Abstract
How do cells distinguish normal genes from transposons? Although much has been learned about RNAi-related RNA silencing pathways responsible for genome defense, this fundamental question remains. The literature points to several classes of mechanisms. In some cases, double-stranded RNA structures produced by transposon inverted repeats or antisense integration trigger endo-siRNA biogenesis. In other instances, DNA features associated with transposons—such as their unusual copy number, chromosomal arrangement, and/or chromatin environment—license RNA silencing. Finally, recent studies have identified improper transcript processing events, such as stalled pre-mRNA splicing, as signals for siRNA production. Thus, the suboptimal gene expression properties of selfish elements can enable their identification by RNA silencing pathways.
Keywords: transposon, genome defense, small RNA, RNA interference, siRNA, piRNA
RNA silencing pathways suppress transposons to protect genome integrity
Transposons have parasitized nearly all eukaryotic genomes, including the human genome, over half of which is derived from transposon sequences. In doing so, transposons have shaped eukaryotic evolution, in part by contributing regulatory and coding information to nearby host genes [1]. In fact, approximately 4% of human protein-coding open reading frames contain transposon-derived sequences [2], as do 25% of promoter regions [3]. Remarkably, host organisms have even co-opted the protein activities encoded by transposon genes, as in the case of RAG1, a transposon-derived endonuclease that now serves to catalyze V(D)J recombination in humans [4]. Despite these positive contributions to host biology, transposons are primarily selfish elements that threaten the integrity of host genomes. Transposon mobilization can disrupt host genes and promote deleterious chromosomal rearrangements [5, 6], and these perturbations contribute to Mendelian disease and cancer in humans [5, 7]. To combat this threat, host organisms have evolved multiple genome defense mechanisms to suppress transposon mobility.
RNAi-related RNA silencing pathways are deeply conserved genome defense mechanisms that act in organisms from protist to human to recognize and suppress transposons [8]. In all of these pathways, ~20–30 nt small RNAs act in complex with Argonaute family proteins to silence complementary transcripts. Depending on the pathway and context, silencing proceeds by a variety of mechanisms, which include RNA degradation, translational repression, and the establishment of repressive histone modifications [9, 10]. The pathways also differ in the means by which they generate small RNAs. In the canonical RNAi pathway, RNaseIII-type Dicer enzymes convert long double-stranded RNA (dsRNA) into small interfering RNA (siRNA). In contrast, PIWI-interacting RNA (piRNA) pathways do not require Dicer, and instead generate small RNA from single-stranded precursors. Still other pathways, such as the endogenous siRNA pathways of worm and plant, require RNA-dependent RNA polymerases (RdRPs) for small RNA biogenesis; these enzymes are thought to produce small RNAs directly or to generate long dsRNA substrates for Dicer. The diversity of small RNA biogenesis mechanisms permits a wide variety of RNA sequences to template the production of small RNAs for genome defense.
The adaptability of RNA silencing pathways, whose targeting depends primarily on the sequences of their small RNA guides, makes them well suited to defend against transposons, which differ extensively in sequence and distribution among eukaryotic species. Indeed, orthologous Argonaute proteins silence different transposon families in different host organisms, suggesting that RNA silencing pathways can adapt to recognize novel transposons that a host has not previously encountered [11, 12]. This adaptability raises a central question in the study of these pathways: how does RNA silencing specifically distinguish transposons from host genes? Here, we review our current understanding of principles by which transposons can be recognized as non-self by RNA silencing pathways of fungi, plants, and metazoans.
The challenges of transposon recognition
RNA silencing pathways must overcome several properties of transposons in order to target them specifically. First, like host genes, transposons reside in the nuclear genome and utilize host gene expression machinery. Thus, transposons are not broadly distinguished by the use of alternative gene expression mechanisms. Second, transposons themselves differ widely in sequence, with many families exhibiting no homology to each other, which hinders sequence-specific transposon recognition mechanisms (Box 1). Finally, even for a single transposon type, individual loci differ extensively, from fully intact, active transposons to degenerated, inactive loci. This means that transposon sequences are not always distinguished from host genes simply by their ability to mobilize. Furthermore, both active and inactive transposon loci might be deleterious to their hosts, since the expression of these abundant sequences, which constitute over 80% of some eukaryotic genomes, confers a metabolic cost and may induce RNA toxicity [13].
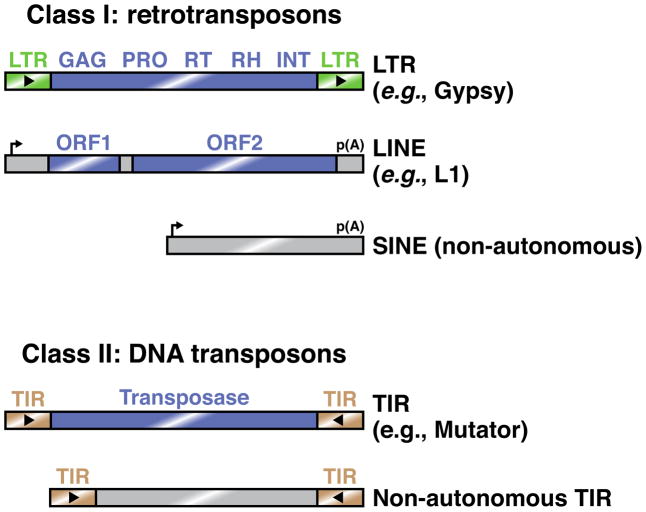
Box 1. Common types of eukaryotic transposable elements.
Transposons are nucleic acid elements that can mobilize to new chromosomal locations. They co-opt the host gene expression machinery to produce their own protein products, which facilitate mobilization. Although common in these respects, transposons are remarkably diverse in sequence and transposition mechanism, which hinders their identification by cellular genome defense pathways.
Transposons can be broadly divided into retrotransposons (Class I) and DNA transposons (Class II) [85, 86]. Retrotransposons mobilize through an RNA intermediate, which is reverse transcribed to allow its integration into the genome. This ‘copy and paste’ mechanism does not alter the original transposon locus and therefore acts to increase transposon copy number. Some retrotransposons encode long terminal repeats (LTRs), which act as promoters and polyadenylation signals. Like their retrovirus relatives, these transposons undergo reverse transcription in virus-like particles in the cytoplasm. Other retrotransposons, such as long interspersed nuclear elements (LINEs), also encode their own promoters and 3′ end formation signals, but lack LTRs. These elements undergo reverse transcription in the nucleus using nicked genomic DNA as a primer. By contrast, short interspersed nuclear elements (SINEs) mobilize in a manner similar to that of LINEs, but do not themselves encode the proteins required for mobilization. They are therefore non-autonomous, and depend on LINE-encoded factors.
DNA transposons do not utilize an RNA intermediate, but instead generally mobilize through a ‘cut and paste’ mechanism in which the original transposon locus is excised and reinserted in a new location. These transposons generally encode terminal inverted repeat (TIR) sequences, which recruit transposase to the transposon DNA locus in order to initiate its excision.
Figure I. Examples of common eukaryotic transposons.
Common Class I transposons include LTR retrotransposons, which generally encode a capsid protein (GAG), protease (PRO), reverse transcriptase (RT), RNaseH (RH), and integrase (INT). By contrast, LINEs typically contain two open reading frames, one of unknown function and one that encodes endonuclease and reverse transcriptase activities. SINEs are non-autonomous elements whose sequence features are recognized by transposon-derived proteins acting in trans. Most Class II elements are TIR transposons, which can mobilize either autonomously or non-autonomously. Blue rectangles indicate protein-coding regions.
Studies of RNA silencing pathways have begun to reveal strategies by which these pathways recognize transposons while avoiding inappropriate silencing of host sequences. These strategies are not mutually exclusive, and an individual RNA silencing pathway sometimes utilizes multiple strategies. One general strategy recognizes transposons by virtue of their tendency to generate dsRNA. A second strategy distinguishes transposons by their unique ability to mobilize, which makes them more likely to exist in unusual chromosomal arrangements or in high copy number. A third class of mechanism exploits the suboptimal gene expression properties of transposons, which might arise due to their distinct evolutionary histories, to distinguish them from host genes. A fourth class of mechanism licenses small RNA production against transposons based on the prior capture of transposon sequences by specialized chromatin niches. These four transposon recognition strategies are discussed in turn in this review.
Transposons are identified by their production of dsRNA
Early studies of Caenorhabditis elegans RNAi indicated that some mutants defective in gene silencing triggered by exogenous dsRNA are also defective in suppressing transposons, raising the possibility that endogenous dsRNA initiates transposon silencing [14, 15]. In such a model, transposon-derived dsRNA is processed by Dicer enzymes to yield siRNA, which then acts to repress homologous transposon sequences throughout the genome. An attractive feature of this model is that transposons exhibit several properties that might increase their likelihood of generating dsRNA, thereby enabling them to be distinguished from host genes [16]. For instance, some transposon families encode repeats and antisense promoters that can produce dsRNA. Furthermore, the mobilization of transposons into existing host transcriptional units may lead to the production of antisense transposon transcripts. Finally, the repetitiveness of transposon sequences in the genome, together with their tendency to undergo rearrangements, may promote the formation of structured loci in which duplicated transposon sequences give rise to transcripts that fold to form dsRNA (Figure 1A).
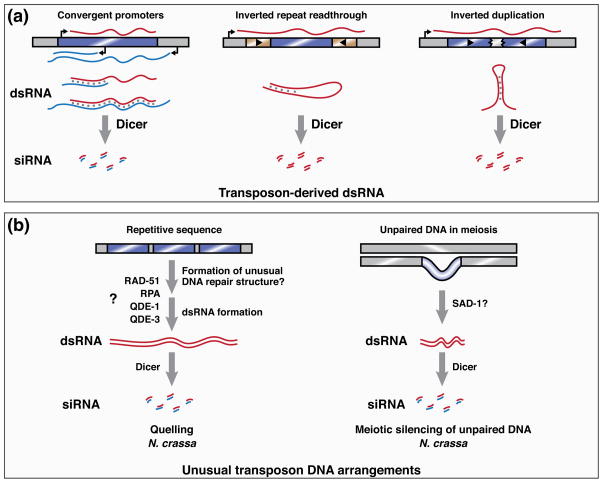
Figure 1. Transposon features recognized by RNA silencing pathways.
A) Transposon-derived dsRNA provides a substrate for Dicer activity and thereby triggers endo-siRNA pathways. Double-stranded RNA can be generated intermolecularly by the action of convergent promoters encoded either by the transposon (blue bar) or by the host. Intramolecular dsRNA is generated from a single transposon when a transcript contains both of its inverted repeat sequences. Inverted duplications of transposons can also cause intramolecular dsRNA formation.
B) Unusual chromosomal arrangements of transposons allow their identification by genome defense pathways. In N. crassa, the quelling pathway (left) silences repetitive sequences. The mechanism by which these sequences are detected and used to template siRNA production is unclear, but may involve the formation of unusual DNA repair intermediates, because quelling requires proteins, such as RAD-51, that mediate homologous DNA recombination. Subsequent siRNA production requires QDE-1—a DNA- and RNA-dependent RNA polymerase—and its binding partners Replication protein A (RPA) and QDE-3. The meiotic silencing of unpaired DNA pathway (right) detects loci that lack a partner during homologous chromosome pairing in meiosis I. siRNA production from these loci requires SAD-1, an RdRP that is paralogous to QDE-1.
Several of the above mechanisms of transposon dsRNA production have been validated experimentally. For instance, the inverted repeats of Tc1 DNA transposons in C. elegans have been shown to form dsRNA in vivo, likely due to intramolecular folding of precursor transcripts that originate from host promoters and read through an entire transposon locus [17]. By contrast, an internal antisense promoter in the human Long Interspersed Element-1 (LINE-1) is required for the production of small RNA targeting this transposon, suggesting that intermolecular dsRNA triggers LINE-1 siRNA production, although the Dicer-dependence of these small RNAs remains to be demonstrated [18]. Finally, unusual tandem arrangements of transposon-derived sequences, such as inverted duplications, have been found in mouse, nematode, plant, and yeast to be hotspots of siRNA production, typically owing to formation of intramolecular dsRNA [12, 17, 19, 20]. Remarkably, a single locus of this type in maize, the Mu killer locus, is sufficient to trigger silencing of Mutator DNA transposons genome-wide [19]. Thus, the capacity of RNA silencing to act in trans enables an entire family of transposons to be recognized, even if only a subset of these loci generates dsRNA.
The above examples suggest that the transposon-derived dsRNA can, in at least some cases, trigger the production of small RNAs for the purpose of genome defense. Nevertheless, the general rules that dictate the in vivo formation of dsRNA, and its subsequent processing into siRNA, remain unclear. For instance, whereas the intramolecular dsRNA produced by the Mu killer locus can induce RNA silencing of homologous Mutator transposons, this effect cannot be achieved by expression of sense and antisense Mutator transcripts from distinct loci [21], suggesting that intermolecular dsRNA comprising these transcripts is either inefficiently formed or poorly processed by Dicer. Similarly, the 31 genomic copies of Tc1 in C. elegans generate both sense and antisense transcripts, but these transcripts do not appear to form intermolecular dsRNA, as assessed by analysis of in vivo dsRNA editing by the ADAR adenosine deaminase [17]. These findings suggest that the initiation of RNA silencing by dsRNA may require a licensing step, in which only a subset of potential dsRNA substrates gains the capacity to trigger RNA silencing. Such a licensing step may also explain why endogenous non-transposon loci that produce complementary transcripts are often poor triggers of siRNA biogenesis, as in Drosophila [22].
Future experiments that systematically address the relationships between cellular ssRNA, dsRNA, and siRNA will help elucidate the rules by which dsRNA triggers small RNA production in vivo. These rules may involve the ability of given RNAs to form dsRNA, which could be influenced by their expression levels, subcellular localizations, or associated proteins. Alternatively, specific features of the dsRNA itself, such as whether the chemistry of its ends allows efficient Dicer processing, could determine the efficiency of siRNA production. A more complete understanding of the rules for dsRNA and siRNA production could explain why only some transposon families are targeted by endogenous siRNA, and why different targeted families exhibit siRNAs corresponding to distinct regions of their sequence [23].
Unusual DNA arrangements distinguish transposons from host genes
Transposons are often found in unusual chromosomal arrangements. The repetitive sequences they encode can cause replication fork slippage, leading to the formation of tandem arrays [24, 25]. Furthermore, their ability to mobilize can lead to the asymmetric distribution of transposon sequences on homologous chromosomes. These unusual arrangements, which represent targets of the two RNA silencing pathways in the filamentous fungus Neurospora crassa, may thus distinguish transposons for detection by host genome defense pathways (Figure 1B). The quelling pathway operates in vegetative cells, where it targets tandem array sequences for silencing. By contrast, the meiotic silencing of unpaired DNA (MSUD) pathway acts in mating cells, where it silences loci that lack a partner during homologous chromosome pairing in meiosis I. Each pathway requires its own RdRP, suggesting that silencing is triggered not by a naturally occurring dsRNA, but rather by the directed action of RdRP activity at particular targets. These targets, however, do not include active transposons, which are not found in the modern-day genome of N. crassa. Instead, RNA silencing pathways in this organism are studied primarily in their capacity to silence transgenes. Nevertheless, N. crassa produces small RNAs corresponding to ancient, degenerated transposon sequences in its genome, pointing to genome defense as a biological role for these pathways [26].
The quelling pathway targets repetitive DNA arrangements
Quelling, one of the first known RNA silencing pathways, was discovered over 20 years ago [27, 28]. In this pathway, repetitive transgenes, which are often oriented in tandem arrays, are used as templates for the production of siRNA, which post-transcriptionally silences homologous loci throughout the genome. Like other endogenous siRNA pathways, quelling requires Dicer enzymes, an Argonaute protein (QDE-2), and an RdRP (QDE-1) [27]. Interestingly, QDE-1 can act not only as an RdRP but also as a DNA-dependent RNA polymerase, and recombinant QDE-1 is sufficient to generate dsRNA from a DNA template [29]. These findings, together with the observation that quelling can be triggered by transgenes lacking Pol II promoters [30], suggest that quelling may be initiated by QDE-1-mediated production of single- and double-stranded RNA from transgene sequences. In such a model, the recruitment of QDE-1 to particular loci would underlie the specificity of quelling for transgenes and other foreign genetic elements.
Recent findings outline a model in which QDE-1 recruitment to transgenic loci is triggered by the repetitiveness of the DNA itself. An initial clue came from the finding that QDE-1 physically interacts with Replication Protein A (RPA) in a manner that requires QDE-3, a quelling factor related to RecQ helicases [29, 31]. Because RPA binds ssDNA to facilitate DNA replication and repair, and RecQ helicases play roles in DNA repair, these findings suggest a functional link between DNA metabolism and quelling. Further studies indicated that factors involved in homologous DNA recombination bind to transgenic loci and are required for quelling, supporting a model in which homologous recombination generates recombination intermediates that recruit QDE-3 and, subsequently, QDE-1 for the purpose of small RNA production [32]. These events may take place more often at repetitive loci because of these loci are more likely to undergo homologous recombination, or because their repetitiveness leads to the production of aberrant recombination structures that are resolved by QDE-3. Consistent with such a model, treatment of cells with hydroxyurea, an agent that induces double-strand DNA breaks, enhances quelling [32]. Although the detailed mechanism of QDE-3 recruitment to and QDE-1 action at repetitive loci remain areas of active investigation, these findings point to homologous recombination as a mechanism to sense the repetitiveness of particular genomic sequences, thereby identifying them as targets for a genome defense pathway.
Meiotic silencing of unpaired DNA
In contrast to quelling, which targets foreign sequences present in multiple genomic copies, the MSUD pathway can recognize even a single-copy sequence as foreign based on its inability to pair with a homologous chromosome during meiosis [33]. This pathway requires a Dicer protein, an Argonaute, and an RdRP (SAD-1), which generate small RNA corresponding specifically to the unpaired region [34]. All of these proteins exhibit perinuclear localization, suggesting a specialized site for small RNA biogenesis [27]. In fact, only a single known MSUD factor localizes to the nucleus: SAD-5, a fungal-specific protein with no predicted domains [35]. This has hindered progress on a central question in the study of meiotic silencing: how precisely are unpaired regions detected in the nucleus and engaged for small RNA production?
Studies to date suggest that the detection of unpaired regions is not restricted to particular DNA sequences or chromosomal regions [33]. Rather, detection efficiency correlates with the size of the unpaired region as well as the degree of nonidentity it shares with its homolog [36]. MSUD can detect even subtle amounts of sequence nonidentity (~5% across 2–4 kb), and its sensitivity is further increased if the unpaired region carries DNA methylation marks, suggesting that not only DNA sequence but also chromatin context may contribute to detection of unpaired regions [37]. Detection proceeds even if the unpaired region lacks a Pol II promoter, further suggesting that the initial detection event involves recognition of some aspect of DNA structure [36]. Presumably, noncanonical transcription produces the RNA transcripts that template dsRNA and siRNA production. An attractive, but so far untested, model is that SAD-1 acts successively as a DNA-dependent and RNA-dependent RNA polymerase—like its paralog, QDE-1, in the quelling pathway—in order to synthesize dsRNA from unpaired loci. Future studies will therefore benefit from characterization of the precursor transcripts that give rise to small RNAs from unpaired regions. Another area of investigation will be to determine whether chromatin states influence the MSUD pathway, either in its detection or small RNA production steps. In this regard, it is interesting to note that a meiotic silencing pathway in C. elegans also requires an RdRP (Ego-1), and is coupled to histone H3 lysine 9 (H3K9) methylation of unpaired loci [38, 39].
Sensing of transposons via the gene expression machinery
Whereas the quelling and MSUD pathways recognize properties of a foreign element’s DNA copy number or chromosomal arrangement, other RNA silencing pathways appear to instead target features of transposon-encoded RNAs. What features could distinguish transposon transcripts from host transcripts, since both are produced and undergo processing by the same host cellular machineries? As discussed further below, recent findings in the yeast Cryptococcus neoformans suggest that transposon-derived transcripts encode suboptimal splicing signals and tend to stall on spliceosomes, which directs their use as templates for dsRNA and siRNA production [40]. Although transposon gene expression signal strength has not been systematically examined in other organisms, plant genome defense pathways mediated by endogenous siRNA may also detect features of transcript maturation, as evidenced by the finding that 5′ and 3′ end formation features can dictate RNA silencing specificity in these systems. Together, these results raise the possibility that gene expression signal strength generally distinguishes transposons from host genes, perhaps because the evolutionary history of transposons differs from that of their host organisms.
The spliceosome is a transposon sensor in a basidiomycetous yeast
In C. neoformans, endo-siRNA mediates post-transcriptional gene silencing and is produced primarily from transposon-derived sequences [40, 41]. Strikingly, these small RNAs correspond to both exons and introns of their targets, suggesting that incompletely spliced mRNA precursors are preferred templates for siRNA production. Furthermore, several C. neoformans siRNA biogenesis factors, including an Argonaute and an RdRP, form a nuclear protein complex (SCANR) that physically interacts with the spliceosome. These findings point to a kinetic competition model for RNA silencing specificity, in which efficiently spliced host transcripts are not targeted for siRNA production, whereas transposon transcripts, by virtue of their poor splicing kinetics, become susceptible to SCANR-mediated siRNA production (Figure 2). Consistent with this model, the transcripts targeted for siRNA production exhibit intron sequence features predictive of poor splicing and tend to accumulate abnormally on spliceosomes in vivo [40].
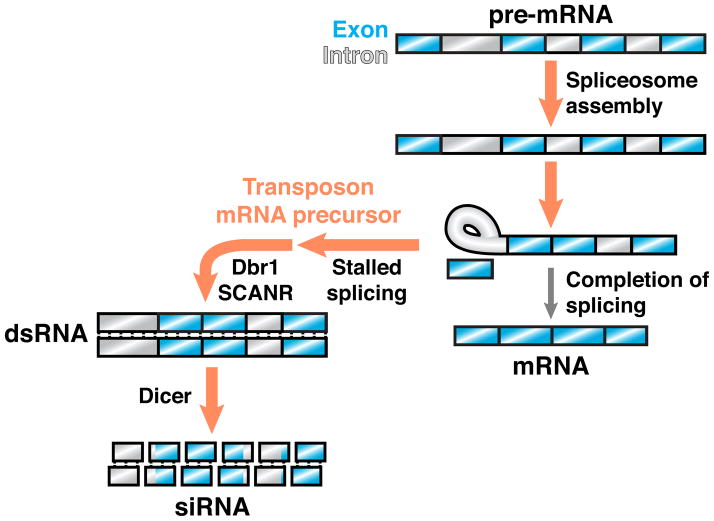
Figure 2. Stalled spliceosomes license RNA silencing in an endo-siRNA pathway.
In the yeast C. neoformans, transcripts targeted by RNA silencing, which primarily include transposons, exhibit sequence features predictive of poor splicing and tend to stall in spliceosomes. The stalled splicing of transposon mRNA precursors is required for siRNA biogenesis mediated by SCANR, a protein complex that contains an RdRP and physically associates with the spliceosome. Lariat debranching enzyme (Dbr1) is also required for siRNA production, suggesting that transposon mRNA precursors in the lariat intermediate stage are linearized to enable dsRNA formation by SCANR. In this hypothetical example, splicing of a transcript’s first intron stalls at the lariat intermediate stage, whereas downstream introns remain partially spliced.
Experimental manipulations of siRNA target transcripts in C. neoformans further indicate that splicing signals dictate RNA silencing specificity. For instance, elimination of a transcript’s introns suppresses its ability to template siRNA biogenesis. Conversely, introduction of a 3′ splice site mutation, which stalls splicing at the stage of lariat intermediate formation, dramatically increases siRNA production from the mutated transcript. Importantly, the effect of a 3′ splice site mutation is suppressed by introduction of a 5′ splice site mutation, which prevents intron engagement with the spliceosome. The effect of a 3′ splice site mutation is also suppressed in the absence of lariat debranching enzyme, further supporting a model in which the processing of stalled splicing intermediates is required for the production of dsRNA and subsequent siRNA. Together, these findings establish stalled spliceosomes as a necessary signal by which transposon-derived transcripts can be distinguished for the purpose of genome defense [40].
Although the reason why siRNA targets in C. neoformans, which are primarily transposon-derived, possess weaker splicing signals than those of host genes is not yet understood, this property may have been shaped by the distinct features of transposon propagation. First, intron-containing retrotransposons must produce both spliced and unspliced transcripts, with the latter used selectively as transposition substrates [42], in order to maintain intron sequences in new genomic copies. Weak splicing signals may thus be favored as a mechanism for producing both transcript types. Second, the capacity of transposons to be horizontally transferred between different organisms may predispose them to possess suboptimal splicing signals. For instance, because gene expression signals, such as splicing signals, differ substantially among species [43, 44], the horizontal transfer of a transposon may insert it into a host, like C. neoformans, with incompatible signal preferences. The limited co-evolution of transposons with their hosts would thus make transposons in general more poorly equipped than host genes to undergo the many steps of eukaryotic gene expression. This potential disparity provides opportunities for the evolution of genome defense pathways, and may even contribute to the evolutionary emergence of host gene expression steps themselves [45].
RNA processing signals impact RNA silencing in plants
Several findings raise the possibility that RNA silencing specificity in plants is influenced, as in C. neoformans, by the RNA processing signals encoded in foreign elements. In plants, two endo-siRNA pathways—each with distinct Dicer, Argonaute, and RdRP proteins—act to generate siRNA against transposons. In the post-transcriptional gene silencing (PTGS) pathway, transposon transcripts produced by RNA Pol II are used as substrates for siRNA production in a manner thought to be initiated by RDR6, an RdRP [46]. In the RNA-directed DNA methylation (RdDM) pathway, transcription of transposon loci by the specialized RNA polymerases Pol IV and Pol V enables siRNA production and DNA methylation, respectively, at these loci [47]. Because DNA methylation not only represses transcription but also recruits additional Pol IV activity, this pathway leads to lasting, heritable silencing, such that most transposons in plant genomes are not normally expressed [48]. Newly encountered transposons can be targeted for methylation if they exhibit sequence similarity to existing RdDM targets, presumably because of the in trans action of RdDM pathway siRNA [48]. But how can a transposon of novel sequence be identified? Recent studies suggest that the initial recognition of transposon transcripts by the PTGS pathway can subsequently engage the RdDM pathway to effect lasting DNA methylation [48, 49]. These findings highlight RDR6 activity as a key factor in transposon recognition and RNA silencing specificity in plants.
Promoter sequences are required for the detection of foreign elements by PTGS, suggesting that RNA signals guide this pathway’s specificity [50]. However, these signals remain largely unknown, in part due to the challenges of manipulating and analyzing repetitive, endogenous transposons. Nevertheless, studies of transgene silencing have provided several clues. First, PTGS can recognize even single-copy transgenes that are not predicted to form dsRNA, supporting the model that RDR6 action on single-stranded, non-self transcripts is an initiating event in this silencing pathway [51, 52]. Second, screens for A. thaliana mutants that enhance PTGS yielded mutations affecting the 5′-3′ RNA exonucleases XRN2/3/4 as well as 3′ end formation factors that mediate transcript cleavage and polyadenylation [53–55]. These mutants accumulate uncapped or non-polyadenylated transgene transcripts, suggesting a model in which ‘aberrant’ transcripts with one or more incompletely processed ends may be particularly good substrates for RDR6. Consistent with this model, siRNA production from a transgene transcript is promoted by the absence of transgene termination signals, and is suppressed by the addition of strong terminators [52]. Interestingly, an XRN4 mutation does not simply increase silencing of appropriate PTGS targets, but also stimulates the production of siRNA from host transcripts not normally targeted by PTGS, further suggesting that 5′ end formation acts to generally suppress RDR6 activity [56].
Several questions regarding the initiation of PTGS by foreign elements remain to be answered. First, it will be important to determine the precise substrate of RDR6 activity in vivo. The results described above highlight transcripts with bare 5′ or 3′ ends as an attractive possibility, and are further supported by the observation that host transcripts, if separated from their 5′ and 3′ end modifications by miRNA-mediated cleavage events, become RDR6 substrates [57]. However, it is unclear whether this model is sufficient to explain the observation that perturbations of several other RNA processing steps—including splicing [53, 58], nonsense mediated decay [59], and exosome-mediated decay [59]—also affect PTGS targeting. It is possible that all of these perturbations act by indirectly influencing a single preferred RDR6 substrate, whose identity remains hidden. Second, it will be important to understand how RDR6 substrate preferences enable PTGS to distinguish self from non-self. Anecdotal evidence suggests that repetitive sequences tend to produce poorly polyadenylated transcripts [52], but whether transposons in plant genomes generally exhibit poor polyadenylation or cap formation has not yet been systematically tested.
Genomic memory of foreign sequences enables transposon detection
RNA silencing pathways of the piRNA class silence transposons and other foreign elements in animal germlines, where they are required for fertility [60]. In these pathways, small RNAs are produced in a Dicer-independent manner, then act with Argonaute proteins of the PIWI clade to effect transcriptional and post-transcriptional silencing of complementary target transcripts [61]. Although the molecular mechanisms of piRNA biosynthesis and target silencing vary among species, piRNA pathways share a common ‘adaptive immune system’ strategy for foreign element recognition. Specifically, these pathways generate a large diversity of piRNA sequences, which survey the transcriptome for complementary targets. Upon target recognition, an amplification step generates additional target-specific small RNAs, which enforce long-term silencing, often in a heritable manner [61]. Thus, piRNA pathway specificity for transposons is determined in large part by the initial selection of piRNA sequences, a process that in Drosophila involves the restriction of piRNA production to specialized genomic loci called piRNA clusters (Figure 3A). Regulation of secondary piRNA amplification provides an additional opportunity to impose silencing specificity, and recent studies suggest that this is a significant source of piRNA specificity in C. elegans (Figure 3B).
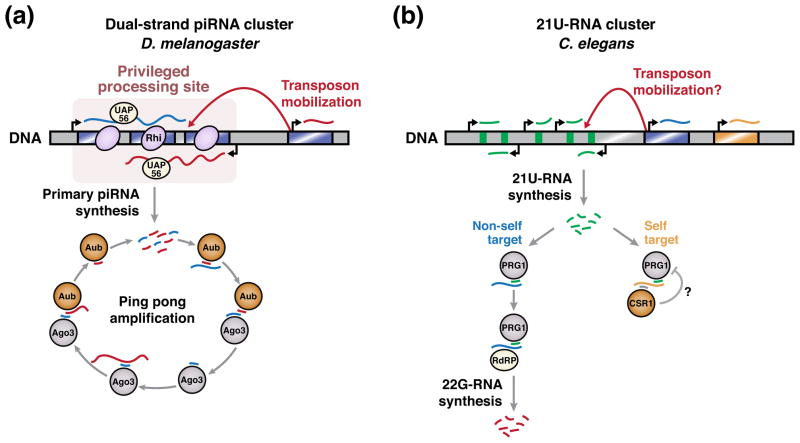
Figure 3. piRNA production occurs at specialized genomic loci.
A) In D. melanogaster, the chromatin environment at dual-strand piRNA clusters enables piRNA production from these loci. This chromatin is associated with H3K9 methylation and the specialized heterochromatin protein 1 (HP1) homolog Rhino (Rhi). Primary transcripts produced from piRNA clusters are bound by UAP56, a DEAD box protein required for piRNA processing specificity [87]. The ping pong amplification cycle subsequently promotes the generation of piRNAs complementary to active transposons (see text for details). Mobilization of transposons into piRNA clusters contributes to the enrichment of foreign sequences at these loci.
B) In C. elegans, 21U-RNAs are encoded in genomic clusters, but each is expressed by its own promoter. 21U-RNA sequences are very diverse, and not strongly enriched in transposon sequences, suggesting that transposon mobilization into 21U-RNA clusters is not a major mechanism by which 21U-RNA specificity is achieved. 21U-RNAs loaded in the PIWI protein PRG-1 are potentially complementary to both non-self and self transcripts. In the former case, PRG-1 triggers the production of repressive 22G-RNAs to silence the non-self transcript. In the latter case, PRG-1-mediated silencing appears to be less efficient, potentially due to a protective mechanism. This protective mechanism is hypothesized to involve targeting of self transcripts by 22G-RNAs loaded in the Argonaute protein CSR-1.
piRNA in D. melanogaster
In Drosophila, germline piRNAs are produced from specialized loci that contain clusters of transposon fragments [62, 63]. Clusters thus represent a genomically encoded memory of prior transposon insertions. Single-stranded transcripts from these clusters undergo processing to generate primary piRNA, which act with PIWI family proteins (Piwi, Aub) to guide cleavage of complementary transposon transcripts originating throughout the genome. This cleavage defines the 5′ end of a secondary piRNA, which is then loaded into a distinct PIWI protein (Ago3) and undergoes 3′ trimming to proper length. Subsequently, mature Ago3 complexes act to repeatedly bind and cleave complementary piRNA cluster transcripts, thereby promoting precursor conversion into mature piRNAs [62, 64, 65]. This amplification step, termed the ping-pong cycle, biases piRNA production towards transcriptionally active transposons, and ensures that most piRNAs are antisense to their targets, and therefore active [65, 66]. These abundant antisense piRNAs then promote both post-transcriptional and transcriptional transposon silencing, with the latter being mediated by H3K9 methylation of target loci [61].
A central question in Drosophila piRNA specificity is to understand how particular genomic regions are marked as sources of piRNA production in such a way that ensures their enrichment for transposon sequences. In one model, piRNA clusters act as passive transposon traps. In this model, the repetitiveness and mobility of transposons make them more likely than host genes to integrate at a defined genomic locus, such as a piRNA cluster [67]. Each integration event would enable piRNA-mediated silencing of the integrated transposon and its relatives, and tend to be fixed in the population. This model is supported by the finding that piRNA clusters can produce piRNAs from a wide variety of experimentally inserted sequences [68], and by the observation that, among Drosophila species, the genomic location of a piRNA cluster is often conserved even when the constituent transposon fragments differ [69]. Furthermore, the initiation of a transposon’s silencing by its mobilization into piRNA clusters has been observed experimentally [70].
Other observations raise the possibility that the selection of sequences for piRNA biogenesis involves additional licensing steps. For instance, transgenes inserted into piRNA clusters template the production of piRNAs in a nonuniform manner [68, 71], suggesting that piRNA precursor processing preferences may dictate selection of particular sequences that are best suited for genome defense. Furthermore, some transposon families appear to mobilize to piRNA cluster loci more often than would be expected by chance [70, 72], suggesting that an active mechanism promotes insertion into a cluster. One possibility is that the specialized chromatin landscape at piRNA clusters, which includes H3K9 methylation and a heterochromatin protein 1 (HP1) homolog (Rhino), promotes local transposon insertions [73, 74]. Another intriguing possibility, suggested by recent work [75, 76], is that the silencing of piRNA target loci leads to the birth of new piRNA clusters at these loci, potentially in a mechanism involving the H3K9 methyl marks deposited during Piwi-mediated silencing [61]. Such a mechanism would enrich transposon and other piRNA target sequences within piRNA clusters. Further testing of these hypotheses awaits a more complete understanding of the specialized chromatin at piRNA clusters, and how its constituent proteins, such as Rhino, enable piRNA production from these loci.
21U-RNA in C. elegans
In C. elegans, the PIWI protein PRG-1 associates with small RNAs of the 21U class [60]. Like Drosophila piRNAs, 21U-RNAs are encoded largely in genomic clusters [77], but they differ in that 21U-RNAs are expressed as autonomous units instead of from long precursor transcripts [78, 79]. Recent findings indicate that 21U-RNAs can trigger repression of transposon and transgene sequences [11, 80–83]. Recognition of a target transcript by a 21U-RNA leads to RdRP-dependent production of 22G siRNA of the worm-specific Argonaute (WAGO) class, which are antisense to the target. Production of 22G siRNA not only serves to amplify the RNA silencing response, but also engages transcriptional silencing mechanisms, which depend on H3K9 methylation and can be stably inherited. However, the pool of 21U-RNAs is vast (>16,000 distinct species) and complex in sequence, raising the question of how genome surveillance by these RNAs can specifically distinguish self from non-self.
The sequences of 21U-RNAs do not appear sufficient to establish specificity for foreign elements. For instance, although 21U-RNA are depleted in sequences that would target host genes, they do not show a strong enrichment for transposon sequences [11]. Furthermore, 21U-RNAs appear to direct PRG-1 to complementary targets even when the targets contain as many as 4 mismatches with the 21U-RNA sequence [11, 82]. Given such relaxed sequence specificity, the 21U-RNA population would be expected to target most host genes [11, 82]. To explain the lack of self-reactivity in 21U-RNA pathways, it has been proposed that an additional mechanism protects self transcripts from 21U-mediated silencing by antagonizing the amplification of repressive 22G siRNA. This mechanism is proposed to involve CSR-1, an Argonaute protein involved not in gene silencing but rather in facilitating chromosome segregation via a class of 22G siRNA that targets host genes expressed in the germline [84]. Consistent with such a model, 21U-RNAs are less efficient in silencing a target transcript that is also targeted by 22G siRNAs in CSR-1 [82]. Furthermore, C. elegans transgenes that escape 21U-mediated silencing can establish a stably active state that dominantly activates silenced, homologous loci in trans, consistent with the idea that diffusible factors, such as CSR-1-bound small RNAs, act to license host gene expression [83]. Together, these findings suggest that piRNA pathway specificity in worms relies not only on a comparison of transcript and 21U-RNA sequences, but also on features of transcript expression history, as potentially indicated by CSR-1 22G RNAs.
Concluding remarks
We have summarized evidence that RNA silencing pathways recognize several distinguishing features of transposons, including their tendency to produce dsRNA, exist in unusual chromosomal arrangements, exhibit suboptimal gene expression properties, and occupy specialized chromatin contexts (Table 1). The extent to which these features are sufficient to distinguish transposons from host genes is unknown. In this regard, it is interesting to note that distinct RNA silencing pathways may act combinatorially to identify non-self elements, as has been suggested for the MSUD and piRNA pathways in C. elegans [81]. Recent observations further suggest that the specificity of RNA silencing pathways for transposons may involve not only distinguishing signals in the transposons themselves but also protective signals possessed by host genes. Efforts to understand the nature of these signals should reveal fundamental principles by which organisms detect and silence genomic parasites while avoiding inappropriate targeting of beneficial genomic sequences.
Table 1.
Proposed mechanisms for licensing of RNA silencing pathways
| RNA silencing pathway | Organism | Proposed licensing mechanism | Ref. |
|---|---|---|---|
| Endogenous siRNA | H. sapiens | Formation of dsRNA substrates for Dicer | [12, 17–20, 23] |
| M. musculus | |||
| D. melanogaster | |||
| C. elegans | |||
| Z. mays | |||
| S. castellii | |||
| Quelling | N. crassa | Unusual DNA repair structure at tandem repeat locus | [32] |
| Meiotic silencing of unpaired DNA | N. crassa | Unpaired DNA in meiosis I | [33, 38] |
| C. elegans | |||
| Endogenous siRNA | C. neoformans | Stalled splicing of target transcript | [40] |
| Post-transcriptional gene silencing | A. thaliana | Improper target transcript 5' or 3' end formation | [52–56] |
| piRNA | D. melanogaster | Privileged chromosomal loci for piRNA production (e.g., specialized chromatin) | [62, 74] |
| C. elegans | Absence of self-protective signals in target (e.g., CSR-1 targeting?) | [82, 83] |
Highlights.
How transposons are recognized by RNAi-based mechanisms is poorly understood
Transcription of some transposons produces dsRNA substrates for Dicer
Transposon DNA arrangement and chromatin context can also license RNA silencing
Defective RNA processing signals encoded by transposons can promote RNA silencing
Acknowledgments
We thank members of the laboratory for helpful discussions, and apologize to authors whose work we could not cite on account of space constraints. This work is supported by the National Institutes of Health (GM71801).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nekrutenko A, Li WH. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 3.Jordan IK, et al. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 4.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Donnell KA, Burns KH. Mobilizing diversity: transposable element insertions in genetic variation and disease. Mob DNA. 2010;1:21. doi: 10.1186/1759-8753-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagijn MP, et al. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drinnenberg IA, et al. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 15.Ketting RF, et al. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 16.Plasterk RH. RNA silencing: the genome's immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 17.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 18.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 19.Slotkin RK, et al. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet. 2005;37:641–644. doi: 10.1038/ng1576. [DOI] [PubMed] [Google Scholar]
- 20.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SH, Walbot V. Deletion derivatives of the MuDR regulatory transposon of maize encode antisense transcripts but are not dominant-negative regulators of mutator activities. Plant Cell. 2003;15:2430–2447. doi: 10.1105/tpc.014605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura K, et al. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998c. [DOI] [PubMed] [Google Scholar]
- 23.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bzymek M, Lovett ST. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A. 2001;98:8319–8325. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed M, Liang P. Transposable elements are a significant contributor to tandem repeats in the human genome. Comp Funct Genomics. 2012;2012:947089. doi: 10.1155/2012/947089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chicas A, et al. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 2004;32:4237–4243. doi: 10.1093/nar/gkh764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang SS, et al. RNA interference pathways in fungi: mechanisms and functions. Ann Rev Microbiol. 2012;66:305–323. doi: 10.1146/annurev-micro-092611-150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee HC, et al. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010;8:e1000496. doi: 10.1371/journal.pbio.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cogoni C, et al. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 31.Nolan T, et al. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 2008;36:532–538. doi: 10.1093/nar/gkm1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, et al. Homologous recombination as a mechanism to recognize repetitive DNA sequences in an RNAi pathway. Genes Dev. 2013;27:145–150. doi: 10.1101/gad.209494.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiu PK, et al. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 34.Hammond TM, et al. Identification of small RNAs associated with meiotic silencing by unpaired DNA. Genetics. 2013;194:279–284. doi: 10.1534/genetics.112.149138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammond TM, et al. Novel proteins required for meiotic silencing by unpaired DNA and siRNA generation in Neurospora crassa. Genetics. 2013;194:91–100. doi: 10.1534/genetics.112.148999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee DW, et al. Properties of unpaired DNA required for efficient silencing in Neurospora crassa. Genetics. 2004;167:131–150. doi: 10.1534/genetics.167.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratt RJ, et al. DNA methylation affects meiotic trans-sensing, not meiotic silencing, in Neurospora. Genetics. 2004;168:1925–1935. doi: 10.1534/genetics.104.031526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maine EM, et al. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Curr Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.She X, et al. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dumesic PA, et al. Stalled Spliceosomes Are a Signal for RNAi-Mediated Genome Defense. Cell. 2013;152:957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, et al. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 2010;24:2566–2582. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang W, et al. BARE retrotransposons are translated and replicated via distinct RNA pools. PLoS One. 2013;8:e72270. doi: 10.1371/journal.pone.0072270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kupfer DM, et al. Introns and splicing elements of five diverse fungi. Eukaryot Cell. 2004;3:1088–1100. doi: 10.1128/EC.3.5.1088-1100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Irimia M, et al. Coevolution of genomic intron number and splice sites. Trends Genet. 2007;23:321–325. doi: 10.1016/j.tig.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Madhani HD. The frustrated gene: origins of eukaryotic gene expression. Cell. 2013 doi: 10.1016/j.cell.2013.10.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voinnet O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 2008;13:317–328. doi: 10.1016/j.tplants.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 48.Nuthikattu S, et al. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013;162:116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mari-Ordonez A, et al. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet. 2013;45:1029–1039. doi: 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- 50.Vaucheret H, et al. A Transcriptionally Active State Is Required for Post-Transcriptional Silencing (Cosuppression) of Nitrate Reductase Host Genes and Transgenes. Plant Cell. 1997;9:1495–1504. doi: 10.1105/tpc.9.8.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalmay T, et al. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 52.Luo Z, Chen Z. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007;19:943–958. doi: 10.1105/tpc.106.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herr AJ, et al. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:14994–15001. doi: 10.1073/pnas.0606536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gazzani S, et al. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 55.Gy I, et al. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Axtell MJ, et al. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 58.Christie M, et al. Intron splicing suppresses RNA silencing in Arabidopsis. Plant J. 2011;68:159–167. doi: 10.1111/j.1365-313X.2011.04676.x. [DOI] [PubMed] [Google Scholar]
- 59.Moreno AB, et al. Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants. Nucleic Acids Res. 2013;41:4699–4708. doi: 10.1093/nar/gkt152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishizu H, et al. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 62.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 63.Senti KA, Brennecke J. The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 65.Li C, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelleher ES, Barbash DA. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol Biol Evol. 2013;30:1816–1829. doi: 10.1093/molbev/mst081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muerdter F, et al. Production of artificial piRNAs in flies and mice. RNA. 2012;18:42–52. doi: 10.1261/rna.029769.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khurana JS, et al. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Vanssay A, et al. Profiles of piRNA abundances at emerging or established piRNA loci are determined by local DNA sequences. RNA Biol. 2013;10:1233–1239. doi: 10.4161/rna.25756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karpen GH, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rangan P, et al. piRNA production requires heterochromatin formation in Drosophila. Curr Biol. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klattenhoff C, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olovnikov I, et al. De novo piRNA cluster formation in the Drosophila germ line triggered by transgenes containing a transcribed transposon fragment. Nucleic Acids Res. 2013;41:5757–5768. doi: 10.1093/nar/gkt310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Vanssay A, et al. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- 77.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 78.Gu W, et al. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Billi AC, et al. A conserved upstream motif orchestrates autonomous, germline-enriched expression of Caenorhabditis elegans piRNAs. PLoS Genet. 2013;9:e1003392. doi: 10.1371/journal.pgen.1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Das PP, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ashe A, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee HC, et al. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shirayama M, et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Claycomb JM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 86.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 87.Zhang F, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]