Abstract
Influenza A and B viruses are responsible for the severe morbidity and mortality worldwide in annual influenza epidemics. Currently circulating influenza B virus belongs to the B/Victoria or B/Yamagata lineage that was diverged from each other about 30–40 years ago. However, a mechanistic understanding of their divergent evolution is still lacking. Here we report the crystal structures of influenza B/Yamanashi/166/1998 hemagglutinin (HA) belonging to B/Yamagata lineage and its complex with the avian-like receptor analogue. Comparison of these structures with those of undiverged and diverged influenza B virus HAs, in conjunction with sequence analysis, reveals the molecular basis for the divergent evolution of influenza B virus HAs. Furthermore, HAs of diverged influenza B virus strains display much stronger molecular interactions with terminal sialic acid of bound receptors, which may allow for a different tissue tropism for current influenza B viruses, for which further investigation is required.
Keywords: Divergent evolution, Hemagglutinin, Influenza B virus, Positive selective pressure, Receptor binding, Sialic acid receptors
Introduction
Influenza A and B viruses of the Orthomyxoviridae family are responsible for seasonal influenza epidemics with severe morbidity and mortality worldwide (Palese and Shaw, 2007; Taubenberger and Morens, 2008; Wright et al., 2007). Annual influenza vaccine has been conventionally used to reduce human suffering and economic burden caused by the viruses. However, the constant genetic and antigenic changes of influenza viruses render them the ability to evade host immune system (Nelson and Holmes, 2007), thus limiting the vaccine effectiveness.
Influenza B virus was first isolated in 1940 and has diverged into two genetically and antigenically distinct lineages since 1983 or earlier (Chen et al., 2007; Shen et al., 2009): the B/Victoria lineage represented by the reference strain B/Victoria/2/87, and the B/Yamagata lineage represented by the B/Yamagata/16/88 strain, respectively (Kanegae et al., 1990; Rota et al., 1990; Shaw et al., 2002). The B/Victoria lineage predominated during the 1980s, while the B/Yamagata lineage predominated in most part of the world during the 1990s (Lin et al., 2004). The B/Victoria lineage re-emerged in Europe and United States in 2001, and the two lineages have co-circulated ever since (Ikonen et al., 2005; Shaw et al., 2002).
As one of the two major surface glycoproteins of influenza virus, hemagglutinin (HA) mediates host entry of the virus and is a primary target for host neutralizing antibodies (Han and Marasco, 2011). The precursor of HA, HA0, is synthesized as a single-stranded polypeptide, which is then cleaved into two disulfide-bonded subunits: HA1 and HA2 (Copeland et al., 1986). HA1 contains the receptor-binding site and harbors the majority of antigenic sites that undergo constant antigenic variations (Knossow and Skehel, 2006). On the other hand, HA2, which contains the fusion peptide at its N-terminus and is responsible for inducing fusion of viral envelope and endosomal membrane, is the most conserved (Vareckova et al., 2003). In an infection, HA first binds to the sialic-acid receptors on the host cell surface, thus triggering the internalization of the virus by endocytosis (Matlin et al., 1981; Skehel and Wiley, 2000). The low-pH environment in the late endosome results in the protonation of multiple negative charged residues located at the HA1–HA1 and HA1–HA2 interfaces, thereby dissociating HA1 and HA2 (Korte et al., 2007; Rachakonda et al., 2007; Wang et al., 2008). The subsequent large-scale conformational change in HA2 (Bullough et al., 1994; Chen et al., 1999) fuses the viral and endosomal membranes and allows the delivery of viral genetic materials into cellular cytosol.
The determination of the first crystal structure of the ectodo-main of influenza virus B/Hongkong/8/1973 (B/HK/73) HA has allowed the mapping of its antigenic structure (Wang et al., 2008). There are four major epitopes on the membrane-distal globular domain of influenza B virus HA: the 120-loop (HA1116–137), the 150-loop (HA1141–150), the 160-loop (HA1162–167), and the 190-helix (HA1194–202) and their respective surrounding regions. All these four epitopes have been demonstrated to be under positive selective pressure in the course of evolution (Nunes et al., 2008; Pechirra et al., 2005; Shen et al., 2009). However, the mechanism by which amino-acid substitutions change the antigenic property of HA remains elusive.
Here we report the crystal structure of influenza B/Yamanashi/166/1998 (B/Yamanashi/98) HA determined to 3.54-Å resolution (Table 1), a strain belonging to the B/Yamagata lineage. Together with the recently determined structure of influenza B/Brisbane/60/2008 (B/Brisbane/08) HA (B/Victoria lineage) and the complex structure of the membrane-distal globular domain of influenza B/Florida/4/2006 (B/Florida/06) HA (B/Yamagata lineage) in the region of HA133–324 with its antibody (Dreyfus et al., 2012), we performed a systematic structural comparison to characterize the evolution of HA antigenicity at the molecular level. In addition, we have also determined the crystal structure of B/Yamanashi/98 HA in complex with avian-like receptor analogue to 2.50-Å resolution. This, in comparison with the structure of B/HK/73 HA complexed with avian-like receptor analogue (Wang et al., 2007), reveals that the diverged influenza B virus HAs have developed a distinct and much more efficient receptor-binding site than early strains.
Table 1.
Data collection and refinement statistics.
| B/Yamanashi/98 HA | B/Yamamashi/98 HA-LSTa | |
|---|---|---|
| Data collection statistics | ||
| Resolution range (Å) | 43.6–3.54 (3.73–3.54) | 39.37–2.50 (2.64–2.50) |
| Space group | C121 | C121 |
| Unit cell | ||
| a, b, c (Å) | 174.9, 101.3, 136.8 | 176.1,101.5, 137.4 |
| α, β, γ (s=deg) | 90, 115.2, 90 | 90, 115.2, 90 |
| Unique reflections | 26,547 | 75,849 |
| Multiplicity | 3.8 (3.8) | 3.8 (3.8) |
| Completeness (%) | 99.98 (100.00) | 99.99 (100.00) |
| Mean l/sigma(l) | 10.6 (2.3) | 11.7 (2.3) |
| Wilson B-factor (Å2) | 109.1 | 46.7 |
| Rmerge (%) | 9.8 (49.7) | 9.1 (49.4) |
| Refinement statistics | ||
| Rcryst (%) | 19.8 (27.2) | 18.8 (26.2) |
| Rfree (%) | 24.4 (30.9) | 22.4 (32.0) |
| Number of atoms | 12,115 | 12,754 |
| Protein | 11,695 | 11,695 |
| Ligands | 420 | 588 |
| Water | 0 | 471 |
| RMSD bond length (Å)/bond angle (°) | 0.004/1.03 | 0.007/0.87 |
| Ramachandran plot | ||
| Favored, allowed, disallowed (%) | 96.3, 3.6, 0.1 | 96.5, 3.4, 0.1 |
| Average B-factor (Å2) | 120.6 | 50.3 |
Statistics for the highest-resolution shell are shown in parentheses. The coordinates and structure factors of B/Yamanashi/98 HA and B/Yamanashi/98 HA-LSTa have been deposited into Protein Data Bank under accession codes of 4M40 and 4M44, respectively.
Results
The overall structure of B/Yamanashi/98 HA
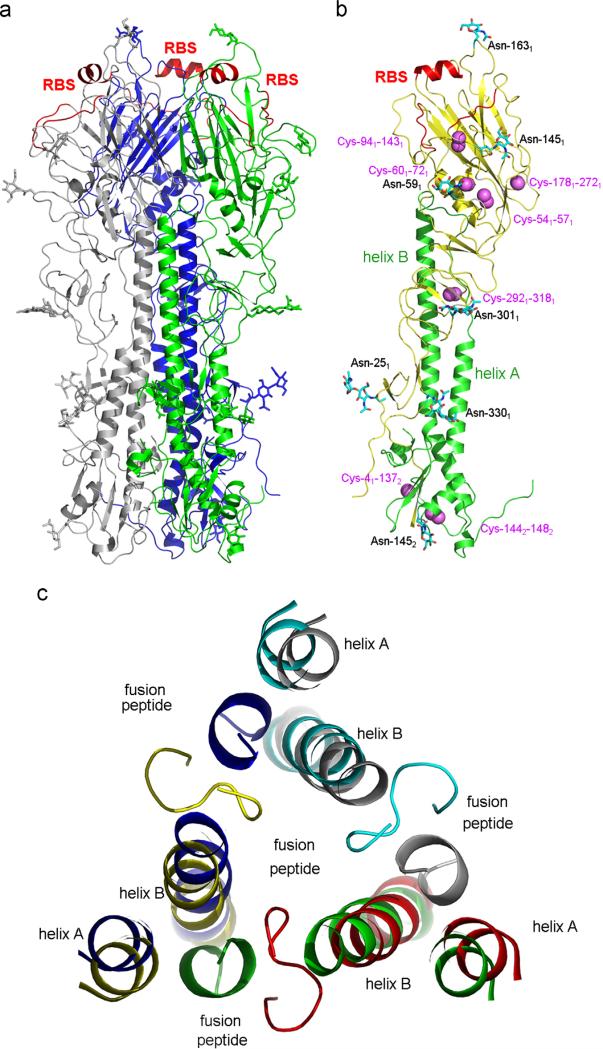
The structure of unliganded B/Yamanashi/HA is determined to 3.54-Å resolution (Table 1). There is one HA trimer in an asymmetric unit (Fig. 1a). Each subunit of B/Yamanashi/98 HA has the same seven disulfide bridges as found in B/HK/73 HA (Wang et al., 2008): Cys41–Cys1372, Cys541–Cys571, Cys601–Cys721, Cys941–Cys1431, Cys1781–Cys2721, Cys2921–Cys3181 and Cys1442–Cys1482 (Fig. 1b).
Fig. 1.
Structure of B/Yamanashi/98 HA. (a) Ribbon diagram of B/Yamanashi/98 HA structure with three subunits differently colored (grey, blue and green). The receptor-binding site (RBS) is highlighted in red color. Glycans resolved in the crystal structure are also shown. (b) Structure of a B/Yamanashi/98 HA subunit. HA1 and HA2 are in yellow and green color, respectively. The two major helices on HA2, helix A and helix B, are also labelled. Disulfide bonds on each subunit are shown as space-filling models in purple, and seven sugar residues are in ball-and-stick models. The RBS is labelled in red. (c) Comparison of the fusion peptide between B/Yamanashi/98 HA and influenza A/H3 virus HA (PDB code: 3HMG). The helix A and helix B and fusion peptide from each subunit are shown in different colors: green, blue and grey for B/Yamanashi/98 HA, and red, yellow and cyan for influenza A/H3 virus HA. It is clear that the fusion peptide of B/Yamanashi/98 HA points away from its helix A and helix B and interacts with the helices from a neighbouring subunit. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
B/Yamanashi/98 HA has nine predicted glycosylation sites (six on HA1: 25, 59, 145, 163, 301, 330; and three on HA2: 145, 171, and 184) on each subunit. All glycans are built in the final structure (Fig. 1a and b) with the exception of HA2171 for which no clear density is observed and HA2184 that was not present in the construct used for crystallization. The glycosylation at HA1145 that first appeared in B/Great Lakes/54 HA has been maintained up to today. On the other hand, the glycosylation at HA1123 as found in B/HK/73 HA (Wang et al., 2008) is completely abolished in HAs of both B/Victoria and B/Yamagata lineages due to the predominant presence of Ile-125 instead of Thr-125 (Table 2). It is not clear why the glycosylation site at HA1123 is so short-lived. One explanation is that since the 120-loop is a strong epitope of influenza B virus HA, different mutational strategies have been used to evade recognition by host immunity. The glycosylation site at HA1123 in B/HK/73 is just one such strategy and did not get populated in subsequent influenza B virus strains. However, the majority of B/Victoria lineage strains have established a new glycosylation site at HA1230, which is present in 50% of early influenza B virus strains, and not present in the majority of B/Yamagata lineage strains (Table 2).
Table 2.
Frequent amino-acid substitutions in influenza B virus HAs.
| Residues at sitea | HA (1972–1982, six sequences) | HA (Victoria, 278 sequences) | HA (Yamagata, 282 sequences) |
|---|---|---|---|
| 120-Loop | |||
| 48 | 50% Q, 50% K | 75% E, 22% K | 58% R, 41% K |
| 56 | 100% N | 95% K, 1% D | 65% D, 20% T, 12% N |
| 71 | 100% K | 98% K, 1% M | 99% M |
| 75 | 83% T, 16% N | 47% K, 46% N, 3% T | 75% T, 18% I, 3% V |
| 116 | 100% N | 94% H, 2% R, 2% N | 73% N, 22% K, 3% H |
| 122 | 100% R | 97% H, 1% Q | 94% Q, 5% H |
| 125 | 66% I, 33% T | 99% I | 99% I |
| 129 | 50% R, 50% T | 75% N, 21% K, 2% T | 89% K, 7% R, 3% N |
| 179-181 | 100% TKG | 97% TEG | 60% TEG, 21% TKE, 11% TKG |
| 150-Loop | |||
| 148–150 | 100% NGN | 96% NGN, 2% NGI, 1% SKI | 47% SKS, 28% SKI, 20% SRS, 1% SRD |
| 160-Loop | |||
| Insertion at 162–163 | - | 51% NDK, 42% NDN, 3% NEN | 88% DN, 9%D, 1%GN, 1% EN |
| 190-Helix | |||
| 195 | 83% E, 17% A | 97% E, 2% K | 98% K |
| 199 | 100% V | 94% A, 3% V | 99% K |
| 206 | 100% K | 97% K, 1% N | 99% N |
| 230 | 100% N | 97% N, 1% D | 87% D, 7% N, 3% A |
| 232 | 50% T, 50% A | 94% T, 4% A | 98% T, 1% K |
| 235 | 50% E, 50% G | 100% G | 99% G |
| RBS | |||
| 136 | 100% I | 97% K, 1% R | 98% R, 1% K |
| HA2 | |||
| 132 | 83% D, 16% E | 97% E, 2% D | 88% D, 11% E |
| 158 | 100% N | 89% D, 10% N | 98% N, 1% D |
The residues are numbered according to that of B/HK/73 HA.
Similar to B/HK/73 HA (Wang et al., 2008), the fusion peptide of B/Yamanashi/98 HA located at the N-terminus of HA2 points away from its own helix A and helix B to interact with those of a neighbouring subunit instead (Fig. 1c).
Antigenic structure of influenza B virus HAs
Known influenza B virus HA structures are of B/HK/73 representing undiverged early strains (PDB codes: 3BT6, 2RFT and 2RFU) (Wang et al., 2008, 2007), B/Brisbane/08 representing the B/Victoria lineage (PDB code: 4FQM) (Dreyfus et al., 2012), B/Florida/06 (in the region of HA133–324, PDB code: 4FQJ) (Dreyfus et al., 2012) and B/Yamanashi/98 belonging to the B/Yamagata lineage. We performed a structure-based analysis of the divergent evolution of these HAs. Since the B/Florida/06 HA is in complex with human monoclonal antibody CR8071 in the region of residues HA137–41, 52–62, 85–90, and 282–287 (Dreyfus et al., 2012), we will ignore the structural variations in these regions in the following comparison.
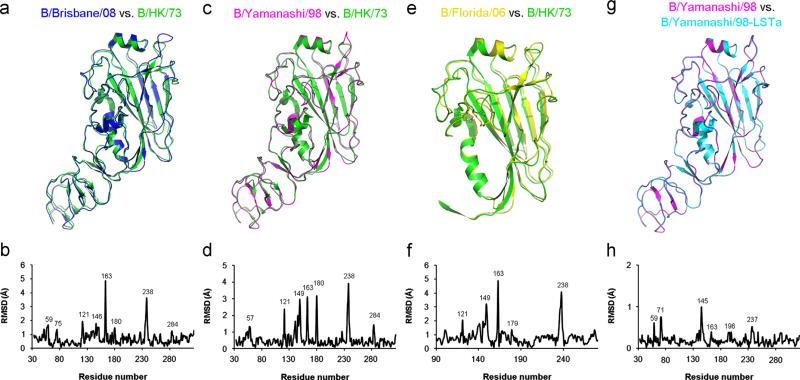
The overall structures of all influenza B virus HAs resemble each other. However, regions with large structural variations, expressed as Cα-root-mean-square differences (Cα-RMSDs), are observed when using the unliganded structure of B/HK/73 HA as a reference (PDB code: 3BT6) (Fig. 2a–f). For instance, there exhibit large structural differences between B/Brisbane/08 and B/HK/73 HAs in the regions of the 120-loop (including residues at or around HA159, 75, 121, 180, 284), 150-loop (at around HA1146), 160-loop (at around HA1163) and 190-helix (at around HA1238) (Fig. 2a and b). Moreover, large structural variations are observed between B/Yamanashi/98 and B/HK/73 HAs in the 120-loop (at around HA157, 121, 180, 284), 150-loop (at around HA1149), 160-loop (at around HA1163) and 190-helix (at around HA1238) (Fig. 2c and d). Similarly, B/Florida/06 and B/HK/73 HAs have large structural changes in all these four regions (Fig. 2e and f). Interestingly, the structural differences in the 150-loop region of B/Yamagata lineage HAs (Fig. 2c–f) are much larger than that of B/Victoria lineage (Fig. 2a and b), consistent with the observation that the 150-loop region appears to be a particularly strong neutralizing epitope for B/ Yamagata lineage strains in recent isolates (Nakagawa et al., 2003; Shen et al., 2009). The overall RMSD in the region of HA191–281 of HAs of recent strains compared to B/HK/73 HA is in the range of 0.840–0.874 Å for Cα-atoms only and 0.917–0.950 Å for main-chain atoms (Table 3).
Fig. 2.
Comparison of known influenza B virus HA structures. Pairwise structural comparison ((a), (c), (e) and (g)) and the corresponding Cα-RMSD ((b), (d), (f) and (h)). The pairs are: (a) and (b) B/Brisbane/08 and B/HK/73 HAs in the range of HA133–324; ((c) and (d)). B/Yamanashi/98 and B/HK/73 HAs in the range of HA133–324; (e) and (f) B/Florida/06 and B/HK/73 HAs in the range of HA191–281; (g) and (h) B/Yamanashi/98 and B/Yamanashi/98-LSTa HAs in the range of HA133–324.
Table 3.
RMSD of known influenza B virus HA structures in the region of residues HA191–281.
| RMSD (Å) | B/Brisbane/08 vs. B/HK/73 | B/Yamanashi/98 vs. B/HK/73 | B/Florida/06 vs. B/HK/73 | B/Yamanashi/98 vs. B/Yamanashi/98-LSTa |
|---|---|---|---|---|
| Cα atoms | 0.874 | 0.840 | 0.854 | 0.214 |
| Main-chain atoms | 0.917 | 0.948 | 0.950 | 0.213 |
We also compared the structures of B/Yamanashi/98 HA and its complex with avian-like receptor analogue LSTa (Fig. 2g and h). The overall RMSD in the region of HA191–281 is 0.214 Å for Cα-atoms only and 0.213 Å for main-chain atoms (Table 3). Although the two structures are highly similar, relatively large structural differences are still observed in regions of the 120-loop (at around HA159, 71), 150-loop (at around HA1145), 160-loop (at around HA1163), and 190-helix (at around HA1196 and 237 (Fig. 2g and h). Among these regions, only the 190-helix is directly in contact with bound receptor analogue. Therefore, the structure of influenza B virus HA is intrinsically flexible in regions of 120-loop, 150-loop, 160-loop and 190-helix regardless of bound receptors.
Previous studies have assigned four major epitopes on the structure of B/HK/73 HA: the 120-loop, 150-loop, 160-loop and 190-helix and their respective surrounding regions (Berton et al., 1984; Berton and Webster, 1985; Hovanec and Air, 1984; Rivera et al., 1995; Wang et al., 2008; Webster and Berton, 1981). Analysis of 278 HA sequences from B/Victoria lineage and 282 HA sequences from B/Yamagata lineage revealed that amino-acid substitutions in the sequences are concentrated on these four epitope regions (Table 2). The high amino-acid substitution frequency and the large structural variations of these regions are probably inter-connected: the high structural flexibility of these regions makes them easily accommodate drifting mutations without compromising the structural integrity of influenza B virus HA, and the drifting mutations introduced therein could easily cause structural changes that are large enough to evade recognition by neutralizing antibodies.
The 120-loop and its surrounding regions
The 120-loop and its surrounding regions have some of the largest deviations from the structure of B/HK/73 HA (Fig. 3a). For instance, the average Cα-RMSD from B/HK/73 HA for the region of HA151–54, 56–59, 73–75, 121–123, 129, 137 is 0.96 Å and 1.29 Å for B/Yamanashi/98 and B/Brisbane/08 HAs, respectively. Since some of these regions in B/Florida/06 HA are involved in binding to the antibody, we omitted the comparison between B/Florida/06 and B/HK/73 HAs. However, the fact that these regions serve as antibody binding site itself confirms their contribution to the antigenicity of influenza BHA. As expected, these regions harbor sites that undergo constant amino-acid substitutions (Fig. 3b, Table 2) and are identified to be under positive selection in previous phylogenetic and antigenic studies, such as HA156, 73, 75, 122, 129, 137 (Nunes et al., 2008; Shen et al., 2009). Overall, the B/Yamagata lineage strains are closer to the early influenza B strains in terms of both structure and residue types in this region (Table 2).
Fig. 3.
Antigenic structure of influenza B virus HA. (a) Surface presentation of the globular domain of B/Yamanashi/98 HA. Residues with large structural variations between B/Yamanashi/98 or B/Brisbane/08 relative to B/HK/73 HA (Cα-RMSD greater than 1.0 Å) are highlighted based on their spatial location (120-loop region in cyan, 150-loop in green, 160-loop in blue and 190-helix region in red). (b)–(e) Amino-acid substitutions in B/Brisbane/08 HA (purple), B/Yamanashi/98 HA (green) relative to B/HK/73 HA (yellow) for the 120-loop region (b), 150-loop (c), 160-loop (d) and 190-helix region (e). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
HA1179–181 in the vestigial esterase domain belongs to the 120-loop epitope (Fig. 3b). This region has the motif of 179TKG181 in early influenza B strains (1972–1982) including B/HK/73 (Table 2). However, this motif is changed to 179TEG181 in 97% of B/Victoria strains. In sharp contrast, in B/Yamagata lineage, the motif of 179TKG181 is kept only in ~11% strains, while 60% strains including B/Florida/06 strain have 179TEG181 motif, and 21% strains including B/Yamanashi/98 strain acquire a new 179TKE181 motif (Table 2). The 179TEG181 in B/Florida/06 and in B/Brisbane/08 has a similar Cα-RMSD from B/HK/73 HA, at 1.24 Å and 1.07 Å, respectively. However, the 179TKE181 motif as found in B/Yamanashi/98 strain has a much larger Cα-RMSD (at 2.27 Å) from B/HK/73 HA. Apparently, the residues in the vicinity of HA1179–181 in the vestigial esterase domain are under much stronger diversifying selective pressure in the B/Yamagata lineage than in the B/Victoria lineage. From a structural point of view, HA1179–181 in B/Yamanashi/98 HA is more protruding than in the structure of B/Brisbane/08 HA (Fig. 3b), resulting in 270 Å2 more exposed surface area in each trimer (from a total solvent accessible surface area of 855 Å2 in B/Brisbane/08 HA to 1,125 Å2 in B/Yamanashi/98 HA), equivalent to an increase from 58% of total solvent accessible surface area to 67%.
Relative to B/HK/73 HA, B/Yamanashi/98 and B/Brisbane HAs have Cα-RMSDs of 1.21 Å and 1.10 Å in the region of HA1283–285, respectively. This region belongs to the 120-loop and is part of the epitope that binds to human monoclonal antibody CR8071 in the complex with B/Florida/06 HA (Dreyfus et al., 2012).
The 150-loop
The 150-loop is a long protruding loop in which single amino-acid substitutions are frequently found to cause altered antigenic properties (Abed et al., 2003; Berton et al., 1984; Berton and Webster, 1985; Hovanec and Air, 1984; Lugovtsev et al., 2007; Nakagawa et al., 2003). The motif of 148NGN150 is conserved in early influenza B strains and in 96% of B/Victoria lineage strains, whereas in B/Yamagata lineage, the motifs of 148SKS150, 148SKI150 and 148SRS150 are found in 47%, 28% and 20% influenza B virus HAs, respectively (Fig. 3c, Table 2). Thus, the 150-loop is likely under strong diversifying selective pressure in B/Yamagata lineage (Shen et al., 2009), which is responsible for the significantly larger structural differences than those of B/Victoria lineage as observed in Fig. 2a–f. For instance, for the residues in the range of HA1 139–142 and 145–151 where large structural differences are noticed relative to B/HK/73 HA, B/Yamanashi/98 and B/Florida/06 HAs (B/Yamagata lineage) have an average Cα-RMSD of 1.65 Å and 1.81 Å, respectively, which is 1.30 Å for B/Brisbane/06 HA (B/Victoria lineage).
The 160-loop
The 160-loop is located at the membrane-distal “tip” of the HA molecule. This is the only region where insertions and deletions are frequently found (Hovanec and Air, 1984; McCullers et al., 1999; Nerome et al., 1998) (Table 2). Comparing with B/HK/73 HA that has the shortest 160-loop, both B/Victoria and B/Yamagata strains have insertions in this region. Relative to the reference B/Victoria/2/87 strain that has a NDN insertion between residues HA1162 and 163, 42% B/Victoria HA sequences have the same motif while 51% sequences including B/Brisbane/08 strain have a NDK insertion instead (Table 2). In contrast, relative to the reference B/Yamagata/16/88 strain that has a single D insertion between residues HA1162 and 163, only 9% HA sequences of 282 recent B/Yamagata strains keep this insertion, 88% sequences including those of B/Yamanashi/98 and B/Florida/06 have a DN insertion and the rest 3% sequences have other two-residue insertions (Fig. 3d, Table 2). The average Cα-RMSD in the region of HA1162–165 relative to B/HK/73 HA is 1.01 Å, 1.51 Å and 1.35 Å (excluding the inserted residues labeled as 163A, B, C in Fig. 3d) for B/Yamanashi/98, B/Florida/06 and B/Brisbane/08 HA, respectively. Therefore, although the 160-loop epitope was once thought to be specific for B/Victoria lineage in antigenic drift (Nakagawa et al., 2001; Nakagawa et al., 2005), the recent B/Yamagata lineage has reclaimed this loop as an epitope.
The 190-helix and its surrounding regions
The 190-helix region (including the 190-helix and 240-loop) is located at the membrane-distal end of the HA molecule and constitutes the top- and left-edge of the receptor-binding site (Fig. 3a). The 190-helix is constantly under strong positive selective pressure in both B/Victoria and B/Yamagata lineages (Nunes et al., 2008; Pechirra et al., 2005; Shen et al., 2009). Two residues in the 190-helix have large structural differences from B/HK/73 HA: HA1194 and HA1206 (Fig. 3a). The average Cα-RMSD for these two residues from that of B/HK/73 HA are 0.46 Å, 0.65 Å and 1.05 Å for B/Yamanashi/98, B/Florida/06 (B/Yamagata lineage) and B/Brisbane/08 (B/Victoria lineage), respectively. In addition, frequent amino-acid substitutions are found in three positions: HA1195, 199 and 200 (Table 2, Fig. 3e). The average Cα-RMSD at these sites with B/HK/73 HA is 0.33 Å, 0.40 Å and 0.58 Å for B/Yamanashi/98, B/Florida/06 (B/Yamagata lineage) and B/Brisbane/08 (B/Victoria lineage), respectively. Therefore, B/Victoria HA is apparently more diverged from B/HK/73 HA in this region (Fig. 3e).
The region of HA1235–240 has the largest Cα-RMSD of all from B/HK/73 HA: at 2.23–2.34 Å for B/Yamanashi/98, B/Florida/06 and B/Brisbane/08 HAs (Fig. 3a), as the result of a large structural rearrangement in this region (Fig. 3e). The residue Glu-235 in B/ HK/73 HA is changed to Gly-235 in 100% B/Victoria lineage and 99% B/Yamagata lineage strains (Table 2). The presence of Gly-235 results in a more extended 240-loop and causes one-residue shift in side-chain orientation (Fig. 3e). For example, the side chain of Pro-238 in B/Yamanashi/98, and B/Brisbane/08 HA structures is placed at a similar location to that of Leu-237 in B/HK/73 HA (Fig. 3e). Consequently, the side chain of Gln-239 in all other influenza B virus HA structures is at the same location as that of Pro-238 in B/HK/73 HA, pointing towards the receptor-binding site (Fig. 3e). The re-orientation of the side-chains in the region of HA1235–240 could profoundly change the antigenic properties of this region.
The receptor-binding site and the B/Yamanashi/98 HA-LSTa complex
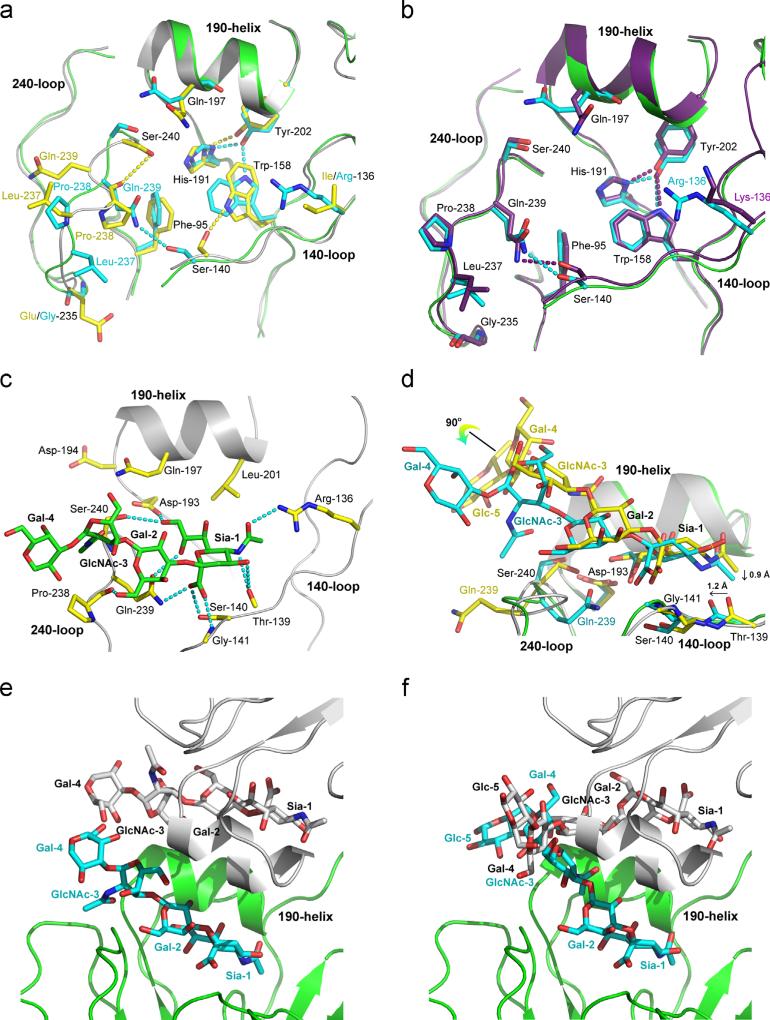
The receptor-binding site of influenza B virus HA is located at the top of the membrane-distal domain (Fig. 1) formed by 190-helix, 140-loop and 240-loop (Wang et al., 2007). Its base is constituted by four absolutely conserved residues: Phe-95, Trp-158, His-191 and Tyr-202 (Fig. 4a). In B/HK/73 HA (Wang et al., 2007), besides π-stacking interactions among these aromatic residues, there are three additional hydrogen bonds that help stabilize the receptor-binding site: between Tyr-202 and His-191, Ser-140 and Trp-158, Pro-238 and Ser-240 (Fig. 4a). In B/Yamanashi/98 HA, the π-stacking interactions are conserved. Although a rotation of the side chain of Trp-158 results in the loss of the hydrogen bond between Ser-140 and Trp-158, a new hydrogen bond between Trp-158 and Tyr-202 is observed. Moreover, the large structural rearrangement in the 240-loop as described above causes the loss of the interaction between Pro-238 and Ser-240, but a hydrogen bond is gained between the side chain of Gln-239 and Ser-140 instead (Fig. 4a). Overall, similar π-stacking interactions and the same number of hydrogen bonds are observed in the receptor-binding site of both B/HK/73 and B/Yamanashi/98 HAs.
Fig. 4.
Receptor binding of influenza B virus HA. (a) Comparison of receptor-binding sites between B/Yamanashi/98 (green) and B/HK/73 (grey) HAs. Important residues are highlighted according to the atom types with the exception that carbon atoms are colored in yellow for B/HK/73 HA and in cyan for B/Yamanashi/98 HA. (b) Comparison of receptor-binding sites between B/Yamanashi/98 (green) and B/Brisbane/08 (purple) HAs. Important residues are highlighted according to the atom types with the exception that carbon atoms are colored in cyan for B/Yamanashi/98 HA and in purple for B/Brisbane/08 HA. (c) Molecular interactions between B/Yamanashi/98 HA and avian-like receptor analogue LSTa. Dashed lines are for hydrogen bonds listed in Table 4. (d) Comparison of LSTa in the receptor-binding sites of B/Yamanashi/98 and B/HK/73 HAs. The coloring scheme is the same as in (a). (e) Crystal contacts around the LSTa ligand in the structure of B/Yamanashi/98 HA. Two neighboring HA molecules are shown in green and light grey, and their bound LSTa in cyan and grey, respectively. It is clear that in this conformation, LSTa does not have crystal clashes. (f) Crystal contracts around the LSTa ligand in the structure of B/Yamanashi/98 HA if LSTa were in a conformation as found in B/HK/73 HA. It is clear that LSTa in this conformation will have steric clashes, providing an explanation why LSTa adopts the unusual conformation as found in B/Yamanashi/98 HA-LSTa. Coloring scheme is the same as in (e). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In the complex structure of B/Yamanashi/98 HA-LSTa, Sia-1, Gal-2, GlcNAc-3 and Gal-4 are clearly visible (Fig. 4c). The Sia-1 moiety makes extensive hydrogen bonding interactions within the receptor-binding site of B/Yamanashi/98 HA (Fig. 4c, Table 4), including one by the main-chain amide of Gly-141, one by each of the side-chains of Ser-140, Asp-193 and Ser-240. These four hydrogen bonds were also observed between LSTa and B/HK/73 HA (Table 4). In addition, B/Yamanashi/98 HA makes five more hydrogen bonds that were not observed in B/HK/73 HA: two hydrogen bonds by the main-chain carbonyl of Thr-139, one hydrogen bond by the side chain of Arg-136, and two hydrogen bonds by the side chain of Gln-239 (Fig. 4c, Table 4).
Table 4.
Hydrogen bonding interactions of B/HK/73 and B/Yamanashi/98 HAs with bound avian-like receptor analogue LSTa.
| LSTa | Interaction pair | B/HK/73 distance (Å) | B/Yamanashi/98 distance (Å) |
|---|---|---|---|
| Sia−1 | O1A......Gly−141 N | 2.9 | 2.7 |
| O1B......Ser−140 OG | 2.8 | 2.9 | |
| O9.........Asp−193 OD2 | 2.9 | 2.6 | |
| O9.........Ser−240 OG | 2.4 | 3.1 | |
| O4.........Thr−139 O | - | 3.4 | |
| N5.........Thr−139 O | - | 3.2 | |
| O1B......Gln−239 NE2 | - | 2.9 | |
| O8.........Gln−239 OE1 | - | 3.1 | |
| O10......Arg−136 NH2 | - | 2.9 | |
| Gal−2 | O6.........Leu−237 O | 2.5 | - |
| O6.........Pro−238 O | - | 2.9 | |
| GlcNAc−3 | O7.........Gln−197 NE2 | 3.4 | - |
| Glc−5 | O6.........Thr−196 OG1 | 2.6 | - |
Hydrogen bonds were identified by PyMol.
The additional hydrogen bonds with Thr-139 are apparently due to the fact that the bound Sia-1 moiety of LSTa sits ~0.9 Å lower in the receptor-binding site, thus is closer to the region of HA1139–141 in B/Yamanashi/98 HA structure (Fig. 4d). Due to the ~ 1.2 Å left shift of the region HA1139–141 at the lower edge of the receptor-binding site of B/Yamanashi/98 HA in relation to B/HK/73 HA, the bound Sia-1 moiety of LSTa is also left-shifted by a similar distance.
The new hydrogen bond with HA1136 is made possible by the substitution of Ile-136→Arg (Fig. 4a). This substitution is found in HA sequences of 98% B/Yamagata strains and 1% B/Victoria strains (Table 2). However, a similar substitution of Ile-136→Lys is found in BHA sequences of 1% B/Yamagata strains and 97% B/Victoria strains. The side chains of Arg-136 in the structure of B/Yamanashi/98 HA and Lys-136 in the structure of B/Brisbane/08 HA are located at a similar position (Fig. 4b). Therefore, the hydrogen bond between the side-chain of Arg-136 and bound Sia-1 moiety is expected to be present in all these B/Yamagata and B/Victoria strains.
The additional two hydrogen bonds with the side chain of Gln-239 are made possible as the consequence of the large structural rearrangement in the 240-loop due to the presence of Gly-235 (Fig. 4a). This new conformation of the 240-loop is shared among B/Yamanashi/98 and B/Brisbane/08 HA structures (Fig. 4b) and the residue Gly-235 is observed in 100% of B/Victoria lineage and 99% of B/Yamagata lineage strains (Table 2). Therefore, the hydrogen bonding interactions by the side-chain of Gln-239 are preserved in almost all of recent influenza B virus HAs.
Altogether, these five new hydrogen bonds observed between B/Yamanashi/98 HA and the Sia-1 moiety of bound LSTa receptor analogue are expected to be preserved in HAs of almost all of currently circulating influenza B virus strains with both human-like and avian-like receptors that share the Sia-1 moiety. The much stronger binding between Sia-1 and HAs of these influenza B viruses of both lineages is indicative of higher receptor-binding affinity. The impact of the enhanced receptor-binding affinity on the pathogenesis and tissue tropism of the virus requires further investigation.
Moreover, Gal-2 of bound LSTa forms one hydrogen bond with the main-chain carbonyl atom of Pro-238 in B/Yamanashi/98 HA, while in the case of B/HK/73 HA, there was one hydrogen bond with the main-chain carbonyl atom of Leu-237 instead. The GlcNAc-3 and Gal-4 moieties of LSTa do not interact with the receptor-binding site of B/Yamanashi/98 HA. This is in sharp contrast to the hydrogen bonding interactions between GlcNAc-3 and the side chain of Gln-197, and between Glc-5 and the side chain of Thr-196 of B/HK/73 HA (Table 4). This difference is mainly due to the different binding mode of LSTa in the receptor-binding sites of B/HK/73 and B/Yamanashi/98 HAs (Fig. 4d). In B/HK/73 HA, the Gal-4 and Glc-5 point upwards and interact with the 190-helix, while in B/Yamanashi/98 HA, the Gal-4 lies above the 240-loop due to the ~90° rotation of GlcNAc-3 (Fig. 4d). The unusual conformation of GlcNAc-3 and Gal-4 of LSTa in B/Yamanashi/98 HA is probably the result of limited space at near the receptor-binding site in the crystal lattice (Fig. 4e). If they were adopting the same conformation as found in B/HK/73 HA, Gal-4 and Glc-5 would have steric clashes with those of a symmetry-related molecule (Fig. 4f). The fact that LSTa has similar conformations in all three subunits of B/ Yamanashi/98 HA in an asymmetric unit with strong electron densities suggests that this is probably one of its most stable conformations. In sharp contrast, the binding of human receptor analogue LSTc is apparently in conflict with the existing crystal packing of B/Yamanashi/98 HA, thus decreasing the resolution to 4–8 Å.
The HA1–HA1 interface
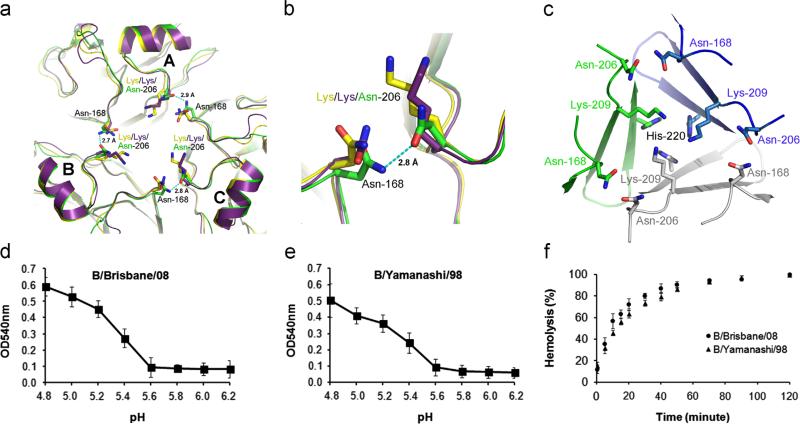
One of the large structural differences on the 190-helix is observed at HA1206 (Fig. 5a–c). In both B/HK/73 and B/Brisbane/08 HA structures, the side chain of residue Lys-206 points towards or a little away from the three-fold axis. However, this residue becomes Asn-206 in B/Yamanashi/98 HA that points towards neighboring HA1 subunits to form strong hydrogen bonds with a neighboring Asn-168 (Fig. 5a and b). The total of three strong hydrogen bonds between Asn-206 and Asn-168 for a trimeric B/Yamanashi/98 HA may help stabilize the neutral-pH HA structure. However, this stabilizing effect is expected to be marginal to the overall stability of B/Yamanashi/98 HA as the HA1–HA1 interface is predominantly stabilized by the interactions among three highly-conserved Lys-209 and His-220 at close to the three-fold axis (Fig. 5c).
Fig. 5.
The Lys206→Asn mutation of HAs in B/Yamagata lineage. (a) The stabilization of the HA1–HA1 interface by three new hydrogen bonds between Asn-206 and Asn-168 shown as dashed lines with the distances labeled. The coloring scheme is the same as in Fig. 3. (b) Zoom-in of the hydrogen bond between Subunits B and C. (c) The residues at the HA1–HA1 interface in B/Yamanashi/98 HA structure. Residues Lys-209 and His-220 interact with each other at the 3-fold axis of the structure and are the major stabilizing force. Three HA subunits are colored as green, blue and light grey, respectively. (d) pH-dependent hemolysis of B/Brisbane/08 virus. Shown are averaged values and standard deviations from at least three independent measurements. (e) pH-dependent hemolysis of B/Yamanashi/98 virus. Shown are averaged values and standard deviations from at least three independent measurements. (f) Comparison of the rate of hemolysis between B/Brisbane/08 virus and B/Yamanashi/98 virus at pH 4.8. Shown are averaged values and standard deviations from at least three independent measurements. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To explore the impact of the new hydrogen bonds by Asn-206 on the structure and stability of influenza B virus HAs experimentally, B/Brisbane/08 and B/Yamanashi/98 viruses were used to study pH-dependent hemolysis and rate of hemolysis at a given pH. The pH-dependent hemolysis reflects how many protons are needed to induce the conformational changes of HA that include: (1) dissociation of HA1–HA1 interfaces; (2) dissociation of HA1–HA2 interfaces; and (3) unfolding and refolding of HA2. (2) and (3) are similar for B/Brisbane/08 and B/Yamanashi/98 strains. For (1), due to the presence of conserved Lys-209 and His-220 common to both strains, the difference in pH at which hemolysis starts would reflect the contribution of the new hydrogen bonds by Asn-206. The fact that significant hemolysis were similarly observed for both strains when pH was lower than 5.6 (Fig. 5d and e) suggests that the impact of the new hydrogen bonds by Asn-206 is not predominant. The rate of hemolysis at a given pH reflects how fast the conformational changes of HA could happen when sufficient protons exist. By fitting the data to first order reaction, we obtained the reaction rate of 0.059±0.007 min–1 and 0.050±0.005 min–1 for B/Brisbane/08 and B/Yamanashi/98 strains, respectively. The about ~20% faster reaction rate for B/Brisbane/08 strain than B/Yamanashi/98 strain indicates that the conformational changes of HA in the former is somewhat easier than the latter, which has been further stabilized by the new hydrogen bonds formed by Asn-206. Its impact on the pathogenesis of B/Yamanashi/98 virus needs to be separately evaluated.
Ionizable residue clusters
Structural analysis of B/HK/73 HA has revealed ionizable residue clusters that are strategically placed at all regions where structural rearrangement is expected when HA is exposed to low pH (Wang et al., 2008). Presumably due to their importance in low-pH-induced membrane fusion that is crucial for successful host invasion by influenza virus, all the ionizable residue clusters identified in the previous study (Wang et al., 2008) are well maintained.
Amino-acid substitutions on HA2
Between the B/Victoria and B/Yamagata lineages, amino-acid substitutions are observed on HA2 at positions 132 and 158 (Table 2). At HA2132, the changes are between negatively charged residues Glu and Asp, and no large structural difference is noticed due to this mutation. The residue Asn at HA2158 in early influenza B virus strains is kept in 98% of B/Yamagata lineage strains, but is changed to Asp-158 in 89% of B/Victoria lineage strains (Table 2). The negative charge at HA2158 in most of B/Victoria lineage strains might affect the conformation of membrane-proximal linker and transmembrane trimer packing of HAs (Kordyukova et al., 2011).
Discussion
The two currently co-circulating lineages of influenza B viruses diverged from the early strains in 1983 or earlier (Chen et al., 2007; Shen et al., 2009). Comparison among the HA structures of representative influenza B virus strains before and after the divergence together with the sequence analysis provides a structural basis for the divergent evolution of influenza B virus over the past several decades.
Antigenic structure of influenza B virus HA
Through a combined systematic structural and sequence analysis of HA of influenza B virus before and after its divergence into two distinct lineages: B/Victoria and B/Yamagata, we found that the largest structural variations among different BHAs are strongly correlated with frequent amino-acid substitutions concentrated at the four major epitopes. Having predominantly loop structures in these four major epitopes makes it possible to easily accommodate amino-acid substitutions that result in structural changes large enough to evade recognition by neutralizing antibodies, but still sufficiently small to maintain the structural integrity of the protein, particularly of the receptor-binding site to ensure efficient binding of cell surface receptors.
Moreover, among the four major epitopes of influenza B virus HAs, the 120-loop, 150-loop, 160-loop and 190-helix regions, differential selective pressures appear to operate on B/Victoria and B/Yamagata lineage strains. For instance, 150-loop appears to be under stronger positive selective pressure in the B/Yamagata lineage than in the B/Victoria lineage. On the other hand, the 160-loop that used to be specific for the B/Victoria lineage is now also being used by the B/Yamagata lineage in recent years. Furthermore, the close spatial locations of the 150-loop, 160-loop and 190-helix, the facts that neutralizing antibodies recognizing the 150- and 160-loops, and the 190-helix compete with each other (Berton and Webster, 1985; Hovanec and Air, 1984) and that they all interact with the human monoclonal antibody CR8033 (Dreyfus et al., 2012) suggest that they most likely constitute a large continuous antigenic site. On the other hand, the lack of cross-reactivity between the antibodies recognizing the 120-loop region and the 160-loop/190-helix (Berton and Webster, 1985; Hovanec and Air, 1984) and the fact that the footprint of the antibody CR8071 is solely on the 120-loop (Dreyfus et al., 2012) suggest that the 120-loop is a separate antigenic site.
Receptor binding of influenza B virus HA
The receptor-binding activity of HA is one of the key factors of the pathogenesis of influenza viruses (Matrosovich et al., 2000; Rogers and Paulson, 1983; Viswanathan et al., 2010). There are two most noticeable changes in the receptor-binding site of HA from recent influenza B virus isolates.
First, recent HAs of both lineages acquired a long-chain basic residue at HA1136 (Arg or Lys) (Table 2). The side-chain of Arg-136 in B/Yamanashi/98 HA contributes one hydrogen bond to the Sia-1 moiety of bound LSTa receptor analogue (Fig. 4c). The side-chain of Lys-136 in unliganded B/Brisbane/08 HA is similarly located in the receptor-binding site (Fig. 4b), suggesting that both Arg-136 (predominant in B/Yamagata HA) and Lys-136 (predominant in B/Victoria HA) will likely make similar hydrogen bonding interactions with Sia-1 of bound human or avian-like receptor analogues.
Second, a large structural shift is observed in the 240-loop, as a consequence of the Glu-235 to Gly-235 mutation in both B/Victoria and B/Yamagata lineages. The structural shift causes re-orientation of the residues on the 240-loop, despite the overall high sequence identity in this region among all influenza B virus HAs. For instance, Gln-239 in B/Yamanashi/98 HA is flipped over to interact with the bound receptors. Therefore, current influenza B virus of both lineages has evolved a receptor-binding site that is distinct from that of the early strains, and capable of binding the Sia-1 moiety of both human and avian-like receptors with a much higher affinity than B/HK/73 HA (Table 4). A stronger binding of receptors might allow for a different tissue tropism, for which further investigation is required.
HA1–HA1 interface of B/Yamagata HAs
The Lys206→Asn mutation of HAs in B/Yamagata lineage introduces three new strong hydrogen bonds between neighboring subunits of HA1 (Fig. 5). This results in a slight decrease in the rate of hemolysis at a given pH, suggesting a small but noticeable contribution to the overall stability of the neutral-pH structure of HAs of B/Yamgata lineage.
Materials and methods
Protein expression and crystallization
The sequence of B/Yamanashi/98 HA (GI: 125663100) was retrieved from NCBI gene bank (Benson et al., 2012) and the gene corresponding to its ectodomain (HA11–342–HA21–176) was assembled from oligonucleotides according to the previously published protocol (Stemmer et al., 1995) and inserted into the pRB21 plasmid containing a trimerizing sequence from the bacteriophage T4 fibritin (foldon) (Stevens et al., 2004) and a His6-tag at the extreme C-terminus. The ectodomain of B/Yamanashi/98 HA was expressed by using vaccinia virus system as previously reported (Blasco and Moss, 1995). Briefly, CV-1 monolayers were first infected with vaccinia virus that lacks the vp37 gene at an MOI of 2.0 for 4 h at 37 °C in a CO2 incubator. The pRB21 plasmid containing the B/Yamanashi/98 HA gene was transfected into the CV-1 cells by Lipofectamine 2000 according to the manufacture′s instruction (Invitrogen). The cells were further incubated for 4 h at 37 °C. The supernatant was changed to Dulbecco′s modified eagle medium with 10% fetal bovine serum and further incubated for 40 h. The infected cells that contain recombinant vaccinia viruses were harvested and lysed by three freeze-and-thaw cycles in ice/ethanol and 37 °C water bath. The recombinant viruses were selected based on their ability to produce large plaques. The plaques were further amplified in CV-1 cells. For expression, CV-1 cells were infected with recombinant vaccinia viruses at an MOI of 1.0 for 40 h. The media of infected CV-1 cells were cleared by centrifugation and dialyzed overnight against the buffer containing 10 mM Tris–HCl (pH 7.2), 100 mM NaCl, and loaded onto the Cobalt resin (Thermo Scientific). The ectodomain of B/Yamanashi/98 HA protein was released from the beads by trypsin digestion (Sigma). The protein was subjected to anion-exchange (mono-Q 4.6/100 PE, GE Healthcare) and gel-filtration (Superdex 200 10/300 GL, GE Health-care) chromatography. The peak corresponding to trimeric HA was collected and concentrated to about 25 mg/mL. Crystals were grown by hanging drop vapor diffusion method in reservoir solution of 0.1 M Tris–HCl (pH 8.5), 0.2 M Tri-methylamine N-oxide, 23% PEG 2000 MME at 290 K. A final concentration of 15% glycerol was included in the reservoir solution as the cryoprotectant. In order to get structures of B/Yamanashi/98 HA with sialic-acid receptor analogues, the crystals were soaked in 16 mM avian-like receptor analogue LSTa or human-like receptor analogue LSTc (Sigma) for 2 h. The soaking of the B/Yamanashi/98 HA crystals with LSTa increased the resolution to 2.50 Å, while in contrast, the soaking with human-like receptor LSTc decreased the resolution to about 4–8 Å, possibly due to the damage to crystal lattice upon the binding of LSTc.
Structure determination and model building
The diffraction data were indexed and integrated by using MOSFLM (Leslie, 2006; Powell, 1999), scaled by SCALA, and truncated to structure factor amplitude by TRUNCATE in CCP4 (Collaborative Computational Project, 1994). Five percent of unique reflections were randomly chosen as the test set for calculating the Rfree-factors. The unliganded B/HK/73 HA structure (Protein Data Bank (PDB) code: 3BT6) (Wang et al., 2008) was used as the search model for molecular replacement and three copies of HA1/HA2 monomer was found by AutoMR module implemented in PHENIX (Adams et al., 2010). The model was re-built by AutoBuild module and refined by Refinement module in PHENIX (Adams et al., 2010). The model and map were checked in COOT for further manual adjustment (Emsley and Cowtan, 2004). Figures for structural snapshots were generated by PyMOL.
Sequence analysis
The full-length influenza B virus HA protein sequences were analyzed in NCBI flu database (Bao et al., 2008). 573 unique sequences from the year 1940 to 2012 were aligned. The phylo-genetic tree was generated by neighbor-joining method (Fig. S1). Three clades were selected in the tree: the early influenza B strains from 1972 to 1982 (six sequences); the B/Victoria lineage (278 sequences); and the B/Yamagata lineage (282 sequences). The amino-acid residue substitution frequency in each of three clades was calculated by Jalview (Waterhouse et al., 2009) (Table 2).
Hemolysis assay
The B/Brisbane/08 and B/Yamanashi/98 viruses were provided by Centers for Disease Control and Prevention (Atlanta, GA). The pH-dependent hemolysis assay was performed as previously reported (Maeda and Ohnishi, 1980; Sato et al., 1983). Briefly, the viruses were mixed with human erythrocytes on ice for 1 h. The erythrocytes that were bound with viruses were pelleted by centrifugation and resuspended in 100 mM citrate buffer at different pH. The reaction was incubated at 37 °C for 1 h and the supernatant was used to record the absorbance at 540 nm. For the time-course of hemolysis assay, the virus-erythrocyte suspension was incubated in citrate buffer at pH 4.8. The sample was taken at different time points and a pre-determined volume of NaOH was immediately added to bring the buffer pH to about 7.0. All the assays were performed at least three times.
Supplementary Material
Acknowledgments
Q.W. thanks support from the National Institutes of Health (R01-AI067839), the Gillson-Longenbaugh Foundation, the Simmons Collaborative Research Fund Awardand and The Welch Foundation (Q-1826). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817).
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2013.07.035.
References
- Abed Y, Coulthart MB, Li Y, Boivin G. Evolution of surface and nonstructural-1 genes of influenza B viruses isolated in the Province of Quebec, Canada, during the 1998–2001 period. Virus Genes. 2003;27(2):125–135. doi: 10.1023/a:1025768308631. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66(2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 2008;82(2):596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;41:D36–D42. doi: 10.1093/nar/gks1195. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berton MT, Naeve CW, Webster RG. Antigenic structure of the influenza B virus hemagglutinin: nucleotide sequence analysis of antigenic variants selected with monoclonal antibodies. J. Virol. 1984;52(3):919–927. doi: 10.1128/jvi.52.3.919-927.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berton MT, Webster RG. The antigenic structure of the influenza B virus hemagglutinin: operational and topological mapping with monoclonal antibodies. Virology. 1985;143(2):583–594. doi: 10.1016/0042-6822(85)90396-4. [DOI] [PubMed] [Google Scholar]
- Blasco R, Moss B. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene. 1995;158(2):157–162. doi: 10.1016/0378-1119(95)00149-z. [DOI] [PubMed] [Google Scholar]
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Nat. Acad. Sci. U.S.A. 1999;96(16):8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Guo YJ, Wu KY, Guo JF, Wang M, Dong J, Zhang Y, Li Z, Shu YL. Exploration of the emergence of the Victoria lineage of influenza B virus. Arch. Virol. 2007;152(2):415–422. doi: 10.1007/s00705-006-0852-6. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50(5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Copeland CS, Doms RW, Bolzau EM, Webster RG, Helenius A. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J. Cell Biol. 1986;103(4):1179–1191. doi: 10.1083/jcb.103.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337(6100):1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Han T, Marasco WA. Structural basis of influenza virus neutralization. Ann. N.Y. Acad. Sci. 2011;1217:178–190. doi: 10.1111/j.1749-6632.2010.05829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanec DL, Air GM. Antigenic structure of the hemagglutinin of influenza virus B/Hong Kong/8/73 as determined from gene sequence analysis of variants selected with monoclonal antibodies. Virology. 1984;139(2):384–392. doi: 10.1016/0042-6822(84)90384-2. [DOI] [PubMed] [Google Scholar]
- Ikonen N, Pyhala R, Axelin T, Kleemola M, Korpela H. Reappearance of influenza B/Victoria/2/87-lineage viruses: epidemic activity, genetic diversity and vaccination efficacy in the Finnish Defence Forces. Epidemiol. Infect. 2005;133(2):263–271. doi: 10.1017/s0950268804003462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegae Y, Sugita S, Endo A, Ishida M, Senya S, Osako K, Nerome K, Oya A. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J. Virol. 1990;64(6):2860–2865. doi: 10.1128/jvi.64.6.2860-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology. 2006;119(1):1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordyukova LV, Serebryakova MV, Polyansky AA, Kropotkina EA, Alexeevski AV, Veit M, Efremov RG, Filippova IY, Baratova LA. Linker and/or transmembrane regions of influenza A/Group-1, A/Group-2, and type B virus hemagglutinins are packed differently within trimers. Biochim. Biophys. Acta. 2011;1808(7):1843–1854. doi: 10.1016/j.bbamem.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Korte T, Ludwig K, Huang Q, Rachakonda PS, Herrmann A. Conformational change of influenza virus hemagglutinin is sensitive to ionic concentration. Eur. Biophys. J. 2007;36(4–5):327–335. doi: 10.1007/s00249-006-0116-0. [DOI] [PubMed] [Google Scholar]
- Leslie AG. The integration of macromolecular diffraction data. Acta Crystal-logr., Sect. D: Biol. Crystallogr. 2006;62(1):48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- Lin YP, Gregory V, Bennett M, Hay A. Recent changes among human influenza viruses. Virus Res. 2004;103(1–2):47–52. doi: 10.1016/j.virusres.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Lugovtsev VY, Vodeiko GM, Strupczewski CM, Ye Z, Levandowski RA. Generation of the influenza B viruses with improved growth phenotype by substitution of specific amino acids of hemagglutinin. Virology. 2007;365(2):315–323. doi: 10.1016/j.virol.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Maeda T, Ohnishi S. Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett. 1980;122(2):283–287. doi: 10.1016/0014-5793(80)80457-1. [DOI] [PubMed] [Google Scholar]
- Matlin KS, Reggio H, Helenius A, Simons K. Infectious entry pathway of influenza virus in a canine kidney cell line. J. Cell Biol. 1981;91(3 Pt 1):601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 2000;74(18):8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, Wang GC, He S, Webster RG. Reassortment and insertion– deletion are strategies for the evolution of influenza B viruses in nature. J. Virol. 1999;73(9):7343–7348. doi: 10.1128/jvi.73.9.7343-7348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Kubota R, Nakagawa T, Okuno Y. Antigenic variants with amino acid deletions clarify a neutralizing epitope specific for influenza B virus Victoria group strains. J. Gen. Virol. 2001;82(9):2169–2172. doi: 10.1099/0022-1317-82-9-2169. [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Kubota R, Nakagawa T, Okuno Y. Neutralizing epitopes specific for influenza B virus Yamagata group strains are in the ‘loop’. J. Gen. Virol. 2003;84(4):769–773. doi: 10.1099/vir.0.18756-0. [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Kubota R, Okuno Y. Variation of the conserved neutralizing epitope in influenza B virus victoria group isolates in Japan. J. Clin. Microbiol. 2005;43(8):4212–4214. doi: 10.1128/JCM.43.8.4212-4214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Holmes EC. The evolution of epidemic influenza. Nat. Rev. Genet. 2007;8(3):196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- Nerome R, Hiromoto Y, Sugita S, Tanabe N, Ishida M, Matsumoto M, Lindstrom SE, Takahashi T, Nerome K. Evolutionary characteristics of influenza B virus since its first isolation in 1940: dynamic circulation of deletion and insertion mechanism. Arch. Virol. 1998;143(8):1569–1583. doi: 10.1007/s007050050399. [DOI] [PubMed] [Google Scholar]
- Nunes B, Pechirra P, Coelho A, Ribeiro C, Arraiolos A, Rebelo-de-Andrade H. Heterogeneous selective pressure acting on influenza B Victoria- and Yamagata-like hemagglutinins. J. Mol. Evol. 2008;67(4):427–435. doi: 10.1007/s00239-008-9154-9. [DOI] [PubMed] [Google Scholar]
- Palese P, Shaw M. Orthomyxoviridae: the viruses and their replications. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. fifth ed. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1648–1690. [Google Scholar]
- Pechirra P, Nunes B, Coelho A, Ribeiro C, Goncalves P, Pedro S, Castro LC, Rebelo-de-Andrade H. Molecular characterization of the HA gene of influenza type B viruses. J. Med. Virol. 2005;77(4):541–549. doi: 10.1002/jmv.20490. [DOI] [PubMed] [Google Scholar]
- Powell HR. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55(10):1690–1695. doi: 10.1107/s0907444999009506. [DOI] [PubMed] [Google Scholar]
- Rachakonda PS, Veit M, Korte T, Ludwig K, Bottcher C, Huang Q, Schmidt MF, Herrmann A. The relevance of salt bridges for the stability of the influenza virus hemagglutinin. FASEB J. 2007;21(4):995–1002. doi: 10.1096/fj.06-7052hyp. [DOI] [PubMed] [Google Scholar]
- Rivera K, Thomas H, Zhang H, Bossart-Whitaker P, Wei X, Air GM. Probing the structure of influenza B hemagglutinin using site-directed muta-genesis. Virology. 1995;206(2):787–795. doi: 10.1006/viro.1995.1001. [DOI] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127(2):361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175(1):59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- Sato SB, Kawasaki K, Ohnishi S. Hemolytic activity of influenza virus hemagglutinin glycoproteins activated in mildly acidic environments. Proc. Nat. Acad. Sci. U.S.A. 1983;80(11):3153–3157. doi: 10.1073/pnas.80.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MW, Xu X, Li Y, Normand S, Ueki RT, Kunimoto GY, Hall H, Klimov A, Cox NJ, Subbarao K. Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000–2001 and 2001–2002 seasons. Virology. 2002;303(1):1–8. doi: 10.1006/viro.2002.1719. [DOI] [PubMed] [Google Scholar]
- Shen J, Kirk BD, Ma J, Wang Q. Diversifying selective pressure on influenza B virus hemagglutinin. J. Med. Virol. 2009;81(1):114–124. doi: 10.1002/jmv.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164(1):49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303(5665):1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu. Rev. Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vareckova E, Mucha V, Wharton SA, Kostolansky F. Inhibition of fusion activity of influenza A haemagglutinin mediated by HA2-specific monoclonal antibodies. Arch. Virol. 2003;148(3):469–486. doi: 10.1007/s00705-002-0932-1. [DOI] [PubMed] [Google Scholar]
- Viswanathan K, Koh X, Chandrasekaran A, Pappas C, Raman R, Srinivasan A, Shriver Z, Tumpey TM, Sasisekharan R. Determinants of glycan receptor specificity of H2N2 influenza A virus hemagglutinin. PLoS One. 2010;5(10):e13768. doi: 10.1371/journal.pone.0013768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Cheng F, Lu M, Tian X, Ma J. Crystal structure of unliganded influenza B virus hemagglutinin. J. Virol. 2008;82:3011–3020. doi: 10.1128/JVI.02477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Tian X, Chen X, Ma J. Structural basis for receptor specificity of influenza B virus hemagglutinin. Proc. Nat. Acad. Sci. U.S.A. 2007;104(43):16874–16879. doi: 10.1073/pnas.0708363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Berton MT. Analysis of antigenic drift in the haemagglutinin molecule of influenza B virus with monoclonal antibodies. J. Gen. Virol. 1981;54(2):243–251. doi: 10.1099/0022-1317-54-2-243. [DOI] [PubMed] [Google Scholar]
- Wright P, Neumann G, Kawaoka Y. Orthomyxoviruses. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1693–1741. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.