S, Hansen E, Straley K, et al. IDH mutations and tumorgenicity. AACR Annual Meeting. 2012 [Google Scholar]
- 21.Levis M, Ravandi F, Wang ES, Baer MR, Perl A, Coutre S, et al. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood. 2011;117:3294–301. doi: 10.1182/blood-2010-08-301796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stone RM FT, Paquette R. A phase 1b study of midostaurin (PKC412) in combination with daunorubicin and cytarabine induction and high-dose cytarabine consolidation in patients under age 61 with newly diagnosed de novo acute myeloid leukemia: Overall survival of patients whose blasts have FLT3 mutations is similar to those with wild-type FLT3. Blood. 2009;114 [Google Scholar]
- 23.Zarrinkar PP, Gunawardane RN, Cramer MD, Gardner MF, Brigham D, Belli B, et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML). Blood. 2009;114:2984–92. doi: 10.1182/blood-2009-05-222034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortes J, Foran J, Ghirdaladze D, DeVetten MP, Zodelava M, Holman P, et al. AC220, a Potent, Selective, Second Generation FLT3 Receptor Tyrosine Kinase (RTK) Inhibitor, in a First-in-Human (FIH) Phase 1 AML Study. ASH Annual Meeting Abstracts. 2009;114:636. [Google Scholar]
- 25.Cortes JE, Perl AE, Smith CC, Kovacsovics T, Dombret H, Dohner H, et al. A Phase II Open-Label, Ac220 Monotherapy Efficacy Study In Patients with Refractory/Relapsed Flt3-Itd Positive Acute Myeloid Leukemia: Updated Interim Results. ASH Annual Meeting Abstracts. 2011;118:2576. [Google Scholar]
- 26.Brunet S, Labopin M, Esteve J, Cornelissen J, Socie G, Iori AP, et al. Impact of FLT3 Internal Tandem Duplication on the Outcome of Related and Unrelated Hematopoietic Transplantation for Adult Acute Myeloid Leukemia in First Remission: A Retrospective Analysis. J Clin Oncol. 2012;30:735–41. doi: 10.1200/JCO.2011.36.9868. [DOI] [PubMed] [Google Scholar]
- 27.Odenike O, Curran E, Iyengar N, Popplewell L, Kirschbaum M, Erba HP, et al. Phase II Study of the Oral MEK Inhibitor AZD6244 in Advanced Acute Myeloid Leukemia (AML). ASH Annual Meeting Abstracts. 2009;114:2081. [Google Scholar]
- 28.Liu H, Diaz-Flores E, Poire X, Odenike O, Koval G, Malnassy G, et al. Combination of a MEK Inhibitor, AZD6244, and Dual PI3K/mTOR Inhibitor, NVP-BEZ235: An Effective Therapeutic Strategy for Acute Myeloid Leukemia. ASH Annual Meeting Abstracts. 2010;116:3978. [Google Scholar]
- 29.Min YH, Eom JI, Cheong JW, Maeng HO, Kim JY, Jeung HK, et al. Constitutive phosphorylation of Akt/PKB protein in acute myeloid leukemia: its significance as a prognostic variable. Leukemia. 2003;17:995–7. doi: 10.1038/sj.leu.2402874. [DOI] [PubMed] [Google Scholar]
- 30.Xu Q, Simpson SE, Scialla TJ, Bagg A, Carroll M. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood. 2003;102:972–80. doi: 10.1182/blood-2002-11-3429. [DOI] [PubMed] [Google Scholar]
- 31.Perl AE, Kasner MT, Tsai DE, Vogl DT, Loren AW, Schuster SJ, et al. A phase I study of the mammalian target of rapamycin inhibitor sirolimus and MEC chemotherapy in relapsed and refractory acute myelogenous leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6732–9. doi: 10.1158/1078-0432.CCR-09-0842. [DOI] [PubMed] [Google Scholar]
- 32.Altman JK, Sassano A, Kaur S, Glaser H, Kroczynska B, Redig AJ, et al. Dual mTORC2/mTORC1 targeting results in potent suppressive effects on acute myeloid leukemia (AML) progenitors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4378–88. doi: 10.1158/1078-0432.CCR-10-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapuis N, Tamburini J, Green AS, Vignon C, Bardet V, Neyret A, et al. Dual inhibition of PI3K and mTORC1/2 signaling by NVP-BEZ235 as a new therapeutic strategy for acute myeloid leukemia. Clin Cancer Res. 2010;16:5424–35. doi: 10.1158/1078-0432.CCR-10-1102. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Manero G, Estey EH, Jabbour E, Borthakur G, Kadia T, Naqvi K, et al. Final Report of a Phase II Study of 5-Azacitidine and Vorinostat in Patients (pts) with Newly Diagnosed Myelodysplastic Syndrome (MDS) or Acute Myelogenous Leukemia (AML) Not Eligible for Clinical Trials Because Poor Performance and Presence of Other Comorbidities. ASH Annual Meeting Abstracts. 2011;118:608. [Google Scholar]
- 35.Garcia-Manero G, Tambaro FP, Bekele NB, Yang H, Ravandi F, Jabbour E, et al. Phase II Trial of Vorinostat With Idarubicin and Cytarabine for Patients With Newly Diagnosed Acute Myelogenous Leukemia or Myelodysplastic Syndrome. J Clin Oncol. doi: 10.1200/JCO.2011.38.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Manero G, Yang AS, Giles F, Faderl S, Ravandi F, Cortes J, et al. Phase I/II Study of the Oral Isotype-Selective Histone Deacetylase (HDAC) Inhibitor MGCD0103 in Combination with Azacitidine in Patients (pts) with High-Risk Myelodysplastic Syndrome (MDS) or Acute Myelogenous Leukemia (AML). ASH Annual Meeting Abstracts. 2006;108:1954. [Google Scholar]
- 37.Cho Eunpi WM, Kowalski Jeanne, Tsai Hua-Ling, Jones Richard J., Smith B. Douglas. Combination Therapy with Entinostat (MS-275) and GM-CSF to Enhance Differentiation In Myeloid Malignancies. Blood. 2010 Nov;116 Abstract 3312. [Google Scholar]
- 38.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cashen AF, Schiller GJ, O'Donnell MR, DiPersio JF. Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. J Clin Oncol. 2010;28:556–61. doi: 10.1200/JCO.2009.23.9178. [DOI] [PubMed] [Google Scholar]
- 43.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Gattermann N, Germing U, et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J Clin Oncol. 2010;28:562–9. doi: 10.1200/JCO.2009.23.8329. [DOI] [PubMed] [Google Scholar]
- 45.Maurillo L, Venditti A, Spagnoli A, Gaidano G, Ferrero D, Oliva E, et al. Azacitidine for the treatment of patients with acute myeloid leukemia: report of 82 patients enrolled in an Italian Compassionate Program. Cancer. 2012;118:1014–22. doi: 10.1002/cncr.26354. [DOI] [PubMed] [Google Scholar]
- 46.Metzeler KH, Walker A, Geyer S, Garzon R, Klisovic RB, Bloomfield CD, et al. DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia. 2011 doi: 10.1038/leu.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lancet JE, Gojo I, Burton M, Quinn M, Tighe SM, Kersey K, et al. Phase I study of the heat shock protein 90 inhibitor alvespimycin (KOS-1022, 17-DMAG) administered intravenously twice weekly to patients with acute myeloid leukemia. Leukemia. 2010;24:699–705. doi: 10.1038/leu.2009.292. [DOI] [PubMed] [Google Scholar]
- 48.Rao R, Fiskus W, Yang Y, Lee P, Joshi R, Fernandez P, et al. HDAC6 inhibition enhances 17-AAG--mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood. 2008;112:1886–93. doi: 10.1182/blood-2008-03-143644. [DOI] [PubMed] [Google Scholar]
- 49.Bali P, George P, Cohen P, Tao J, Guo F, Sigua C, et al. Superior activity of the combination of histone deacetylase inhibitor LAQ824 and the FLT-3 kinase inhibitor PKC412 against human acute myelogenous leukemia cells with mutant FLT-3. Clin Cancer Res. 2004;10:4991–7. doi: 10.1158/1078-0432.CCR-04-0210. [DOI] [PubMed] [Google Scholar]
- 50.Pellagatti A, Jadersten M, Forsblom AM, Cattan H, Christensson B, Emanuelsson EK, et al. Lenalidomide inhibits the malignant clone and up-regulates the SPARC gene mapping to the commonly deleted region in 5q- syndrome patients. Proc Natl Acad Sci U S A. 2007;104:11406–11. doi: 10.1073/pnas.0610477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escoubet-Lozach L, Lin IL, Jensen-Pergakes K, Brady HA, Gandhi AK, Schafer PH, et al. Pomalidomide and lenalidomide induce p21 WAF-1 expression in both lymphoma and multiple myeloma through a LSD1-mediated epigenetic mechanism. Cancer Res. 2009;69:7347–56. doi: 10.1158/0008-5472.CAN-08-4898. [DOI] [PubMed] [Google Scholar]
- 52.Fehniger TA, Uy GL, Trinkaus K, Nelson AD, Demland J, Abboud CN, et al. A phase 2 study of high-dose lenalidomide as initial therapy for older patients with acute myeloid leukemia. Blood. 2011;117:1828–33. doi: 10.1182/blood-2010-07-297143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–50. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 54.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 55.Schimmer AD, Estey EH, Borthakur G, Carter BZ, Schiller GJ, Tallman MS, et al. Phase I/II trial of AEG35156 X-linked inhibitor of apoptosis protein antisense oligonucleotide combined with idarubicin and cytarabine in patients with relapsed or primary refractory acute myeloid leukemia. J Clin Oncol. 2009;27:4741–6. doi: 10.1200/JCO.2009.21.8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, et al. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113:6206–14. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, Hladnik LM, et al. A phase I/II study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood. 2012 doi: 10.1182/blood-2011-10-383406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersdorf S, Kopecky K, Stuart RK, Larson RA, Nevill TJ, Stenke L, et al. Preliminary Results of Southwest Oncology Group Study S0106: An International Intergroup Phase 3 Randomized Trial Comparing the Addition of Gemtuzumab Ozogamicin to Standard Induction Therapy Versus Standard Induction Therapy Followed by a Second Randomization to Post-Consolidation Gemtuzumab Ozogamicin Versus No Additional Therapy for Previously Untreated Acute Myeloid Leukemia. ASH Annual Meeting Abstracts. 2009;114:790. [Google Scholar]
- 59.Burnett AK, Hills RK, Milligan D, Kjeldsen L, Kell J, Russell NH, et al. Identification of patients with acute myeloblastic leukemia who benefit from the addition of gemtuzumab ozogamicin: results of the MRC AML15 trial. J Clin Oncol. 2011;29:369–77. doi: 10.1200/JCO.2010.31.4310. [DOI] [PubMed] [Google Scholar]
- 60.Castaigne S, Pautas C, Terre C, Raffoux E, Bordessoule D, Bastie JN, et al. Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): a randomised, open-label, phase 3 study. Lancet. 2012;379:1508–16. doi: 10.1016/S0140-6736(12)60485-1. [DOI] [PubMed] [Google Scholar]
- 61.Burnett AK, Hills RK, Hunter AE, Milligan D, Kell WJ, Wheatley K, et al. The Addition of Gemtuzumab Ozogamicin to Intensive Chemotherapy in Older Patients with AML Produces a Significant Improvement in Overall Survival: Results of the UK NCRI AML16 Randomized Trial. ASH Annual Meeting Abstracts. 2011;118:582. [Google Scholar]