Abstract
The Gram-negative intracellular pathogen Brucella abortus is the causative agent of brucellosis, which is among the most common zoonoses globally. The B. abortus RicA protein binds the host-expressed guanosine nucleotide-binding protein, Rab2, and modulates B. abortus infection biology. We have solved the first X-ray crystal structure of RicA to 2.7 Å resolution, and have quantified the affinity of RicA binding to human Rab2 in its GDP bound and nucleotide free forms. RicA adopts a classic γ-carbonic anhydrase (γ-CA) fold containing a left-handed β-helix followed by a C-terminal α-helix. Two homotrimers of RicA occupy the crystallographic asymmetric unit. Though no zinc was included in the purification or crystallization buffers, zinc is contained within the RicA crystals as demonstrated by X-ray fluorescence spectroscopy. Electron density for a Zn2+ ion coordinated by three histidine residues is evident in the putative active site of RicA. However, purified RicA preparations do not exhibit carbonic anhydrase activity, suggesting that Zn2+ may not be the physiologically relevant metal cofactor, or that RicA is not a bona fide carbonic anhydrase enzyme. Isothermal titration calorimetry (ITC) measurements of purified RicA binding to purified human Rab2 and GDP-Rab2 revealed similar equilibrium affinities (KD ≈ 35 µM and 40 µM, respectively). This study thus defines RicA as a Zn2+ binding, γ-carbonic anhydrase-like protein that binds the human membrane fusion/trafficking protein, Rab2, with low micromolar affinity in vitro. These results support a model in which γ-CA family proteins may evolve unique cellular functions while retaining many of the structural hallmarks of archetypal γ-CA enzymes.
Carbonic anhydrases (CA) are broadly conserved metalloenzymes 1–5 that function to reversibly hydrate carbon dioxide to bicarbonate (CO2 + H2O <−> HCO3− + H+). CAs are required for many cellular processes including photosynthesis, respiration, and CO2 transport and fixation 3, 4, 6–8. Five independent CA classes have been defined on the basis of their three-dimensional structures: α, β, γ, δ and ζ. Though the structures of these enzymes are distinct, all five catalyze the same chemical transformation; in all cases catalysis involves a divalent metal-containing active site 1–5.
γ-class CAs are among the most ancient and broadly conserved members of the family 3, 7. In their monomeric form, γ-CAs are composed of a left-handed, seven-stranded parallel β-helix. An α-helix is appended to the C-terminus and positioned anti-parallel to the axis of the β-helix. Structural studies of γ-CAs have demonstrated that the monomers assemble into a trimer to form a triangular motif 3, 7, 9. In its trimeric active form, three Zn2+-containing active sites are formed. Each active site spans a monomer-monomer interface; the catalytic zinc ion is coordinated by three histidines from two adjacent monomers 7, 9, 10. In some cases, Fe2+ and not Zn2+ is the physiologically relevant active site cation 11–13.
The Cam and Cam-homologue (CamH) proteins of the archaeon Methanosarcina thermophila have been extensively characterized biochemically, and represent the archetypes of the γ-CA class 9–11, 14, 15. However, despite broad conservation very few γ-CAs have demonstrated carbonic anhydrase activity. The function of proteins classified as γ-CAs thus remains largely undefined. Indeed, only γ-CAs from M. thermophila (CamH and Cam) 12, 14, 15, the γ-CA domain of CcmM from Thermosynechococcus elongatus 16 and the γ-CA of Porphyromonas gingivalis (PgiCA) 17 have measurable activities in vitro. γ-CA-related proteins YrdA and Cap of Escherichia coli 18 and Pyrococcus horikoshii 19, respectively, and γCA2 of Arabidopsis thaliana 20, 21 have no reported enzymatic activity, despite significant homology to M. thermophila CamH. Sequence differences in the active sites, or the presence of non-physiological bound metal ions may explain the lack of anhydrase activity in these proteins 7, 10, 18–20, 22, 23, though the molecular basis of disparity in the activities of these closely related proteins remains unclear. Together, these results raise the possibility that γ-CA-related proteins may possess other functions beyond the chemical transformation of carbon dioxide and bicarbonate.
In the Gram-negative intracellular pathogen Brucella abortus 24, the first secreted effector was recently described 25. This protein, RicA (Rab2 interacting conserved protein A), is related to γ-CA-like proteins at the sequence level25, 26. RicA is secreted through an unknown mechanism and is reported to interact with the GDP-bound form of the mammalian host protein, Rab2 25, 26. Rab2 functions in membrane trafficking between the Golgi apparatus and the endoplasmic reticulum 27, and has been previously described as playing a role in the B. abortus infection and intracellular replication process 25, 28, 29. Thus, protein-protein interaction between RicA and Rab2 is postulated to directly modulate B. abortus trafficking inside the macrophage 25. These results suggest an entirely new function for a γ-CA family protein.
In this study, we report the crystal structure of RicA at 2.7 Å resolution and biochemically characterize its ligand binding properties. RicA adopts a classic γ-CA fold consisting a left-handed β-helix followed by a C-terminal α-helix and assembles into a homotrimer, of which two are present in the crystallographic asymmetric unit. Each homotrimer contains three zinc-binding sites. Enzyme activity assays did not yield measurable carbonic anhydrase activity from purified RicA samples under a variety of conditions. Thus B. abortus RicA is part of a growing list of γ-CA-related proteins without measurable carbonic anhydrase activity. We have further quantified the interaction of RicA with human Rab2 in its GDP-bound and unbound forms by isothermal titration calorimetry (ITC). RicA and Rab2 interact with an affinity in the tens of micromolar; this interaction does not depend on the bound nucleotide state of Rab2 under the assayed in vitro conditions. Our results thus provide direct experimental evidence that RicA is a structural homolog of γ-CA family proteins, and that this protein contains bound zinc ions in the putative enzyme active sites yet has no measurable carbonic anhydrase activity under the tested conditions. The effect of ricA mutations on B. abortus intracellular replication 25, 28, and RicA binding to Rab2 in a physiologically-relevant affinity range provides evidence that γ-CA family proteins may evolve unique cellular functions while retaining many of the structural hallmarks of γ-carbonic anhydrases.
Experimental Procedures
Construction of expression plasmids
The sequence encoding residues M1 to A175 of Brucella abortus RicA (gene number BruAb1_1263) was amplified by PCR from B. abortus genomic DNA. Primers used for amplification were RicA-UP, ATATCATATGCCGATCTATGCATATAACGG (NdeI) and RicA-LO, ATATAAGCTTTCAGGCAGGCTCCATGCCG (HindIII), for cloning into pET28c (Novagen). The sequence encoding the Homo sapiens Rab2 protein (Homo sapiens Gene ID: 5862) was synthesized by Integrated DNA Technologies (Coralville, IA) and reamplified with the following specific primers: Rab2-UP, ATATCATATGGCGTATGCCTACCTCTTTA (NdeI) and Rab2-LO, ATATAAGCTTCAACAACAGCCACCACCG (HindIII), for cloning into pET28c.
PCR products were resolved on an agarose gel and specific bands were isolated and purified by gel extraction (Omega Bio-tek), and digested with the corresponding restriction enzymes (New England Biolabs). Digested PCR products were repurified and ligated using T4 DNA ligase (New England Biolabs) into the corresponding sites of predigested and purified pET28c plasmid. pET28c plasmids encoding RicA (1–175) and Rab2 (1–212) with N-terminal His6 tags were transformed into chemically competent Top10 cells (Invitrogen). Strains FC2113 and FC2114 respectively carry pET28-ricA and pET28-rab2 plasmids.
RicA and Rab2 over-expression and purification
Recombinant His6-tagged RicA and Rab2 proteins were expressed in E. coli Rosetta(DE3)pLysS (Novagen) (strain numbers FC2115 and FC2116, respectively). A 50 ml overnight culture in Luria-Bertani (LB) medium supplemented with 50 µg/ml kanamycin (LabScientific) (LB-Kan50) was used to inoculate 2 liters of LB-Kan50; this culture was incubated at 37°C in a rotary shaker at 220 rpm. Transcription of recombinant proteins was induced at a culture density of 0.8 (monitored spectrophotometrically at 600 nm) by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, GoldBio Technology). After 4 h of induction, the cells were harvested by centrifugation at 12,000 × g for 20 min at 4°C. Cell pellets were resuspended in 30 ml of lysing/binding buffer (10 mM Tris pH 7.4, 150 mM NaCl, 10 mM imidazole supplemented with DNaseI (Sigma-Aldrich) and Phenyl-Methyl-Sulfonyl-Fluoride (PMSF; Sigma-Aldrich).
Cells were disrupted by three passages in a French pressure cell (Thermo Scientific), and the cell debris was removed by centrifugation for 20 min at 25,000 × g. The supernatant was loaded onto a Ni2+ Sepharose affinity column (GE Life Sciences) pre-equilibrated with the binding buffer. Two washing steps were performed using 10 mM and 75 mM of imidazole (Fisher Scientific) followed by two elution steps with 200 mM and 1 M imidazole in the binding buffer. Protein purity was estimated at 95 % as assessed by 14 % SDS-PAGE stained with Coomassie brilliant blue. The protein solution was then serially dialyzed against 10 mM Tris pH 7.4, 150 mM NaCl buffer to remove imidazole. All purification steps were carried out at 4°C.
Crystallization of RicA
Purified RicA was concentrated using a centrifugal filter (3 kDa MWCO, Amicon-Millipore). Initial crystallization screening was carried out using the sitting-drop, vapor-diffusion technique in 96-well microplates (Nunc). Trays were set up using a Mosquito robot (TTP LabTech) and commercial crystallization kits (Nextal-Qiagen). The hanging drops were set up by mixing equal volumes (0.1 µl) of the protein and the precipitant solutions equilibrated against 75 µl of the precipitant solution. In all trials, the protein concentration was ~ 40 mg/ml. In approximately one week at 19°C, small crystals appeared in condition 5 of the PEGs Suite crystallization kit (Qiagen). All manual crystallization attempts were carried out using the hanging-drop, vapor-diffusion technique in 24-well plates (Hampton). The drops were set up by mixing different volumes (1:1, 1:2 and 2:1 µl) of the protein (at 10 mg/ml) and the crystallization solutions equilibrated against 500 µl of the precipitant solution. After manual refinement, the best crystals were obtained at 19°C by mixing 1 µl of the protein solution (at 10 mg/ml) with 2 µl of the following crystallization solution: 100 mM Sodium Acetate (pH 4.6) (Fisher Scientific), 25 % (w/v) PEG 1000 (Acros Organics), 200 mM NaCl (Fisher Scientific) and 30 mM Ammonium Sulfate (Fisher Scientific). Crystals grew to their final size in one to two weeks. Before flash freezing with liquid nitrogen, crystals were cryo-protected by soaking in the crystallization solution containing 25 % glycerol (Fisher Scientific).
Crystallographic data collection and data processing
Diffraction from the majority of crystals was poor: diffraction was highly mosaic and anisotropic. The best crystal diffracted to dmin = 2.73 Å; a single dataset (200 images) was collected from this crystal at a temperature of 100 K using a 1 degree oscillation range on beamline 21-ID-F (LS-CAT, Advanced Photon Source, Argonne, Illinois). Images were collected on a MAR Mosaic 225 detector. Diffraction images were processed using the xia2 data reduction suite 30 applying the -3da XDS 31 and Aimless 32 options, which were accessed through the SBGrid consortium 33. Geometric refinement and examination of the scaled amplitudes revealed that RicA crystals belong to the monoclinic space group P21, with cell dimensions a = 102.43 A, b = 57.44 Å, c = 127.16 Å (β = 91.8°) (see Table 1). The structure was solved by molecular replacement in Phenix 34 using a model of RicA that was based on the structure of carbonic anhydrase from Pyrococcus horikoshii (PDB code: 1V3W). For purposes of the molecular replacement search, the number of RicA molecules in the asymmetric unit was estimated using the Matthews Probability Calculator web server (http://www.ruppweb.org/mattprob/). The RicA search model was generated using the Expasy Swiss-model Workspace server35.
Table 1.
Crystallographic Data and Refinement Statistics.
| Data Collection Statistics | |
| X-ray energy (keV) | 12.66 |
| X-ray wavelength (Å) | 0.979 |
| Resolution range (Å) | 20 - 2.73 (2.83 - 2.73) |
| Unique Reflections | 39784 |
| Rmergea | 0.08 |
| I / σ (I) | 9.44 (1.80) |
| Redundancy | 4.2 (3.8) |
| Completeness | 99.8 (99.8) |
| Refinement Statistics | |
| Space group | P21 |
| a, b, c (Å) | 102.4, 57.4, 127.6 |
| β (°) | 91.8 |
| Rcrystc | 0.205 |
| Rfreed | 0.254 |
| Wilson B factor (Å2) | 47.0 |
| RMSDe bond lengths (Å) | 0.01 |
| RMSD bond angles (º) | 1.40 |
| Ramachandran analysis | |
| Preferred (%) | 96 |
| Allowed (%) | 4.1 |
| Disallowed (%) | 0.2 |
Rmerge = ∑hkl∑i |Ii−〈I〉| / ∑hkl∑iIi, for all data I/σ(I) > 3
Rcryst = ∑hkl ||Fobs|−|Fcalc|| / ∑hkl |Fobs|, includes all data
Rfree uses 2005 total reflections for cross-validation.
rmsd, root-mean-square deviation.
We located two RicA trimers within the crystallographic asymmetric unit. The initial structural model was manually examined and corrected; ions, PEG and waters were added, and refinement of the structure was conducted iteratively using Coot 36 and phenix.refine 34. The final structural model was refined to an Rwork of 20.5 % and Rfree of 25.4 %. Coordinates of B. abortus RicA have been deposited in the Protein Data Bank (PDB code: 4N27). Crystallographic data and refined model statistics are presented in Table 1.
Carbonic anhydrase activity assay
The carbonic anhydrase activity of RicA was assessed using a colorimetric method adapted from the Wilbur–Anderson assay 37, 38. Purified RicA was serially dialyzed overnight against 2 liters of Tris-NaCl buffer to remove imidazole and concentrated to 40 mg/ml. In a 1 cm cuvette, dialyzed RicA (final concentration 5 µM) was mixed with 500 µl of a Tris-NaCl phenol red buffer (20 mM Tris pH8.8, 150 mM NaCl, 1 mM ZnCl2, 200 µM phenol red) and 500 µl of water saturated with CO2 using dry ice. Immediately after addition of CO2 saturated water, the decrease of pH was indirectly monitored at 558 nm by measuring the red to yellow color shift of the phenol red buffer.
As a control, the same experiment was performed with the dialysis buffer alone and with a commercial carbonic anhydrase from bovine erythrocytes (final concentration 5 nM, Sigma-Aldrich). A UV-1650pc Shimadzu spectophotometer was used for measurements. All measurements were done in triplicate at room temperature. Carbonic anhydrase activity (Wilbur–Anderson Units, WAU) per milligram was calculated according to the following equation:
where t0 and t are the times that are required for uncatalyzed (buffer control) and catalyzed (with enzyme) reactions to drop to the transition point of the dye, respectively 37.
Isothermal titration calorimetry binding measurements
All samples (proteins and buffers) were degassed for 20 min prior to ITC measurements, and final sample dilutions were carried out using the final degassed dialysis buffer (10 mM Tris pH7.4, 150 mM NaCl). Rab2-GDP bound (2 mM Rab2 in Tris-NaCl buffer supplemented with 10 mM GDP and 10 mM MgCl2) was injected into a 200 µl sample cell containing 200 µM of RicA in the same buffer. ITC was performed at 25°C with a 2 µl injection volume every 180 s. GDP-Rab2 was titrated into ITC buffer alone, and the resulting heat of dilution was subtracted from the experimental curve. The equivalent experiment without GDP was performed in the same conditions. ITC was conducted using an iTC200 microcalorimeter (MicroCal, GE Healthcare). Data were analyzed and fitted using the Microcal Origin software suite. A ‘One Set of Sites’ curve-fitting model was used 39, 40. Each titration was performed three times.
X-ray Fluorescence measurement
To assay for the presence of zinc in RicA, we scanned a single crystal at Advanced Photon Source beamline 21-ID-D (LS-CAT) at an energy range of 9.62 to 9.69 keV, which spans the K-edge absorption of zinc. X-ray fluorescence as a function of energy was measured on a Bruker-AXS XFlash 1001 SDD detector. Anomalous scattering curves f’ and f” were calculated from the measured fluorescence data.
Sequence alignment and protein visualization methods
The Dali server (http://ekhidna.biocenter.helsinki.fi/dali_server/) 41 was used to compare the RicA structure to structures in the Protein Data Bank (PDB). Protein sequence alignments were done with T-Coffee Expresso sever (http://www.tcoffee.org/) 42 and shaded in Boxshade (http://www.ch.embnet.org/software/BOX_form.html). RicA ribbon rendering and γ-carbonic anhydrase structural alignment were performed with PyMOL (Version 1.3 Schrödinger, LLC). The PDBsum server (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html) was used to define the molecular interaction map between the different RicA molecules in the trimer.
Results
RicA has the secondary, tertiary and quaternary structural properties of a γ-carbonic anhydrase and contains bound zinc ions
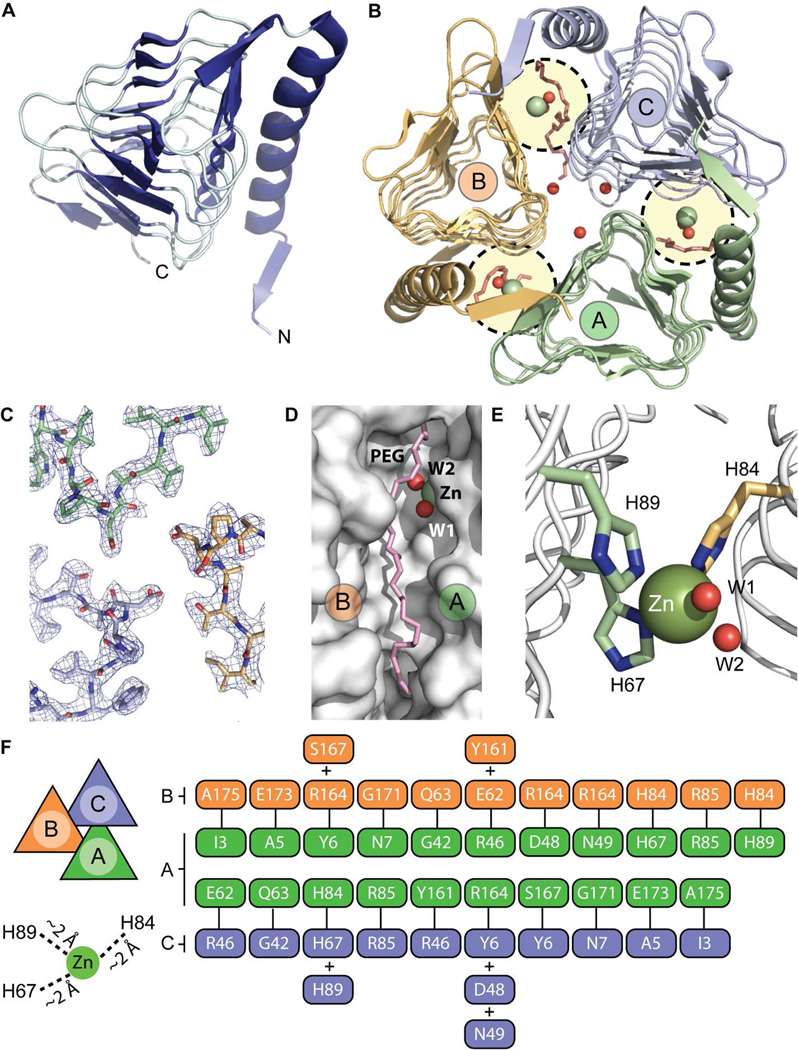
An amino terminal His6 fusion of B. abortus RicA was expressed in E. coli and purified by Ni2+ affinity chromatography. His6-RicA (1–175), hereafter referred to simply as RicA, was crystallized and formed monoclinic crystals of space group P21 with cell dimensions of 102.4, 57.4, and 127.6 Å (β=91.8°). The X-ray crystal structure of RicA was solved to 2.7 Å resolution by molecular replacement using a structural homology model based on the γ-carbonic anhydrase of P. horikoshii (Cap; PDB code : 1V3W). Six molecules of RicA (A, B, C, D, E and F) are contained within the asymmetric unit, organized as two homotrimers (Figure 1A and B). We observed no major structural differences between the two homotrimers in the asymmetric unit in the refined structure. However, we noted small unidentified difference density (adjacent to residue Y161) near the active site in all chains, which we could not unambiguously define at this resolution. Statistics relating to crystallographic data and the final refined model are summarized in Table 1. A simulated annealing composite omit electron density map (contoured at 2σ) of the interface region between the three RicA molecules is shown Figure 1C.
Figure 1.
Structural characterization of RicA (PDB code : 4N27). (A) RicA monomer is a left-handed β-helix containing a C-terminal α-helix. (B) Trimeric form of RicA. Three monomers of RicA (A, B and C) interact with each other to form a closed, triangular, homotrimer. Between two monomers, one active site is present (circled in yellow) which contains one zinc ion (green spheres), two water molecules (red spheres), one PEG molecule (in red). Additional water molecules present at the center of the trimer are shown as red spheres. (C) Simulated annealing composite omit electron density map of the interface between monomer A (in green), monomer B (in orange) and C (in blue). (D) Zoom view of the cleft between molecule A and B containing a PEG molecule (in pink) and the zinc-binding site (zinc ion is green; water molecules are red). (E) Zoom view of the zinc-binding site; residues H89 and H67 (from molecule A; pale green) and residue H84 (from molecule B; pale orange) are involved in coordination of the zinc ion (green sphere). Two water molecules near the zinc site are highlighted in red. (F) Hydrogen bond interaction map between residues of monomers A and B and monomers A and C. Hydrogen bond cut-off was defined as < 3.25 Å. Distances between the zinc ion and the histidine side chains are also labeled.
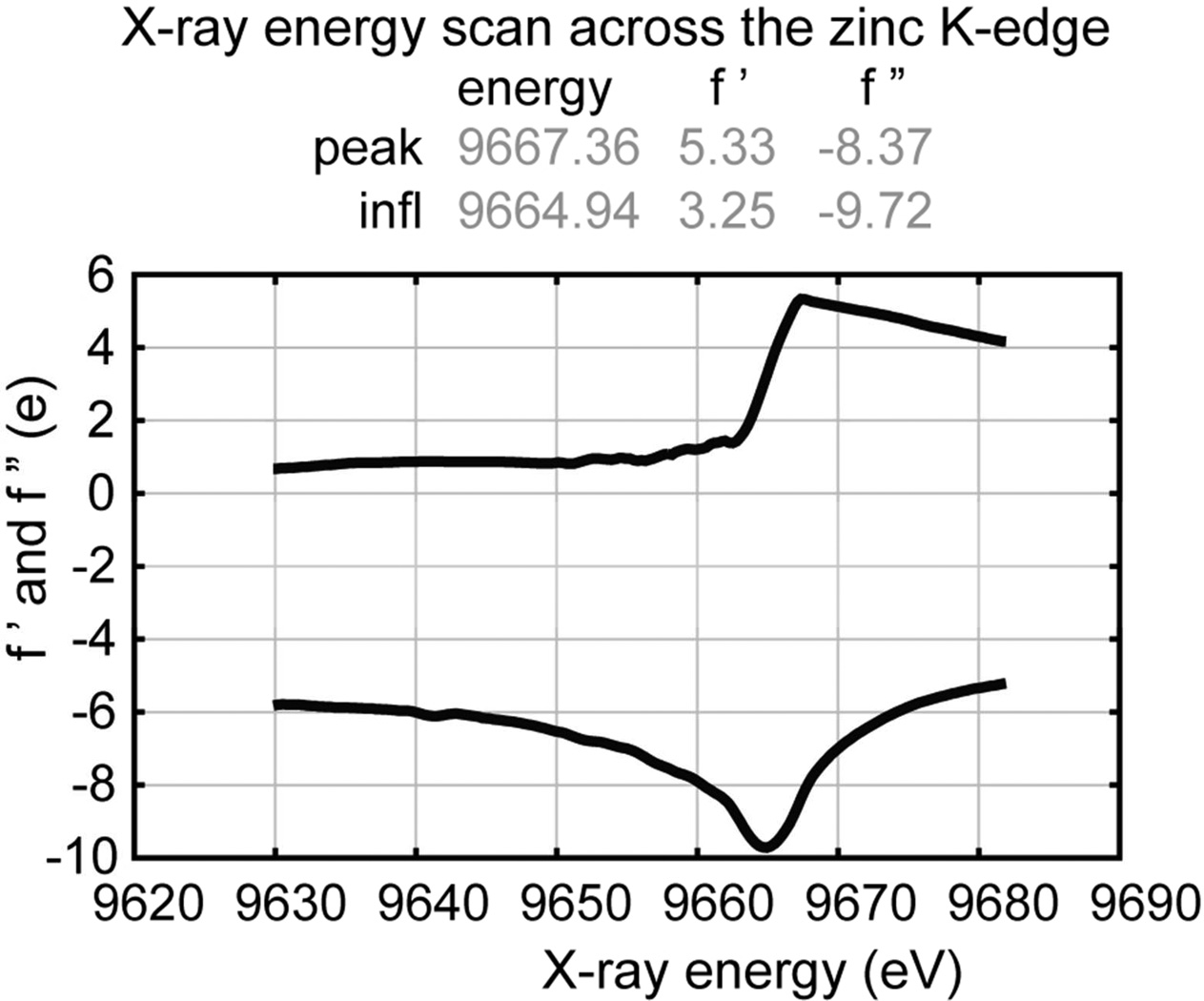
Each monomer contains 175 amino acids; the His6 affinity tag is not visible in the electron density map (Figure 1A). Six additional zinc ions, six polyethyleneglycol (PEG) molecules, eighteen water molecules were also modeled into the electron density (see Figure 1B, D, E). In the RicA homotrimer, each monomer (A, B and C) consists of a left-handed parallel β-helix and a long C-terminal α-helix running anti-parallel to the axis of the β-helix (Figure 1A). At the interface between each monomer (A–B, B–C and C–A), 13 hydrogen bonds (39 hydrogen bonds total in the homotrimer) are formed (Figure 1F). A putative γ-CA active site is formed by two monomers facing each other (A–B, B–C and C–A) (Figure 1B and E). In each of three putative γ-CA active sites, a zinc ion is coordinated by three histidines (H67 and H89 from one monomer and H84 from and adjacent monomer) (Figure 1E). The identity of this bound metal ion as zinc is consistent with the anomalous scattering curves f’ and f”, which were calculated from X-ray fluorescence data measured from a single RicA crystal scanned at an energy range of 9.62 to 9.69 keV. This energy range spans the zinc K absorption edge, and thus provides a unique spectral signature that distinguishes zinc from other elements 43–45 (Figure 2). We did not add zinc to the purification or crystallization buffers, so we presume that zinc co-purified with RicA from the heterologous E. coli expression system.
Figure 2.
X-ray fluorescence scan of a single RicA crystal from 9620 to 9690 electron volts. Anomalous scattering coefficients f' and f" are plotted as a function of incident X-ray energy.
Also present in the vicinity of the zinc ion are two water molecules and one PEG molecule (Figure 1B, D and E). Equivalent water molecules have been described in Cap 19, Cam 9, 10 and CcmM 16 structures as important for carbonic anhydrase catalytic activity 10. Overall, B. abortus RicA has high structural similarity (average RMSD : 0.53 Å) to published structures of γ-CA family proteins 9, 10, 16, 18, 19.
RicA has no measurable carbonic anhydrase activity
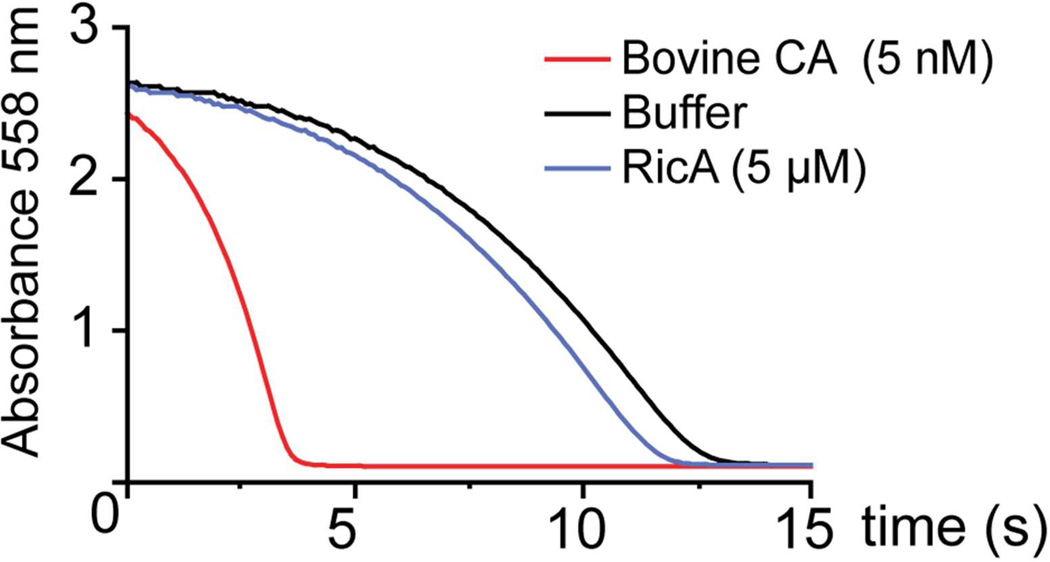
Given its high structural homology to γ-carbonic anhydrases, we sought to determine whether purified preparations of RicA exhibited carbonic anhydrase activity. We assayed enzyme activity using a standard pH dye indicator method 38. The rate of pH change in a Tris-NaCl phenol red buffer saturated with CO2 was monitored after addition of purified RicA at concentrations up to 5 µM (measuring phenol red absorbance at 558 nm), and compared to a buffer only control. The rate of pH change upon addition of RicA was equivalent to the buffer only control (Figure 3). An activity of less than −0.7 ± 0.1 Wilbur–Anderson units per milligram (WAU) was measured. As a positive control, we performed the same experiment with purified bovine erythrocyte β-carbonic anhydrase (final concentration 5 nM), which catalyzed rapid hydration of carbon dioxide and acidification of the buffer (Figure 3); an activity of 13383 ± 3016 Wilbur–Anderson units per milligram (WAU) was measured. We therefore conclude that, under the assayed conditions, RicA does not have carbonic anhydrase activity. To examine whether RicA has particular structural features that may explain its lack of activity, we compared our 2.7 Å RicA crystal structure to structures of bona fide γ-carbonic anhydrases deposited in the Protein Data Bank.
Figure 3.
Carbonic anhydrase assay. CO2 hydration (CO2 + H2O <−> HCO3− + H+) by RicA (5 µM) was indirectly evaluated by measuring decrease in pH in a phenol red buffer (blue line). Change in absorbance at 558 nm was recorded and compared to a negative control (buffer alone, black line) and to a positive control (5 nM Bovine carbonic anhydrase, red line).
A comparative structural analysis of B. abortus RicA to γ-carbonic anhydrase family proteins
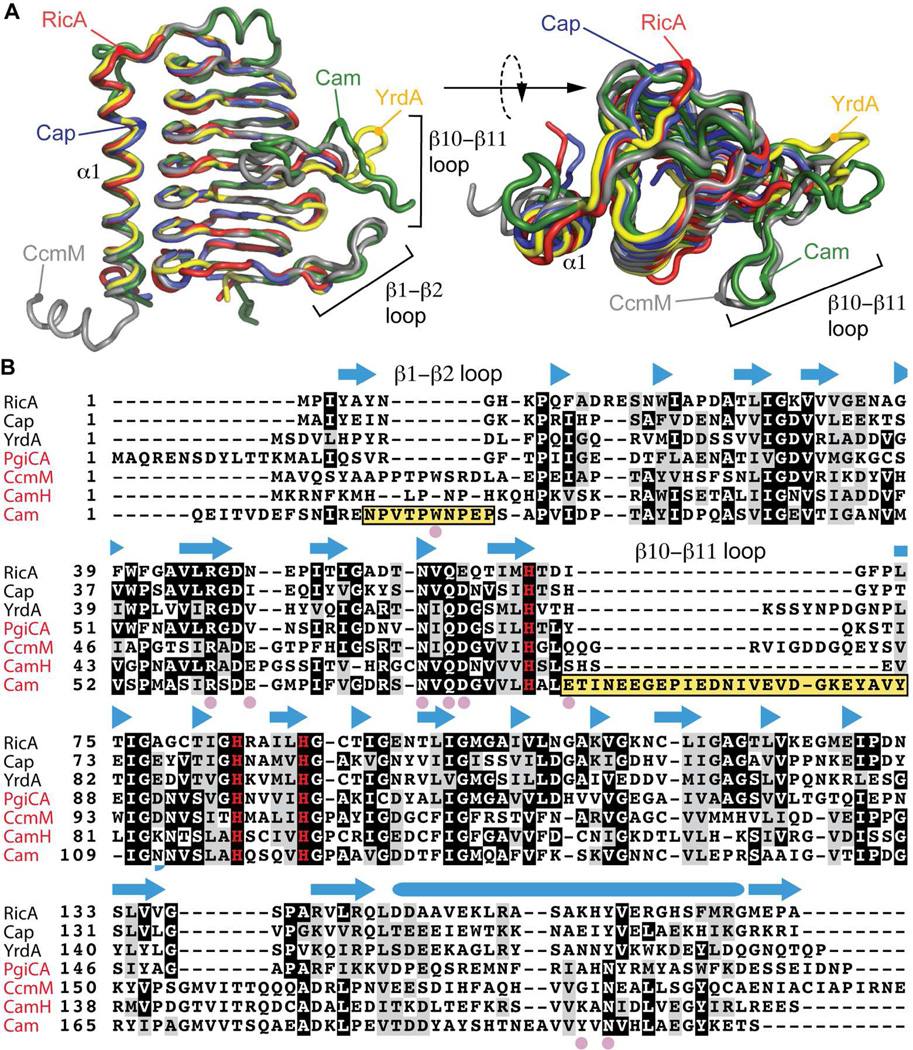
Compared to structurally characterized γ-CAs (Cam, CcmM, Cap and YrdA), RicA has a highly related molecular structure (average RMSD : 0.53 ± 0.14 Å) (Figure 4A). RicA has marginally higher overall sequence and structural similarity to the non-active γ-CA homologs YrdA (overall amino acid sequence identity: 35%; overall RMSD : 0.43 Å) and Cap (identity : 41%, RMSD : 0.48 Å). Structural features directly related to carbonic anhydrase activity in Cam (identity : 21%; RMSD : 0.51 Å) and CcmM (identity : 25%, RMSD : 0.74 Å), are missing in the structure of B. abortus RicA (Figure 4A and B). These features include the loop β1–β2 as well as the loop β10–β11, which contribute to the formation of the Cam 9, 22 and CcmM 16 active sites. However, the loops β1–β2 as well as the loop β10–β11are also absent in the active γ-carbonic anhydrases PgiCA (identity : 31%) and CamH (identity : 27 %). Thus, absence of these loops is not sufficient to explain the lack of measurable carbonic anhydrase activity in purified preparations of RicA.
Figure 4.
Structural comparison of RicA to related γ-CA family proteins. A) Structural alignment of RicA (red) to YrdA (yellow, PDB code : 3TIO), Cap (blue, PDB code : 2FKO), CcmM (grey, PDB code : 3KWC) and Cam (green, PDB code : 1QRE). B) Amino acid sequence alignment of RicA to γ-CA proteins from Escherichia coli (YrdA), Pyrococcus horikoshii (Cap), Porphyromonas gingivalis (PgiCA), Thermosynechococcus elongatus (CcmM) and Methanosarcina thermophil (CamH and Cam). Secondary structure of RicA is annotated above the amino acid sequence. β-strands are represented with blue arrows; α-helix by blue cylinder. Pink solid circles highlight residues in Cam present in the active site and important for carbonic anhydrase activity. Yellow boxes highlight the β1–β2 and β10–β11 loops of Cam. The three histidines (H67, H84 and H89) involved in zinc coordination are shown in red. Names of the proteins with a reported carbonic anhydrase activities are in red.
At the amino acid sequence level, residues described as important for activity of the archetypal CA, Cam 9, 10, 22, 23, are conserved in RicA with the exception of W19 (present in the β1–β2 loop), E62, D76, E84, Y200 and N202 (Figure 4B). Residue D76 is conserved in all known γ-CA and γ-CA-related structures; the general consequence of the conservative substitution to glutamic acid at this position in RicA is unknown (Figure 4B). Residues including E62, E84 and Y200 are also variable at equivalent positions of other active γ-carbonic anhydrases (Figure 4B), suggesting that they are not generally required for γ-carbonic anhydrase activity. An asparagine residue at position 202 (N202) is found exclusively in the active γ-CA enzymes, and may thus play an important role in CA activity. In conclusion, we observed no differences at the secondary and tertiary structural levels that obviously explain the lack of measurable CA activity in purified RicA preparations under the tested conditions. Given its established role as a Rab2-interacting effector protein 25, we next sought to quantitatively characterize the molecular interaction of purified B. abortus RicA with purified human Rab2.
RicA binds human Rab2 in its GDP-bound and GDP-free forms with micromolar affinity
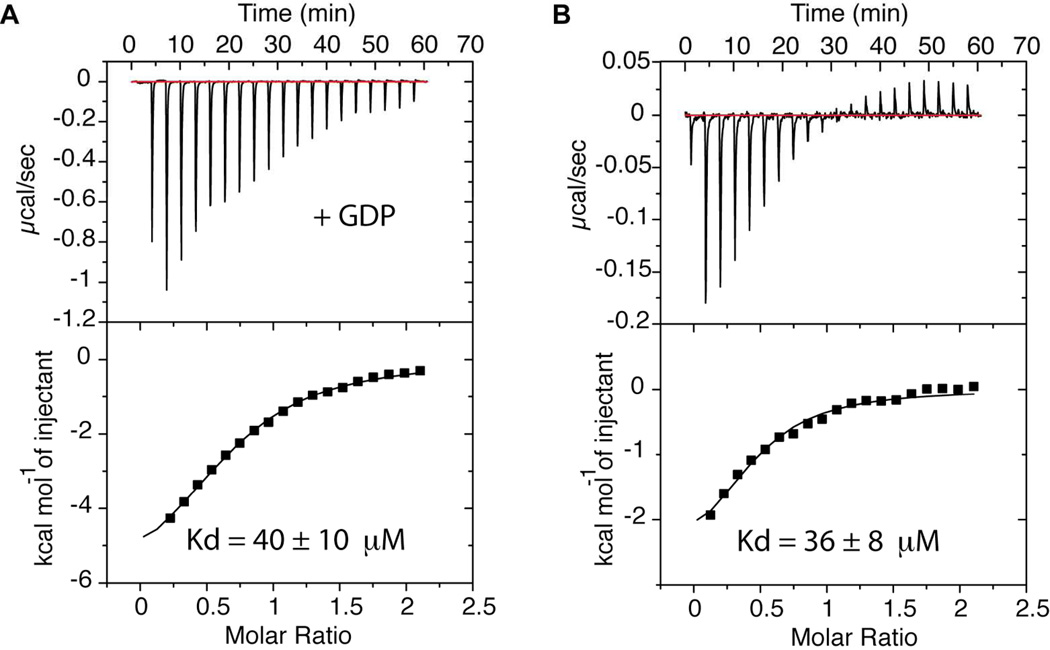
To quantify the interaction between human Rab2 and B. abortus RicA, we conducted binding measurements using isothermal titration calorimetry (ITC). For ITC, 2 mM human Rab2 was preincubated in a buffer composed of 10 mM Tris pH 7.4, 150 mM NaCl, 10 mM MgCl2, and 10 mM GDP. 2 µl of Rab2 were injected into a cell containing 200 µM RicA in an identical buffer. Under these conditions, the two proteins interact with an apparent affinity of 40 ± 10 µM. We then conducted the same experiment without GDP in the buffer, which yielded an apparent binding affinity of 36 ± 8 µM (Figure 5A and 5B).
Figure 5.
Quantification of RicA-Rab2 binding. A) ITC assay performed on purified His6-RicA and GDP-Rab2. GDP-Rab2 (2 mM) was injected stepwise into a cell containing RicA (200 µM) and the heat of binding was measured for every injection. B) ITC assay performed on purified His6-RicA and Rab2. Rab2 (2 mM) was injected stepwise into a cell containing RicA (200 µM) and the heat of binding was measured for every injection.
Discussion
We have solved the first X-ray crystal structure of the B. abortus RicA protein, and have defined its cofactor and ligand-binding properties. The protein adopts a classic γ-CA fold with a left-handed β-helix and a C-terminal α-helix (Figure 1), thus validating structural predictions based on amino acid sequence 26. RicA crystallizes as a homotrimer that contains zinc, as determined by X-ray fluorescence spectroscopy (Figure 2); the zinc ions occupy a position at the putative carbonic anhydrase active site within the RicA structure. However, despite the structural and chemical similarity to known active γ-CA proteins, we did not detect measurable carbonic anhydrase activity in purified RicA preparations (Figure 3). Overall, our results demonstrate that RicA can be clearly classified as a structural homolog of the γ-CA family (specifically, part of the CamH subclass) (Figure 4).
For select γ-CA enzymes, it has been suggested that zinc may not be the physiologically relevant metal cofactor. For example, anaerobically purified M. thermophila CamH contains Fe2+ in its active site and is 3-fold more active in comparison to the zinc-bound form of this protein 11–13. Oxidation of Fe2+ to Fe3+ during the purification process could result in substitution of Zn2+ in the active site, which is more stable than Fe3+ 7. We cannot exclude the possibility that B. abortus RicA primarily functions under conditions that are sufficiently reducing to support binding to Fe2+, and that such ferrous iron is required for RicA CA activity. However, we note that γ-CAs including Cam, CamH, and PgiCA can be purified aerobically, contain zinc, and still have measurable catalytic activities 12, 14, 17. Perhaps the most remarkable feature of the γ-CA homolog, RicA, is that it interacts with the eukaryotic membrane trafficking protein, Rab2 25, 26. Rab2 binding is proposed to modulate the interaction between B. abortus and its mammalian host cell. This provides evidence that a γ-CA protein (RicA) has evolved a unique function, beyond the reversible hydration of CO2. To further quantify the biochemical properties of RicA, we measured the interaction between RicA and human Rab2 by ITC. Heterologously-expressed and purified B. abortus RicA and human Rab2 bind with an equilibrium affinity in the tens of micromolar range regardless of the presence of GDP in the buffer (Figure 5). However, it has been reported that the GDP bound form of Rab2 binds RicA with higher affinity 25. Under the conditions in our in vitro assay, we were unable to detect a GDP-dependence on the RicA-Rab2 interaction. Other cellular factors or post-translational modifications absent from our ITC assay on purified components may explain why we do not observe a GDP-dependence on binding; we elaborate on this possibility below.
At the molecular level, the interaction between RicA and Rab2 is moderate affinity (> 10 µM). That stated, physiologically-relevant moderate or low affinity protein-protein interactions have been previously described in other systems. Examples include human CD8αα binding to the MHC class I molecule HLA-A2 (KD ≈ 0.2 mM) 46, and binding of the UBA domain of Cbl-b ubiquitin ligases to ubiquitin (KD ≈ 0.2 mM) 47. In the case of RicA-Rab2 interaction, the measured affinity reported in this study suggests that the local concentration of these two proteins in the Brucella containing vacuole is high. Alternatively, our in vitro data do not exclude cellular factors or post-translational modifications to RicA or Rab2 that substantially modify binding affinity between these two proteins in the context of the host cell. Indeed, post-translational modification (prenylation) of Rab proteins is known to enhance Rab binding to the GDP-dissociation inhibitor protein (GDI) in yeast, increasing affinity from the micromolar to nanomolar range 48–51.
We also note that RicA could possibly bind different Rab proteins with different affinities. For example, the Legionella pneumophila effector LidA binds both GDP- and GTP-bound Rab1 with high affinity (~ 8 nM) but has also been reported to interact with Rab2 with lower affinity (~ 8 µM) 52.
Further molecular characterization of RicA in the context of a mammalian host is necessary to relate our structural and biochemical studies to the role of this protein in B. abortus infection. In particular, it is important to understand the mechanism by which RicA interaction with Rab2 in the macrophage affects the proliferation of B. abortus. Our clear identification of RicA as a metalloprotein raises the possibility that the metal composition of the bacterium or the host could serve as a possible signal that modulates that activity of RicA as an effector, or as a bona fide carbonic anhydrase enzyme. Such a model is congruent with an increasing appreciation of the importance of metals in Brucella virulence 53 and in bacterial infection biology generally 54.
Acknowledgements
We thank Spencer Anderson of the Life Sciences Collaborative Access Team (LS-CAT) for protein X-ray fluorescence measurements.
Funding source: S.C. acknowledges support for this study by the National Institutes of Health (U19 AI107792) and by the Region V ‘Great Lakes’ RCE (NIH grant U54 AI057153). The Advanced Photon Source is supported by the Department of Energy Office of Basic Energy Sciences (Contract DE-AC02-06CH11357). LS-CAT is supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Abbreviations
- RicA
Rab2 interacting conserved protein
- RMSD
root mean square deviation
- CA
carbonic anhydrase
- GDP
guanosine diphosphate
- PEG
polyethyleneglycol
- ITC
isothermal titration calorimetry
- IPTG
isopropyl β-D-1-thiogalactopyranoside.
References
- 1.Tripp BC, Smith K, Ferry JG. Carbonic anhydrase: new insights for an ancient enzyme. J Biol Chem. 2001;276:48615–48618. doi: 10.1074/jbc.R100045200. [DOI] [PubMed] [Google Scholar]
- 2.Smith KS, Jakubzick C, Whittam TS, Ferry JG. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc Natl Acad Sci U S A. 1999;96:15184–15189. doi: 10.1073/pnas.96.26.15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith KS, Ferry JG. Prokaryotic carbonic anhydrases. FEMS Microbiol Rev. 2000;24:335–366. doi: 10.1111/j.1574-6976.2000.tb00546.x. [DOI] [PubMed] [Google Scholar]
- 4.Esbaugh AJ, Tufts BL. The structure and function of carbonic anhydrase isozymes in the respiratory system of vertebrates. Respir Physiol Neurobiol. 2006;154:185–198. doi: 10.1016/j.resp.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Cherniad'ev II, Terekhova IV, Komarova Iu M, Doman NG, Goronkova OI. Role of carbonic anhydrase in photosynthesis. Dokl Akad Nauk SSSR. 1975;223:501–503. [PubMed] [Google Scholar]
- 6.Supuran CT. Carbonic anhydrases--an overview. Curr Pharm Des. 2008;14:603–614. doi: 10.2174/138161208783877884. [DOI] [PubMed] [Google Scholar]
- 7.Ferry JG. The gamma class of carbonic anhydrases. Biochim Biophys Acta. 2010;1804:374–381. doi: 10.1016/j.bbapap.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferry JG. Carbonic anhydrases of anaerobic microbes. Bioorg Med Chem. 2013;21:1392–1395. doi: 10.1016/j.bmc.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC. A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 1996;15:2323–2330. [PMC free article] [PubMed] [Google Scholar]
- 10.Iverson TM, Alber BE, Kisker C, Ferry JG, Rees DC. A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry. 2000;39:9222–9231. doi: 10.1021/bi000204s. [DOI] [PubMed] [Google Scholar]
- 11.Macauley SR, Zimmerman SA, Apolinario EE, Evilia C, Hou YM, Ferry JG, Sowers KR. The archetype gamma-class carbonic anhydrase (Cam) contains iron when synthesized in vivo. Biochemistry. 2009;48:817–819. doi: 10.1021/bi802246s. [DOI] [PubMed] [Google Scholar]
- 12.Zimmerman SA, Tomb JF, Ferry JG. Characterization of CamH from Methanosarcina thermophila, founding member of a subclass of the {gamma} class of carbonic anhydrases. J Bacteriol. 2010;192:1353–1360. doi: 10.1128/JB.01164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripp BC, Bell CB, 3rd, Cruz F, Krebs C, Ferry JG. A role for iron in an ancient carbonic anhydrase. J Biol Chem. 2004;279:6683–6687. doi: 10.1074/jbc.M311648200. [DOI] [PubMed] [Google Scholar]
- 14.Alber BE, Ferry JG. A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc Natl Acad Sci U S A. 1994;91:6909–6913. doi: 10.1073/pnas.91.15.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alber BE, Ferry JG. Characterization of heterologously produced carbonic anhydrase from Methanosarcina thermophila. J Bacteriol. 1996;178:3270–3274. doi: 10.1128/jb.178.11.3270-3274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pena KL, Castel SE, de Araujo C, Espie GS, Kimber MS. Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, CcmM. Proc Natl Acad Sci U S A. 2010;107:2455–2460. doi: 10.1073/pnas.0910866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Prete S, Vullo D, De Luca V, Carginale V, Scozzafava A, Supuran CT, Capasso C. A highly catalytically active gamma-carbonic anhydrase from the pathogenic anaerobe Porphyromonas gingivalis and its inhibition profile with anions and small molecules. Bioorg Med Chem Lett. 2013;23:4067–4071. doi: 10.1016/j.bmcl.2013.05.063. [DOI] [PubMed] [Google Scholar]
- 18.Park HM, Park JH, Choi JW, Lee J, Kim BY, Jung CH, Kim JS. Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch. Acta Crystallogr D Biol Crystallogr. 2012;68:920–926. doi: 10.1107/S0907444912017210. [DOI] [PubMed] [Google Scholar]
- 19.Jeyakanthan J, Rangarajan S, Mridula P, Kanaujia SP, Shiro Y, Kuramitsu S, Yokoyama S, Sekar K. Observation of a calcium-binding site in the gamma-class carbonic anhydrase from Pyrococcus horikoshii. Acta Crystallogr D Biol Crystallogr. 2008;64:1012–1019. doi: 10.1107/S0907444908024323. [DOI] [PubMed] [Google Scholar]
- 20.Parisi G, Perales M, Fornasari MS, Colaneri A, Gonzalez-Schain N, Gomez-Casati D, Zimmermann S, Brennicke A, Araya A, Ferry JG, Echave J, Zabaleta E. Gamma carbonic anhydrases in plant mitochondria. Plant Mol Biol. 2004;55:193–207. doi: 10.1007/s11103-004-0149-7. [DOI] [PubMed] [Google Scholar]
- 21.Martin V, Villarreal F, Miras I, Navaza A, Haouz A, Gonzalez-Lebrero RM, Kaufman SB, Zabaleta E. Recombinant plant gamma carbonic anhydrase homotrimers bind inorganic carbon. FEBS Lett. 2009;583:3425–3430. doi: 10.1016/j.febslet.2009.09.055. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman S, Domsic JF, Tu C, Robbins AH, McKenna R, Silverman DN, Ferry JG. Role of Trp19 and Tyr200 in catalysis by the gamma-class carbonic anhydrase from Methanosarcina thermophila. Arch Biochem Biophys. 2013;529:11–17. doi: 10.1016/j.abb.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tripp BC, Tu C, Ferry JG. Role of arginine 59 in the gamma-class carbonic anhydrases. Biochemistry. 2002;41:669–678. doi: 10.1021/bi010768b. [DOI] [PubMed] [Google Scholar]
- 24.Moreno E, Moriyon I. The genus Brucella. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The prokaryotes : A Handbook on the Biology of Bacteria : Proteobacteria : Alpha and Beta Subclasses. 3rd ed. Springer; 2006. pp. 315–456. [Google Scholar]
- 25.de Barsy M, Jamet A, Filopon D, Nicolas C, Laloux G, Rual JF, Muller A, Twizere JC, Nkengfac B, Vandenhaute J, Hill DE, Salcedo SP, Gorvel JP, Letesson JJ, De Bolle X. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell Microbiol. 2011;13:1044–1058. doi: 10.1111/j.1462-5822.2011.01601.x. [DOI] [PubMed] [Google Scholar]
- 26.Nkengfac B, Pouyez J, Bauwens E, Vandenhaute J, Letesson JJ, Wouters J, De Bolle X. Structural analysis of Brucella abortus RicA substitutions that do not impair interaction with human Rab2 GTPase. BMC Biochem. 2012;13:16. doi: 10.1186/1471-2091-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tisdale EJ, Balch WE. Rab2 is essential for the maturation of pre-Golgi intermediates. J Biol Chem. 1996;271:29372–29379. doi: 10.1074/jbc.271.46.29372. [DOI] [PubMed] [Google Scholar]
- 28.de Bolle X, Letesson JJ, Gorvel JP. Small GTPases and Brucella entry into the endoplasmic reticulum. Biochem Soc Trans. 2012;40:1348–1352. doi: 10.1042/BST20120156. [DOI] [PubMed] [Google Scholar]
- 29.Fugier E, Salcedo SP, de Chastellier C, Pophillat M, Muller A, Arce-Gorvel V, Fourquet P, Gorvel JP. The glyceraldehyde-3-phosphate dehydrogenase and the small GTPase Rab 2 are crucial for Brucella replication. PLoS Pathog. 2009;5:e1000487. doi: 10.1371/journal.ppat.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Cryst. 2009;43:186–190. [Google Scholar]
- 31.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Wilbur KM, Anderson NG. Electrometric and colorimetric determination of carbonic anhydrase. J Biol Chem. 1948;176:147–154. [PubMed] [Google Scholar]
- 38.Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem. 1971;246:2561–2573. [PubMed] [Google Scholar]
- 39.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 40.ITC Data Analysis in Origin. Tutorial Guide Version 7.0. 2004 [Google Scholar]
- 41.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 43.Cromer DT. Anomalous Dispersion Corrections Computed from Self-Consistent Field Relativistic Dirac-Slater Wave Functions. Acta Cryst. 1965;18:17–23. [Google Scholar]
- 44.Hoyt JJ, de Fontaine D, Warburton WK. Determination of the Anomalous Scattering Factors for Cu, Ni and Ti Using the Dispersion-Relation. J Appl Cryst. 1984;17:344–351. [Google Scholar]
- 45.Kissel L, Pratt RH. Corrections to Tabulated Anomalous-Scattering Factors. Acta Cryst A. 1990;46:170–175. [Google Scholar]
- 46.Wyer JR, Willcox BE, Gao GF, Gerth UC, Davis SJ, Bell JI, van der Merwe PA, Jakobsen BK. T cell receptor and coreceptor CD8 alphaalpha bind peptide-MHC independently and with distinct kinetics. Immunity. 1999;10:219–225. doi: 10.1016/s1074-7613(00)80022-9. [DOI] [PubMed] [Google Scholar]
- 47.Peschard P, Kozlov G, Lin T, Mirza IA, Berghuis AM, Lipkowitz S, Park M, Gehring K. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol Cell. 2007;27:474–485. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 48.Pylypenko O, Rak A, Durek T, Kushnir S, Dursina BE, Thomae NH, Constantinescu AT, Brunsveld L, Watzke A, Waldmann H, Goody RS, Alexandrov K. Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling. EMBO J. 2006;25:13–23. doi: 10.1038/sj.emboj.7600921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu YW, Tan KT, Waldmann H, Goody RS, Alexandrov K. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc Natl Acad Sci U S A. 2007;104:12294–12299. doi: 10.1073/pnas.0701817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci. 2005;62:1657–1670. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shapiro AD, Pfeffer SR. Quantitative analysis of the interactions between prenyl Rab9, GDP dissociation inhibitor-alpha, and guanine nucleotides. J Biol Chem. 1995;270:11085–11090. doi: 10.1074/jbc.270.19.11085. [DOI] [PubMed] [Google Scholar]
- 52.Cheng W, Yin K, Lu D, Li B, Zhu D, Chen Y, Zhang H, Xu S, Chai J, Gu L. Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state. PLoS Pathog. 2012;8:e1002528. doi: 10.1371/journal.ppat.1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roop RM., 2nd. Metal acquisition and virulence in Brucella. Anim Health Res Rev. 2012;13:10–20. doi: 10.1017/S1466252312000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]