Abstract
Self-tumor Ags that elicit antitumor immune responses in responses to IFN-α stimulation remain poorly defined. We screened a human testis cDNA library with sera from three polycythemia vera patients who responded to IFN-α and identified a novel Ag, MPD6. MPD6 belongs to the group of cryptic Ags without conventional genomic structure and is encoded by a cryptic open reading frame located in the 3′-untranslated region of myotrophin mRNA. MPD6 elicits IgG Ab responses in a subset of polycythemia vera patients, as well as patients with chronic myelogenous leukemia and prostate cancer, suggesting that it is broadly immunogenic. The expression of myotrophin-MPD6 transcripts was upregulated in some tumor cells, but only slightly increased in K562 cells in response to IFN-α treatment. By using bicistronic reporter constructs, we showed that the translation of MPD6 was mediated by a novel internal ribosome entry site (IRES) upstream of the MPD6 reading frame. Furthermore, the MPD6-IRES-mediated translation, but not myotrophin-MPD6 transcription, was significantly upregulated in response to IFN-α stimulation. These findings demonstrate that a novel IRES-mediated mechanism may be responsible for the translation of unconventional self-Ag MPD6 in responsive to IFN-α stimulation. The eliciting antitumor immune response against unconventional Ag MPD6 in patients with myeloproliferative diseases suggests MPD6 as a potential target of novel immunotherapy.
Interferon-α, a pleiotropic cytokine, is widely used in cancer therapy (1). IFN-α therapy induces a clinical remission with documented reversal of clonal hemopoiesis to polyclonal hemopoiesis (2) in polycythemia vera (PV)3 patients (3), thus making PV a model to study the antigenic mechanism of the cytokine-enhanced immune responses (1). Similarly, ~25% of patients with chronic myeloid leukemia (CML), another myeloproliferative disease (MPD), treated with IFN-α undergo a cytogenetic remission (4). In addition to its direct cytotoxic effects on tumors, IFN-α has been shown to enhance antitumor immune response (1). Cytogenetic response to IFN-α therapy in CML is often associated with therapy-related autoimmunity (5), suggesting that anti-self Ag immune responses induced by IFN-α may play an important role in controlling these diseases. The mechanism of IFN-α regulation of the expression of self-Ags (4) remains largely unknown.
We previously identified two broadly immunogenic tumor Ags that elicit immune responses associated with CML remission (6), CML66L (7) and CML28 (8), by serological analysis of tumor Ags through screening an expression cDNA library (SEREX) (9). These two Ags, similar to a class of cancer-testis Ags, have restricted expression limited to the testis and are highly expressed in a variety of tumor cells (10). Specific Ab responses to CML66L and CML28 can also be detected in patients undergone PV remission responding to IFN-α therapy, suggesting that cancer-testis-like Ag-elicited immune responses may mediate PV remission (11). Identification of high titers of IgG Ab responses against these Ags suggests that these Ags could also elicit T cell immune responses, because the elicitation of IgG responses requires Th cell function (〈www.cancerimmunity.org/SEREX/〉), as demonstrated for the SEREX Ag NY-ESO-1 in patients with melanoma (12).
Most of the tumor Ags identified so far are conventional Ags, which are defined as protein Ags encoded by genes with conventional genomic exon-intron organization and translated by primary open reading frame (ORF) in mRNAs (13). Some tumor Ags are unconventional cryptic peptide Ags, encoded by introns of genes (14), alternative reading frames (15), suboptimal reading frame initiated at non-ATG translation initiation codons including AUU coding for isoleucine (16, 17), or cryptic reading frame located in the untranslated regions (UTRs) of mRNA transcripts (17). Importantly, previous studies demonstrated that those tumor Ags could elicit T cell responses (18). Whether the unconventional cryptic peptide Ags can also elicit humoral antitumor immune responses remains unknown.
Recently, we demonstrated that alternative splicing was the dominant mechanism for increased immunogenicity of tumor Ag CML66L (19). In addition, we proposed a “stimulation-responsive splicing” model in which dominant noncanonical alternative splicing provides a structural basis for generation of untolerized self-Ag epitopes (20, 21). Furthermore, we identified an unconventional Ag MPD5 encoded in the antisense strand of the intron 1 of NEK6 gene (Z. Xiong, E. Liu, Y. Yan, R. T. Silver, S. Zhang, Y. Yang, S. Verstovsek, F. Yang, I. H. Chen, F. J. Segura, et al., submitted for publication). To determine the novel mechanisms underlying self-Ag immunogenicity, we focused on RNA transcription and processing. However, the role of translational regulation of self-Ag immunogenicity remains unclear (18). The canonical scanning mechanism has been used for the translation initiation of >90% mRNAs in eukaryotic cells in recruiting ribosomes at the capped 5′ end of mRNAs (22). Internal ribosome entry sites (IRESs) are highly structured regions located within the UTR that enable ribosomes to initiate translation effectively (22). It is estimated that up to 10% of all mRNAs have the capability to initiate translation by this mechanism (23). These mRNAs can use IRES to promote the translation of downstream cryptic cistron (22). It has become clear that IRESs are a very important component of protein expression in various essential organismal and cellular processes including development, cell cycle, and apoptosis (23). However, it remains unknown whether IRESs participate in the translation of cryptic ORFs located in UTRs that are involved in the elicitation of antitumor responses.
In this study, we tested the hypothesis that IRES mediates the translation of unconventional Ags and elicits IgG Ab immune responses that are enhanced by IFN-α. We searched for tumor Ags by SEREX (9) to identify aberrantly expressed and translated Ags (24) using sera from three PV patients who underwent IFN-α-induced remission, and we identified IRES translated unconventional Ag, MPD6. Our results demonstrate that the enhancement of IRES-mediated translation of MPD6 by IFN-α is a novel mechanism of IFN-α-enhanced antitumor immune responses.
Materials and Methods
Serum samples
Serum samples were obtained from the patients with PV and CML receiving IFN-α therapy enrolled into Temple University, Baylor College of Medicine, Cornell Medical Center, and M. D. Anderson Cancer Center Institutional Review Board-approved protocols (2). Serum samples from patients with hormone refractory advanced prostate cancer were generously provided by Dr. P. Kantoff (Dana-Farber Cancer Institute, Harvard University, Boston, MA) (7).
Transcriptional based X chromosome inactivation clonality assay (2)
To examine the clonality of hemopoietic cells in patients with PV before and after IFN-α therapy, the mRNA expression of five X chromosome exonic polymorphic genes (MPP1, IDS, G6PD, BTK, and FHL1) in platelets and granulocytes from the peripheral blood was detected using single-stranded conformation polymorphism analysis as previously described (2).
Human testis cDNA library screening by SEREX
The library screening was performed as described previously (6) (Z. Xiong, E. Liu, Y. Yan, R. T. Silver, S. Zhang, Y. Yang, S. Verstovsek, F. Yang, I. H. Chen, F. J. Segura, et al., submitted for publication). DNA sequencing was performed by SeqWright.
Bioinformatic analyses
Sequence analyses were performed using the National Center for Biotechnology Information (NCBI)-GenBank databases (〈www.ncbi.nlm.nih.gov/〉), NCBI-conserved domain databases, and the PROSITE analysis (〈http://us.expasy.org/cgi-bin/scanprosite〉) to determine whether cloned sequences were related or identical with genes, proteins, or protein domains in the databases (20). The gene organizations, such as intron-exon structure and chromosome location, were analyzed by searching in the NCBI-AceView website (20). The expression of studied genes was determined by the Northern blots. The cis-acting regulatory elements in 3′-UTR were analyzed using the IRES (〈http://ifr31w3.toulouse.inserm.fr/IRESdatabase/〉) and the UTR (〈www2.ba.itb.cnr.it/UTRSite/〉) websites (25) with generous support of Dr. S. Liuni (Bioinformatics and Genomic Group, Milan, Italy). The secondary structures of RNAs were predicted by using the MFOLD-Zuker (〈www.bioinfo.rpi.edu/applications/mfold/old/rna/〉) (26) and the Vienna RNA (〈http://rna.tbi.univie.ac.at/〉) (27) web-based algorithms.
Northern blot
Multiple tissue Northern blots were performed with purified polyadenylated RNA (Ambion) as previously reported (7).
Peptide synthesis and peptide ELISA
The MPD6 specific peptide, NH2-IVQIQHLNIPSSSSTHSSPF-COOH, was synthesized at Sigma-Genosys. ELISA was performed as previously described (7).
Construction of bicistronic reporter vectors and the reporter assay
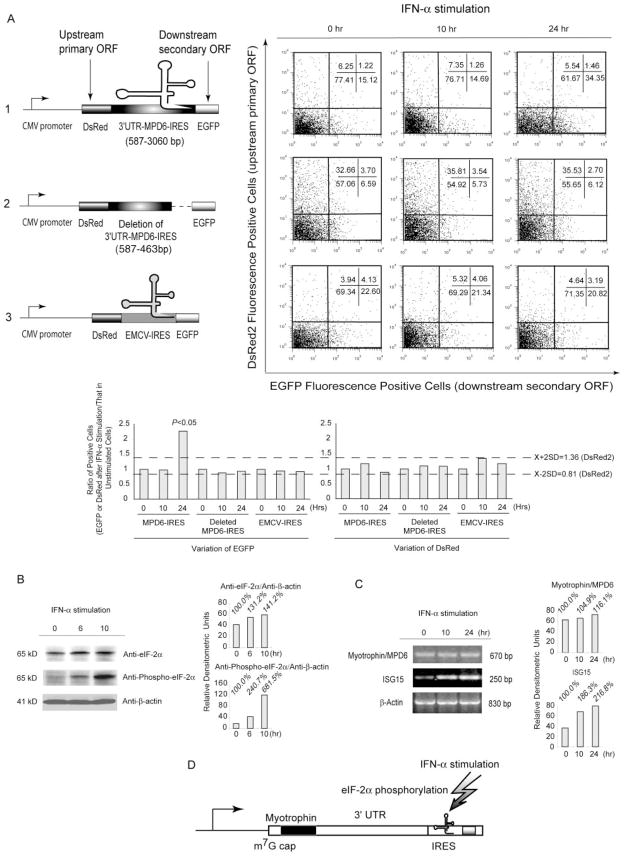
Three bicistronic reporter vectors were constructed: 1) the vector, DsRed-3′-UTR-MPD6-IRES-EGFP (see Fig. 3A1); 2) the vector, DsRed-3′-UTR-deleted MPD6-IRES-EGFP (A2); and 3) DsRed-EMCV-IRES-EGFP (A3). The reporter vector DsRed-EMCV-IRES-EGFP (see Fig. 3A3) was constructed by subcloning DsRed coding sequence (EcoRI-BamHI fragment) into the multiple cloning sites of the commercial bicistronic reporter vector pIRES2-EGFP (BD Clontech). The EcoRI-BamHI DsRed coding fragment was obtained by high-fidelity PCR (BD Clontech) using the DsRed-EcoRI mRNA sense primer specific for the 5′ end of the DsRed ORF (Table I) and the DsRed-BamHI antisense primer (Table I). The reporter vector DsRed-3′-UTR-MPD6-IRES-EGFP (see Fig. 3A1) was constructed by replacing EMCV-IRES with the 3′-UTR-MPD6-IRES (BamHI-BstXI fragment), which was prepared by high-fidelity PCR using the sense primer specific for the 5′ end of the myotrophin 3′-UTR (Table I) and the antisense primer specific for the 3′ end of MPD6-IRES (Table I). The reporter construct DsRed-3′-UTR-deleted MPD6-IRES-EGFP (see Fig. 3A2) was constructed by replacing the EMCV-IRES fragment with the partially deleted 3′-UTR-MPD6-IRES fragment in the pIRES2-EGFP vector. The deleted 3′-UTR-MPD6-IRES fragment was prepared by high-fidelity PCR using the myotrophin 3′-UTR mRNA sense primer to pair with the deleted BstXI antisense primer specific for the myotrophin 3′-UTR region 597 bp upstream of MPD6-IRES (Table I). K562 cells were transfected using the X-tremeGENE Q2 Transfection Reagent (Roche Diagnostics) with the above-described three bicistronic vectors, respectively. Stably transfected cells were selected by the resistance to the neomycin analog G418 (400 μg/ml; Invitrogen Life Technologies). To detect the potential up-regulation of DsRed and enhanced GFP (EGFP) fluorescence induced by IFN-α and avoid the potential expression variation of individual transfected clones, we used transfected cells that are resistant to G418 for additional experiments.
FIGURE 3.
Upregulated translation of EGFP directed by MPD6-IRES in response to IFN-α stimulation. A, Following bicistrionic vectors with DsRed as the upstream primary ORF and with EGFP as the downstream secondary ORF were constructed (upper left panel), including a complete sequence (myotrophin 3′-UTR plus MPD6-IRES) between myotrophin ORF and MPD6 ORF placed between DsRed ORF and EGFP ORF (A1); a deletion mutant with the 5′ portion of the myotrophin 3′-UTR but having no MPD6-IRES sequence in between DsRed ORF and EGFP ORF (A2); and a bicistronic vector control with EMCV-IRES as an documented IRES positive control (A3). The nontransfected cells were used as a negative control for DsRed and EGFP expression (data not shown). In the upper right panels, the bivariate plots show the DsRed fluorescence-positive cells (ordinate) and the EGFP fluorescence-positive cells (abscissa), and percentages of cells in each quadrant in the K562 cells stably transfected with each bicistronic reporter construct, and stimulated with IFN-α for 0, 10, and 24 h. The experiments were repeated three times, and representative results are shown. In the lower right panel, the confidential intervals (mean ± 2SD, 0.81–1.36; shown in the dashed lines) of the ratio of the percentage of DsRed-positive cells after IFN-α stimulation over that before IFN-α stimulation were generated. Similarly, in the lower left panel, the ratio of the percentage of EGFP-positive cells after IFN-α stimulation over that before IFN-α stimulation were calculated. The ratio of the percentage of EGFP-positive cells in MPD6-IRES transfected group 24 h after IFN-α stimulation over that before IFN-α stimulation was 2.3, which was significantly higher than the upper limit (1.36) of the DsRed confidential intervals (p < 0.05). B, The expression of eIF-2α and phosphorylated eIF-2α in K562 myeloid leukemia cells stimulated with IFN-α detected by specific Abs. The upregulation of the expression of eIF-2α and phosphorylated eIF-2α in responses to IFN-α stimulation in myeloid leukemia K562 cells was measured by Western blots with specific Abs to eIF-2α and phosphorylated eIF-2α, respectively, at the time points as indicated. The Western blot with Ab to β-actin was performed as a housekeeping protein control and a no-response control for IFN-α stimulation. In the right panel, the relative densitometric units were calculated by normalizing the densities of the eIF-2α and phosphorylated eIF-2α with that of β-actin in the same sample. The relative densitometric units for the expression of eIF-2α and phosphorylated eIF-2α after IFN-α stimulation over that before IFN-α stimulation were calculated and shown as the percentages. C, The MPD6 expression in K562 myeloid leukemia cells stimulated with IFN-α detected by semiquantitative RT-PCR. The upregulation of MPD6 transcripts in responses to IFN-α stimulation in myeloid leukemia K562 cells was measured by semiquantitative RT-PCR at the time points as indicated (left panel). The RT-PCR amplification of β-actin transcripts was performed as a housekeeping gene control for RT-PCR and a no-response control for IFN-α stimulation. The RT-PCR amplification of ISG15 was used as a positive control for IFN-α stimulation. In the right panel, the relative densitometric units were calculated by normalizing the densities of the PCR products of MPD6 and ISG15 with that of β-actin in the same sample. The relative densitometric units for the expression of myotrophin/MPD6 and ISG15 transcripts after IFN-α stimulation over that before IFN-α stimulation were calculated and shown as the percentages. D, The proposed working model of MPD6 translation mediated by MPD6-IRES. MPD6-IRES is found to be capable in mediation of upregulated translation of MPD6 in response to IFN-α stimulation, which may result from IFN-α-mediated phosphorylation of eIF-2α.
Table I.
Sequences of primers
| Name | Sequence |
|---|---|
| MPD6 ORF5 | 5′-GGCCGCGAATTCCTTTTGTATTAAATCAGTCATTTCA-3′ |
| MPD6 ORF3 | 5′-GGCCGCGAATTCTTACCATTCGGATGTACATGAACT-3′ |
| MPD6RT5 | 5′-AGTGCAACAGGGTGTTTTGA-3′ |
| MPD6RT3 | 5′-TGAAGCTGCAAGGGAGACTT-3′ |
| ISG5 | 5′-GAGAGGCAGCGAATTCATCT-3′ |
| ISG3 | 5′-AAGGGGGACCCTGTCCTG-3′ |
| HB-actin5 | 5′-ATCTGGCACCACACCTTCTACAATGAGCTGCG-3′ |
| HB-actin3 | 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′ |
| DsRed ORF5 | 5′-GGCCGCGAATTCATGGCCTCCTCCGAGAACGTCA-3′ |
| DsRed-BamHI3 | 5′-GGCCGCGGATCCCTACAGGAACAGGTGGTGGC-3′ |
| Myotrophin 3′-UTR5 | 5′-GGCCGCGGATCCGGATGGATGGACTGATAACTCC-3′ |
| MPD6-IRES3 | 5′-GGCCGCCCATGGTTGTGGACAAAACTACAGAACATGCAAAAT-3′ |
| MPD6-IRES de13 | 5′-GGCCGCCCATGGTTGTGGAGAGTGCCCTCCATTTTCAA-3′ |
For IFN-α stimulation assay, transfected K562 cells and mock-transfected K562 control cells were treated with human IFN-α at a concentration of 1000 U/ml for the time course of 0, 10, and 24 h. The treated K562 cells were then washed with PBS twice. The expression of EGFP and DsRed was analyzed on a FACSCalibur flow cytometer (BD Biosciences). Mock-transfected K562 cells were used for negative controls to set up the background gate. Data acquirement and analysis was performed using CellQuest Pro software (BD Biosciences). EGFP was measured in the FL1 channel (the green fluorescence at 508 nm) and DsRed, in the FL2 channel (the red fluorescence at 583 nm) (28).
Western blot
Western blot procedures were performed as described (7). Briefly, proteins in K562 cell lysates were loaded on gradient SDS-PAGE (Invitrogen Life Technologies), analyzed via Western blots with 1/1000 diluted eIF-2α Ab (Cell Signaling Technology), phospho-eIF-2α (Ser51) Ab (Cell Signaling), and anti-β-actin (1/2000) (Santa Cruz Biotechnology), respectively, and revealed by chemiluminescent substrate (Pierce Biotechnology) after exposure on x-ray film (Eastman Kodak).
Semiquantitative RT-PCR and PCR cloning
Human IFN-α (1 × 105 U/100 μl) was purchased from PBL Biomedical Laboratories. K562 cells (a human myeloid leukemia cell line purchased from American Type Culture Collection) were cultured in RPMI 1640 medium (Invitrogen Life Technologies) supplemented with 10% FBS in a humidified atmosphere of 5% carbon dioxide at 37°C. The K562 cells were stimulated with 1000 U/ml IFN-α for the time indicated (29). RNA preparation, RT-PCR, and PCR cloning were performed as previously described (30). A sense primer (MPD6RT5) specific for the 5′ sequence of myotrophin-MPD6 and an antisense primer (MPD6RT3) specific for the 3′ sequence of MPD6 were used for PCR (Table I). The PCR with 25–35 cycles was used for semiquantitation of MPD6 fragment. The 670-bp PCR product was cloned into pCRII-TOPO vector (Invitrogen Life Technologies) and confirmed by DNA sequencing. As an internal control for cDNA preparation, the housekeeping gene β-actin was examined by PCR (sense primer, HB-actin5, and antisense primer, HB-actin3) (Table I). The ethidium bromide staining signals of PCR products were analyzed with a documentation system (Eastman Kodak) and normalized as relative densitometric units by comparing to β-actin amplified in the same cDNA preparations (30). ISG15, as a positive control for the genes upregulated by IFN-α stimulation (29), was amplified by PCR with a sense primer ISG5 and an antisense primer ISG3 (Table I).
Results
Identification of a novel MPD-associated SEREX Ag, MPD6
The human testis expression cDNA library was screened by SEREX using diluted sera collected from the patients with PV who responded to IFN-α therapy (2). Testis is an immune-privileged site where self-Ags are not presented to host immune system (31, 32). As reported by studies in other tumors (24, 33, 34), screening a testis expression cDNA library using SEREX with sera from various tumor patients has proved to be a useful approach to identifying the tumor-associated Ags that are aberrantly expressed in tumors but not normal tissues. We chose the sera from PV patients for cDNA library screening based on two criteria: 1) the patients had undergone remission in response to IFN-α treatment as judged by conversion from monoclonal to polyclonal hemopoiesis determined by X chromosome transcriptional polymorphism analyses (2); and 2) normalization of hemoglobin concentration and platelet count on IFN-α therapy (2, 3). Two of the three PV females with favorable response to IFN-α treatment were previously reported (2). Initial screening of 1 × 106 λ recombinant phage clones was followed by several rounds of the purification of positive phage plaques and further confirmation on their Ag specificities. A unique cDNA clone was identified.
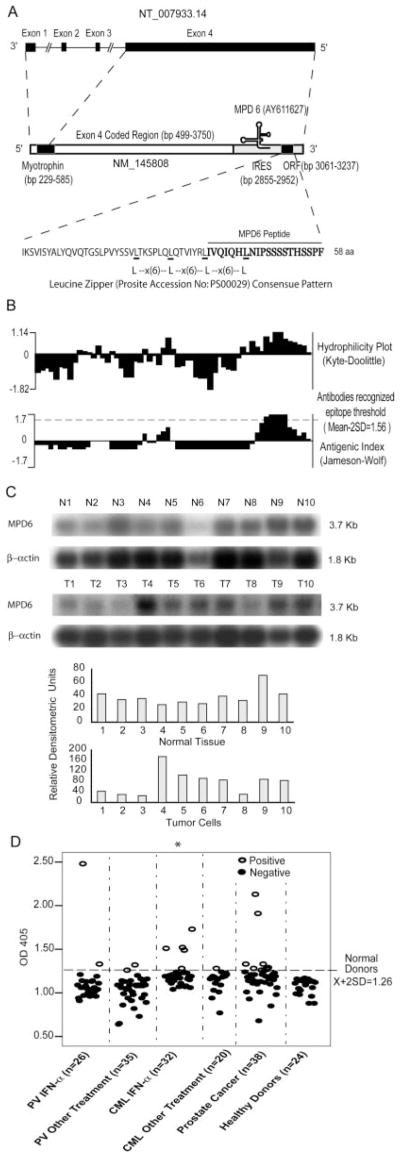
The isolated clone (710-bp insert) was identical with myotrophin mRNA (Fig. 1A). Myotrophin interacts with rel/NF-κB (35, 36) and converts NF-κB p50-p65 heterodimers to homodimers (37). In contrast to DC6 encoded by the cDNA sense strand of tumor Ag CML66 that we previously reported (19), the coding sequence of the isolated clone was located in the mRNA-sense strand of myotrophin mRNA; however, the coding sequence was in the 3′-UTR (bp 3061–3237) of myotrophin mRNA (35) (Fig. 1A). Sequence analysis revealed an ORF that encoded 58 aa located in frame 1 (the same as that of myotrophin) (Fig. 1B). Because the size of the predicted protein encoded by this ORF was 6.4 kDa, we referred it to as MPD6. MPD6 could not result from a mechanism of termination codon readthrough (38), because there were 40 in-frame stop codons between the myotrophin ORF and that of MPD6 (data not shown). The myotrophin-MPD6 gene locus spans 50.7 kb in human chromosome 7q33 with four exons. Exon 4 encodes an mRNA from bp 499-3750, which covers the C-terminal 29 aa of myotrophin and a 3′-UTR that also includes the MPD6 reading frame (Fig. 1A). Thus, MPD6 and myotrophin share the same mRNA transcript, which is independent of alternative splicing (Fig. 1A). Therefore, MPD6 cannot be generated by the following two mechanisms: 1) encoding by a separated exon potentially transcribed by a cryptic promoter as shown for transcription of self-tumor Ag CML66L isoform, CML66S (19), and 2) integration into the C terminus of myotrophin by exon skipping mechanism (39).
FIGURE 1.
Molecular features of MPD6. A, Schematic representation of the location of unconventional Ag MPD6 gene in the 3′-UTR of myotrophin mRNA (GenBank accession no. NM_145808) as well as the genomic structure of the myotrophin-MPD6 gene locus (GenBank accession no. NT_007933.14). The MPD6 ORF (GenBank accession no. AY611627) is located in the region of bp 3061–3237 in the 3′-UTR of myotrophin, whereas the primary ORF myotrophin is located in the region of bp 229–585. An IRES is also located in the region of the 3′-UTR of myotrophin, bp 2855–2952 upstream of MPD6 ORF. B, Feature of MPD6 protein sequence. The MPD6 ORF encodes a 58-aa protein. The start codon is an unconventional start codon AUU (isoleucine), rather than the conventional start codon AUG (methionine). Hydrophilicity plot analysis using by Kyte-Dolittle method indicated that MPD6 has a C-terminal hydrophilic region, which corresponds to the region achieving the higher Jameson-Wolf antigenic index. The Jameson-Wolf antigenic index in the MPD6 C-terminal region is higher than that of previously characterized Abs recognized epitope threshold (mean − 2SD, 1.56). MPD6 peptide used in the ELISA was synthesized according to the MPD6 sequence from aa 39 to 58. C, Higher expression of myotrophin-MPD transcripts in some tumor cells detected by Northern blots. In the upper panel, the lanes N1 to N10 indicate various normal tissues in the order of brain (N1), liver (N2), placenta (N3), small intestine (N4), colon (N5), thymus (N6), spleen (N7), prostate (N8), testis (N9), and ovary (N10), respectively. In the middle panel, the lanes T1 to T10 indicate various tumor cells in the order of acute T cell leukemia (Jurkat cells) (T1), Burkitt’s lymphoma (CA46) (T2), breast cancer (MDA-MD-453) (T3), Burkitt’s lymphoma (Namalwa) (T4), epidermal carcinoma (A-431) (T5), uterine carcinoma (MES-SA) (T6), Burkitt’s lymphoma (Raji) (T7), osterosarcoma (MG-63) (T8), histiocytic lymphoma (U-937) (T9), and cervical adenocarcinoma (Hela S3) (T10), respectively. The hybridization analyses of the normal tissue and tumor cell expression (BD Clontech) with 32P-labeled specific probes, as indicated, were performed, respectively. The transcript sizes are indicated in kilobases. The ratio of the hybridization signal density of MPD6 transcript over the hybridization signal density of β-actin in the same sample was calculated as relative densitometric unit as presented in the lower panels. D, The IgG Ab responses to the C-terminal antigenic epitope (from the aa 39 to 58) of MPD6 detected by peptide ELISA. The experiments were repeated three times, and the representative results were shown. The mean + 2 SD of the OD405 ratios of the peptide over the coating control from 24 healthy donors were calculated as the upper limit of the normal range of Ab responses to MPD6 peptide (mean + 2 SD, 1.26). The detection rates of the IgG Ab responses to MPD6 peptide in the group of CML patients treating with IFN-α are statistically higher than that of CML patients treating with other therapies (χ2 goodness-of-fit test; *, p < 0.05).
The MPD6 reading frame had neither a canonical start codon (AUG) nor an optimal Kozak consensus sequence (A/GN NAUGG) (40). However, this sequence started with the unconventional start codon AUU encoding isoleucine (16, 17), one of the start codons that was previously demonstrated to initiate cryptic Ag peptide translation (18, 41). Moreover, this reading frame matched one (the slip frame) of the three forward fusion frames of the library vector pTriplEx, suggesting that MPD6 sequence fused in the library vector was recognized by Abs in the sera of patients during SEREX screening. The predicted MPD6 amino acid sequence showed no significant homologies to any known proteins in the NCBI-GenBank databases and the SEREX database (〈www2.licr.org/CancerImmunomeDB/SEREX_SeqSimilaritySearch.php〉), indicating that it is a novel protein. The following features of MPD6 supported our contention that it was a human protein-encoding sequence: 1) the frequencies of amino acid codon usage including those amino acid residues with multiple codons (42) were identical with those used in human proteins; 2) the analysis of protein domains and motifs on the PROSITE database demonstrated that two overlapped regions of MPD6, including the first one from aa 17 to 38, and the second one from aa 24 to 45, were identical with the leucine zipper pattern (Leu-x(6)-Leu-x(6)-Leu-x(6)-Leu) (the PROSITE ID no. PS00029) (Fig. 1B), which mediates the dimerization of transcription factors and other regulatory proteins (43). The two overlapping leucine zippers in MPD6 could not have been generated randomly, because the chance of occurrence of leucine at any given position in proteins is 0.09 (44) and the random chance of the frequency for MPD6 to have the simultaneous occurrence of leucine at these five defined positions is extremely low (0.095 = 5.9 × 10−6). These characteristics suggest that the nonrandom leucine zipper patterns may fulfill a specific cellular function. Thus, MPD6 has the characteristics of short protein-encoding ORFs as previously reported (44).
Up-regulated expression of myotrophin-MPD6 transcripts in some tumor cells
Detection of IgG Abs to MPD6 in patients with tumors suggested that expression of myotrophin-MPD6 transcripts might be up-regulated in some tumors. Because MPD6 is always located in the same 3.77-kb transcripts with myotrophin ORF, therefore, the 3.77-kb myotrophin-MPD6 mRNAs detected by Northern blots confirm the authenticity of MPD6. The Northern blots examining the expression of MPD6 transcripts in 10 normal tissues are depicted in the upper panel of Fig. 1C. To avoid possible variation in sample preparation and loading, the ratio of the density of MPD6 transcript over the density of β-actin in the same sample was calculated as relative densitometric unit, as we reported (30). High levels of expression (>75 relative densitometric units) were found in brain (N1), spleen (N7), testis (N9), and ovary (N10). In contrast, MPD6 expression was significantly upregulated (>85 relative densitometric units) in a variety of tumor cells (Fig. 1C, lower panel), including Burkitt’s lymphoma (Namalwa) (T4), epidermal carcinoma (A-431) (T5), uterine carcinoma (MES-SA) (T6), Burkitt’s lymphoma (Raji) (T7), histiocytic lymphoma (U-937) (T9), and cervical adenocarcinoma (Hela S3) (T10).
Detection of MPD6-specific IgG Abs in patients with PV, CML, and prostate cancer
As shown in the lower panels of Fig. 1B, MPD6 had one hydrophilic region and also achieved high antigenic index scores (45), suggesting that this region has potential to be a binding epitope for Ab interactions. We previously showed that the mean ± 2SD of the Jameson-Wolf antigenic scores of 43 experimentally verified, Ab-recognized antigenic epitopes of self-Ags (46) was in the range of 1.56 to 4.36 (21). These analyses suggested that one antigenic epitope in MPD6 with the Jameson-Wolf Ag index scores of 1.7 had good potential ( p < 0.05) in the elicitation of specific Ab responses. To verify whether MPD6 Ag epitope is immunogenic in vivo, a MPD6 peptide was synthesized using the amino acid sequence of the second Ag epitope of MPD6 from aa 39 to 58. The ELISA using the MPD6 peptide showed that the MPD6 epitope-specific IgG Ab responses could be detected in 7.7 and 5.7% of PV patients receiving either IFN-α or other therapies, respectively (Fig. 1D). In addition, MPD6-specific IgG Ab responses could be detected in 15.6 and 5.0% of CML patients receiving either IFN-α or other therapies, respectively. Furthermore, anti-MPD6 IgG Abs were also found in 23.7% of patients with prostate cancer. These results suggested that MPD6 was not only immunogenic in patients with PV, but also broadly immunogenic in patients with other tumors. Previous studies showed that the incidence of IgG Ab immune responses to the aberrantly expressed self-tumor Ags ranges between 5 and 50%, depending on the tumor type and the respective Ag (47). Therefore, the Ab responses to MPD6 in patients with tumors were in the range similar to that of other self-tumor Ags. It is noteworthy that the detection rates of anti-MPD6 IgG Abs were much higher in CML patients receiving IFN-α than in CML patients receiving other therapies ( p < 0.05). This suggests that IFN-α therapy may enhance the anti-MPD6 immune responses either by upregulation of MPD6 expression or by enhancement of immune responses to MPD6 Ag. Enhancement of immune responses to MPD6 was not observed in PV patients receiving IFN-α in comparison to that in PV patients receiving other therapies, which may suggest a disease-specific effect of IFN-α. It is well accepted that Western blots are a good indicator of specific Ag-Ab interaction. Based on this principle, detection of specific Abs to the C-terminal peptide of MPD6 in patients using SEREX suggests that: 1) MPD6 was expressed in a subset of patients with tumors; and 2) the full-length MPD6 may be expressed because the C terminus of MPD6 is the last part of MPD6 sequence to be translated in protein synthesis.
The translation of MPD6 is mediated by an IRES, which is enhanced by IFN-α stimulation
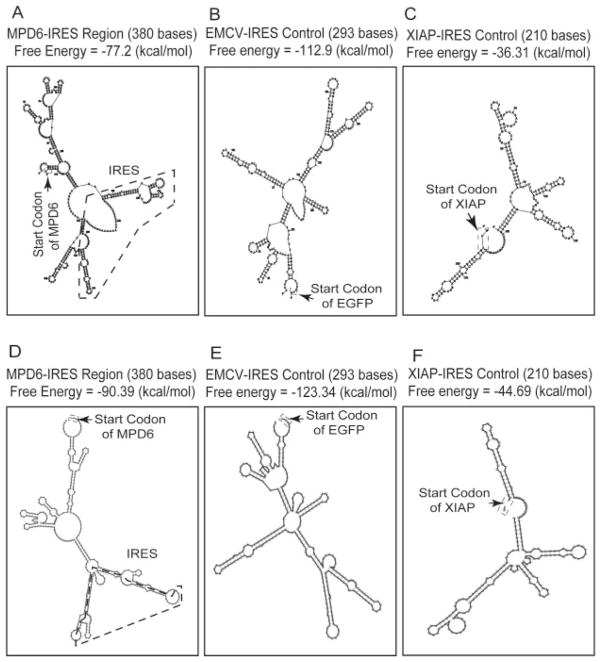
The mechanism of translation of the cryptic ORF located in the 3′-UTR of myotrophin into a protein Ag remained to be determined. We examined the possibility that potential IRESs located in the upstream of MPD6 promoted the translation of MPD6. The analysis of the UTR database (25) showed an IRES-containing region located upstream of MPD6, as shown in Figs. 1A and 2, A and D. Because some cellular IRESs may contain a common Y-type stem-loop structural motif (48), we searched the sequences upstream of MPD6 for stable stem-loop structures by applying two prediction algorithms for RNA secondary structure, MFOLD (26) and RNAfold (27) for the prediction accuracy higher than that using one algorithm. As shown in Fig. 2, the encephalomyocarditis virus (EMCV) IRES region from the widely used bicistronic expression vector IRES2-EGFP had a stable stem-loop structure with a low free energy, i.e., −112.9 kcal/mol (the MFOLD algorithm, Fig. 2B), or −123.34 kcal/mol (RNAfold algorithm, Fig. 2E). We also show that the IRES-containing region located upstream of MPD6 has a stable stem-loop structure with the free energy of either −77.2 kcal/mol calculated by the MFOLD algorithm (Fig. 2A) or −90.39 kcal/mol calculated by the RNAfold algorithm (Fig. 2D). It is noteworthy that we found that the experimentally verified XIAP IRES (49) (Fig. 2, C and F) had a free energy higher than that of the MPD6 IRES region (Fig. 2, A and D) by using the same MFOLD and RNAfold algorithms (26), which suggest that MPD6 IRES may have a stem-loop structure more stable than that of the experimentally verified XIAP IRES. Because a lower free energy of the RNA secondary structure is associated with higher stability of the structure (26, 27), these analyses suggest that the IRES-containing region located upstream of MPD6 is remarkably stable for fulfilling IRES function (26, 27).
FIGURE 2.
The predicted stem-loop structure in MPD6-IRES. The cis-acting regulatory elements in 3′-UTR were analyzed using the IRES website (〈http://ifr31w3.toulouse.inserm.fr/IRESdatabase/〉) and the UTR website (〈www2.ba.itb.cnr.it/UTRSite/〉) with generous support by Dr. S. Liuni at the Bioinformatics and Genomic Group in Italy. The secondary structures of MPD6-IRES (A and D), EMCV-IRES (B and E), and XIAP-IRES (C and F) were predicted by using two web-based algorithms, MFOLD-Zuker (〈www.bioinfo.rpi.edu/applications/mfold/old/rna/〉) (A–C) and RNAfold (〈http://rna.tbi.univie.ac.at/〉) (D–F). The free energy of the secondary structure of these IRES regions was also calculated with both algorithms. The start codon and IRES region of MPD6 are indicated.
To verify that the IRES-containing structure upstream of MPD6 has IRES function, we constructed a set of bicistronic reporter gene vectors with DsRed as the upstream primary ORF and EGFP as the downstream secondary ORF; the latter was translated under the direction of IRES (Fig. 3A). We detected the translation of the downstream secondary ORF EGFP by measuring green fluorescence with flow cytometry, and the translation of the upstream primary ORF DsRed by measuring red fluorescence. As shown in Fig. 3A1, 0 h, the MPD6 IRES region mediated significant translation (2.3-fold) of the downstream EGFP (15.12%) in the absence of IFN-α stimulation, in comparison to the deletion of IRES control (6.59%) (Fig. 3A2, 0 h) and the positive control EMCV-IRES (22.60%) (Fig. 3A3, 0 h). These results suggest that similar to EMCV-IRES, MPD6-IRES upstream MPD6 ORF has the IRES function to mediate the translation of downstream ORF.
The higher rates of anti-MPD6 IgG Abs in patients with CML treated by IFN-α (Fig. 1D) and the IRES-mediated translation of MPD6 raised a possibility that IRES-mediated MPD6 translation may be enhanced by treatment of IFN-α. To test this possibility, we treated K562 cells stably express bicistronic vectors with IFN-α for 0, 10, and 24 h before examining the expression of DsRed and EGFP. For examination of transfection efficiency, as shown in Fig. 3A2, 0 h (in the absence of MPD6-IRES), the protein translation of DsRed was 36.36% (DsRed single-positive + DsRed-EGFP double-positive = 32.66 + 3.70%), suggesting that transfection efficiency after G418 selection was high. We observed that DsRed and EGFP double-positive cells were low throughout all of the experimental groups. Moreover, deletion of MPD6-IRES resulted in increased expression of DsRed from 6.25% in Fig. 3A1, 0 h, to 32.66% in A2, 0 h. The interference of EGFP translation over DsRed translation is not unique to the DsRed-MPD6-IRES-EGFP-transfected cells, because the same effect was observed in the control DsRed-EMCV-IRES-EGFP-transfected cells (Fig. 3A3, 0 h). These results suggest that relative translation interference between the two reading frames occurs, similar to that reported previously (50). The molecular mechanism underlying the interference is not clear. Regardless of interference of EGFP translation over DsRed translation, due to the inclusion of appropriate controls, we could use this system to address two issues: 1) potential translation function of MPD6-IRES; and 2) potential responses of MPD6-IRES to IFN-α stimulation. The MPD6-IRES-mediated EGFP expression was significantly upregulated (2.3-fold) from 15.12% (Fig. 3A1, 0 h) to 34.35% (Fig. A1, 24 h) in comparison with the nearly unchanged percentages of the green fluorescence-positive cells in the MPD6-IRES deletion control-transfected cells after IFN-α treatment (6.59% in Fig. 3A2, 0 h, and 6.12% in A2, 24 h). Because the deletion of MPD6-IRES vector still remained a large portion of myotrophin 3′-UTR (Fig. 3A2), the deletion of MPD6-IRES completely abolished the response to IFN-α stimulation, indicating that the response of the MPD6-IRES-transfected cells with upregulated EGFP expression to IFN-α treatment was MPD6-IRES sequence dependent. In contrast, EMCV IRES-mediated EGFP expression remained stable from 0 h (22.60% in Fig. 3A3, 0 h) to 24 h (20.82% in Fig. A3, 24 h) after IFN-α stimulation, suggesting that the response of IRES-mediated protein translation to IFN-α stimulation was MPD6-IRES specific. Of note, EGFP downstream of MPD6-IRES was the secondary ORF in the bicistronic mRNA transcript that did not contain any introns and splicing signals (51). Therefore, the EGFP translation (Fig. 3A1) could not be mediated by m7G cap-dependent mechanism or regulated by RNA alternative splicing. In addition, we considered the possibility that the response of MPD6-IRES-mediated EGFP translation to IFN-α stimulation might result from the response of the CMV promoter to IFN-α stimulation. Thus, we measured the translation of the upstream primary ORF DsRed. As shown in Fig. 3A2, 0, 10, and 24 h, the protein translation of DsRed was in the range of 32.66–35.81%, suggesting that both CMV promoter activities and the m7G cap-dependent translation of the upstream primary ORF were not affected by IFN-α stimulation, which served as the no-response controls. As shown in the lower right panel of Fig. 3A, the confidential intervals (mean ± 2SD, 0.81–1.36) of the ratio of the percentage of DsRed-positive cells after IFN-α stimulation over that before IFN-α stimulation were generated as the confidential intervals for no-response variations. Similar to reported above, the ratio of the percentage of EGFP-positive cells in MPD6-IRES transfected group 24 h after IFN-α stimulation over that before IFN-α stimulation was 2.3, which was significantly higher than the upper limit (1.36) of the DsRed confidential intervals ( p < 0.05) (Fig. 3A, lower left panel). It was of concern that enhanced expression of MPD6-IRES-mediated EGFP expression by IFN-α stimulation may have resulted from decreased degradation of EGFP. However, when we exchanged DsRed in the secondary ORF position and placed EGFP in the primary ORF position, the results stayed the same as shown in Fig. 3A1 (data not shown). These results showed that MPD6-IRES has IRES function, which is enhanced by IFN-α stimulation.
As we observed previously (Z. Xiong, E. Liu, Y. Yan, R. T. Silver, S. Zhang, Y. Yang, S. Verstovsek, F. Yang, I. H. Chen, F. J. Segura, et al., submitted for publication), phosphorylated eIF-2α was upregulated (681.5% at 10 h) in response to IFN-α stimulation, whereas eIF-2α was only up-regulated in a modest manner (141.2% at 10 h) (Fig. 3B). Because the unconventional leucine start codon is enhanced in the presence of phosphorylated eIF-2α (16), the up-regulated phosphorylated eIF-2α in response to IFN-α stimulation (Z. Xiong, E. Liu, Y. Yan, R. T. Silver, S. Zhang, Y. Yang, S. Verstovsek, F. Yang, I. H. Chen, F. J. Segura, et al., submitted for publication) might enhance MPD6 translation, which is mediated by the unconventional isoleucine start codon.
We also determined whether IRES-mediated translation or the transcription of myotrophin-MPD6 is the major mechanism for up-regulation of MPD6 after IFN-α stimulation. As shown in Fig. 3C, as a positive control, the expression of ISG15 RNA transcripts was significantly up-regulated (116.8%) in response to IFN-α stimulation (29), suggesting the cells were appropriately stimulated with IFN-α. Under the same conditions, the transcription of myotrophin-MPD6 mRNA was increased only 16.1% after IFN-α stimulation. In contrast, the MPD6-IRES-mediated translation of EGFP was significantly increased by 130% (Fig. 3A1, 24 h, and lower left panel of Fig. 3A). Taken together, these results suggest that MPD-IRES-mediated translation is the major mechanism for up-regulation of MPD6 after IFN-α stimulation.
It is noteworthy that MPD6-IRES-mediated enhanced protein expression by IFN-α (Fig. 3A1) did not correlate well with lower detection rates of MPD6-specific Abs in patients who received IFN-α therapy (Fig. 1D). This discrepancy was expected because the up-regulated expression of tumor Ags is not the only determining mechanism for their increased immunogenicity (〈www.cancerimmunity.org/SEREX/〉).
Discussion
IFN-α has been shown to enhance antitumor immune response (4). However, the self-Ag targets and the mechanism of IFN-α inducing antitumor immune response remain poorly defined. The reversion of clonal to polyclonal hemopoiesis in PV patients who responded to IFN-α therapy has been reported (2). These studies have laid a foundation for definition of therapeutic immune responses in patients with PV. Furthermore, current studies suggest that multiple genetic defects may be involved in the pathogenesis of PV (52), reflecting tumor heterogeneity and presumably Ag heterogeneity (53). Therefore, it would be beneficial for the Ag-specific based vaccine and other therapeutic approaches to encompass a broader array of tumor Ags to affect broad subpopulations of tumor cells that may express different tumor Ags (53). The gain-of-function acquired somatic mutation (V617F) of the tyrosine kinase JAK2 has been identified in most patients with PV and other MPDs (54–58). It remains to be determined whether the activating mutation of JAK2 may modulate the expression of tumor Ags and antitumor immune responses, a subject currently under investigation (59).
Development of SEREX has led to rapid identification of a large number of tumor Ags deposited in the SEREX database. In contrast, a modest number of tumor Ags have been targeted in tumor vaccines and immunotherapies (13). Several steps of analysis are mandatory to evaluate SEREX-defined Ags before they become new target Ags for active immunotherapy, including expression analysis, and serological analysis with sera from tumor patients and normal individuals, etc. (60). As a result, a few tumor Ags have yet be exploited in immunotherapy. Demand for new Ag-specific immunotherapies and current technical problems all call for urgent development of new, high-throughput technology in mapping immunodominant T cell Ag epitopes, characterization of more clinically targetable Ags from the database, and elucidation of novel mechanisms underlying the immunogenicity of tumor Ags, as we reported here.
Two previous reports have used the SEREX approach to examine self-tumor Ag repertoire and their expression enhanced by IFN-γ focusing on conventional rather than cryptic Ags (61–63). By applying the SEREX technique to screen a human testis expression cDNA library with sera from PV patients, we have identified a novel SEREX Ag that elicits potent humoral immune responses in a subset of patients with MPD. The cryptic Ag peptides encoded by introns or UTRs can elicit T cell responses (18, 64), but we also demonstrated that unconventional cryptic Ag peptides can elicit IgG Ab responses (Z. Xiong, E. Liu, Y. Yan, R. T. Silver, S. Zhang, Y. Yang, S. Verstovsek, F. Yang, I. H. Chen, F. J. Segura, et al., submitted for publication). Here, we report a novel cryptic Ag MPD6 and show that its expression is IRES mediated. Because these unconventional Ags are small peptides, they require only minimal processing to yield the peptide sizes suitable for MHC class I-, class II-restricted, and Ab-recognized Ag epitopes to effectively expand self-Ag repertoire (17). In addition, although short peptide Ags may have the disadvantage of being presented in fewer MHC allelic molecules, they may possess an advantage of being not very immunogenic, which may lead to Ag-specific anergy in patients with tumors (65). The up-regulation of MPD6-IRES-initiated translation by IFN-α may provide new insight into the potential mechanism of regulating the self-Ag repertoire in response to IFN-α. To our knowledge, the demonstration of tumor-associated Ab responses elicited by a novel IRES-mediated translation of unconventional Ag is the first such study in tumor immunology (66).
IFN-α treatment induces up-regulation of numerous genes in tumor cells (67) and other cells (68). However, proteins up-regulated by IFN-α may not necessarily all become self-tumor Ags (13). As we discussed previously (19, 69), the overexpression of self-Ags must overcome the “threshold” of Ag concentration at which an immune response is initiated as Zinkernagel et al. (70, 71) recently suggested. In addition, overexpressed Ags must access the Ag presentation pathway and immune system by the following mechanisms (72). First, overexpressed Ags may be released from damaged tumor cells due to spontaneous necrosis or apoptosis, and then become available in the extracellular environment for attack by the immune cells, potentially through cross-presentation (73, 74); second, tumor-expressed Ags can translocate across the intracellular membranes (75) via binding to heat shock protein 70 and enter the membrane exosome for MHC class II Ag presentation pathway (76). Moreover, some other factors contributing to the immunogenicity of autoantigens and self-tumor Ags have also been proposed (77).
The novel Ag MPD6, as well as our previously identified MPD associated Ags MPD5 and eIF-2α (Z. Xiong, E. Liu, Y. Yan, R. T. Silver, S. Zhang, Y. Yang, S. Verstovsek, F. Yang, I. H. Chen, F. J. Segura, et al., submitted for publication), was identified using sera from female patients; however, myotrophin-MPD6 gene loci is located on human autosomal chromosomes 7q33 (78) rather than on the Y chromosome, suggesting that antitumor immune responses enhanced by IFN-α therapy in CML but not PV, are at least partially mediated by novel self-tumor Ags, rather than Y chromosome-encoded male-specific Ags (78). Correlation of Ag-specific IgG immune responses with the remission in PV and CML patients (3, 79) suggested that immune responses mediated by both unconventional and conventional Ags may contribute to the MPD remission.
There are two mechanisms used by eukaryotic cells to initiate translation, the classical 7methyl guanosine cap-dependent scanning mechanism and IRES (22). IRESs are diverse cis-acting RNA sequences that are able to mediate internal entry of the 40S ribosomal subunit directly onto an AUG or other start codons (16) of eukaryotic and viral mRNAs (80). IRESs are often found in essential mRNAs encoding regulatory proteins (transcription factors, growth factors, and kinases) (23). IRES activity can be modulated in response to mitotic stimuli, hypoxia, and other stimuli, p38 MAPK signaling (81), GM-CSF, and IL-3 via the PI3K pathway (82), which suggest that IRES-containing transcripts (83) are important determinants of cellular proliferation and/or differentiation (80).
Identification of novel cryptic self-Ag peptides improves our understanding of the self-Ag repertoire (18). The activation of PKR by IFN-α could induce hepatitis C virus IRES-dependent mRNA translation from dicistronic constructs (84), analogous to our observation that the MPD6-IRES-mediated EGFP translation was increased in responses to IFN-α stimulation. IFN-α-induced, dsRNA-activated PKR is a key mediator of the antiviral and antiproliferative effects of IFN (85); thus, IFN-α may activate IRES-dependent translation of MPD6. However, whether IFN-α therapy initiates the IRES-mediated translation of the cryptic Ag peptide or promotes the existing translation of the cryptic peptide was not previously shown, the evidence for which we now submit in this report.
Taken together, our results suggest that the novel IRES-mediated translation of the unconventional cryptic Ag peptide of MPD6, but not transcription of RNA transcripts, may be increased by IFN-α, which may provide a novel mechanism underlying the expansion of the self-Ag repertoire that mediates both autoimmune as well as antitumor immune responses.
Acknowledgments
We are grateful to Drs. B. Ashby at Temple University (Philadelphia, PA), P. Kantoff at Harvard University (Boston, MA), M. Talpaz at University of Michigan (Ann Arbor, MI), U. Certa at Roche Center for Genomics (Basel, Switzerland), and S. Liuni in the Bioinformatics and Genomic Group in the National Research Council Italian EMBnet Node (Milan, Italy) for their assistance.
Footnotes
This work was partially supported by National Institutes of Health Grant AI054514 (to X.-F.Y.), and Leukemia and Lymphoma Society and the Myeloproliferative Disorders Foundation (to X.-F.Y.), and funds from the Cancer Research and Treatment Fund (to R.T.S.).
Abbreviations used in this paper: PV, polycythemia vera; CML, chronic myeloid leukemia; MPD, myeloproliferative disease; ORF, open reading frame; UTR, untranslated region; IRES, internal ribosome entry site; EMCV, encephalomyocarditis virus; EGFP, enhanced GFP.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Belardelli F, Ferrantini M, Proietti E, Kirkwood JM. Interferon-α in tumor immunity and immunotherapy. Cytokine Growth Factor Rev. 2002;13:119–134. doi: 10.1016/s1359-6101(01)00022-3. [DOI] [PubMed] [Google Scholar]
- 2.Liu E, Jelinek J, Pastore YD, Guan Y, Prchal JF, Prchal JT. Discrimination of polycythemias and thrombocytoses by novel, simple, accurate clonality assays and comparison with PRV-1 expression and BFU-E response to erythropoietin. Blood. 2003;101:3294–3301. doi: 10.1182/blood-2002-07-2287. [DOI] [PubMed] [Google Scholar]
- 3.Lengfelder E, Berger U, Hehlmann R. Interferon-α in the treatment of polycythemia vera. Ann Hematol. 2000;79:103–109. doi: 10.1007/s002770050563. [DOI] [PubMed] [Google Scholar]
- 4.Fujii S. Role of interferon-α and clonally expanded T cells in the immunotherapy of chronic myelogenous leukemia. Leuk Lymphoma. 2000;38:21–38. doi: 10.3109/10428190009060316. [DOI] [PubMed] [Google Scholar]
- 5.Sacchi S, Kantarjian H, O’Brien S, Cohen PR, Pierce S, Talpaz M. Immune-mediated and unusual complications during interferon-alfa therapy in chronic myelogenous leukemia. J Clin Oncol. 1995;13:2401–2407. doi: 10.1200/JCO.1995.13.9.2401. [DOI] [PubMed] [Google Scholar]
- 6.Wu CJ, Yang XF, McLaughlin S, Neuberg D, Canning C, Stein B, Alyea EP, Soiffer RJ, Dranoff G, Ritz J. Detection of a potent humoral response associated with immune-induced remission of chronic myelogenous leukemia. J Clin Invest. 2000;106:705–714. doi: 10.1172/JCI10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XF, Wu CJ, McLaughlin S, Chillemi A, Wang KS, Canning C, Alyea EP, Kantoff P, Soiffer RJ, Dranoff G, Ritz J. CML66, a broadly immunogenic tumor antigen, elicits a humoral immune response associated with remission of chronic myelogenous leukemia. Proc Natl Acad Sci USA. 2001;98:7492–7497. doi: 10.1073/pnas.131590998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang XF, Wu CJ, Chen L, Alyea EP, Canning C, Kantoff P, Soiffer RJ, Dranoff G, Ritz J. CML28 is a broadly immunogenic antigen, which is overexpressed in tumor cells. Cancer Res. 2002;62:5517–5522. [PMC free article] [PubMed] [Google Scholar]
- 9.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu CJ, Biernacki M, Kutok JL, Rogers S, Chen L, Yang XF, Soiffer RJ, Ritz J. Graft-versus-leukemia target antigens in chronic myelogenous leukemia are expressed on myeloid progenitor cells. Clin Cancer Res. 2005;11:4504–4511. doi: 10.1158/1078-0432.CCR-05-0036. [DOI] [PubMed] [Google Scholar]
- 11.Yang XF, Yan Y, Phan L, Xiong Z, Jelinek J, Prchal J. Upregulation of cancer-testis antigens in polycythemia vera cells suggests their potential role in immune responses against myeloproliferation. Blood. 2002;100:348b. [Google Scholar]
- 12.Jager E, Nagata Y, Gnjatic S, Wada H, Stockert E, Karbach J, Dunbar PR, Lee SY, Jungbluth A, Jager D, et al. Monitoring CD8 T cell responses to NY-ESO-1: correlation of humoral and cellular immune responses. Proc Natl Acad Sci USA. 2000;97:4760–4765. doi: 10.1073/pnas.97.9.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F, Yang XF. New concepts in tumor antigens: their significance in future immunotherapies for tumors. Cell Mol Immunol. 2005;2:331–341. [PubMed] [Google Scholar]
- 14.Guilloux Y, Lucas S, Brichard VG, Van Pel A, Viret C, De Plaen E, Brasseur F, Lethe B, Jotereau F, Boon T. A peptide recognized by human cytolytic T lymphocytes on HLA-A2 melanomas is encoded by an intron sequence of the N-acetylglucosaminyltransferase V gene. J Exp Med. 1996;183:1173–1183. doi: 10.1084/jem.183.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang RF, Parkhurst MR, Kawakami Y, Robbins PF, Rosenberg SA. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J Exp Med. 1996;183:1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwab SR, Shugart JA, Horng T, Malarkannan S, Shastri N. Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol. 2004;2:e366. doi: 10.1371/journal.pbio.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malarkannan S, Horng T, Shih PP, Schwab S, Shastri N. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity. 1999;10:681–690. doi: 10.1016/s1074-7613(00)80067-9. [DOI] [PubMed] [Google Scholar]
- 18.Shastri N, Schwab S, Serwold T. Producing nature’s gene-chips: the generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 19.Yan Y, Phan L, Yang F, Talpaz M, Yang Y, Xiong Z, Ng B, Timchenko NA, Wu CJ, Ritz J, et al. A novel mechanism of alternative promoter and splicing regulates the epitope generation of tumor antigen CML66-L. J Immunol. 2004;172:651–660. doi: 10.4049/jimmunol.172.1.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng B, Yang F, Huston DP, Yan Y, Yang Y, Xiong Z, Peterson LE, Wang H, Yang XF. Increased non-canonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes. J Allergy Clin Immunol. 2004;114:1463–1470. doi: 10.1016/j.jaci.2004.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang F, Chen IH, Xiong Z, Yan Y, Wang H, Yang X-F. Model of stimulation-responsive splicing and strategies in identification of immunogenic isoforms of tumor antigens and autoantigens. Clin Immunol. doi: 10.1016/j.clim.2006.06.007. In press. [DOI] [PubMed] [Google Scholar]
- 22.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 23.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 24.Gure AO, Tureci O, Sahin U, Tsang S, Scanlan MJ, Jager E, Knuth A, Pfreundschuh M, Old LJ, Chen YT. SSX: a multigene family with several members transcribed in normal testis and human cancer. Int J Cancer. 1997;72:965–971. doi: 10.1002/(sici)1097-0215(19970917)72:6<965::aid-ijc8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 25.Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, Duarte J, Saccone C, Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawley TS, Telford WG, Ramezani A, Hawley RG. Four-color flow cytometric detection of retrovirally expressed red, yellow, green, and cyan fluorescent proteins. BioTechniques. 2001;30:1028–1034. doi: 10.2144/01305rr01. [DOI] [PubMed] [Google Scholar]
- 29.Smith JK, Siddiqui AA, Krishnaswamy GA, Dykes R, Berk SL, Magee M, Joyner W, Cummins J. Oral use of interferon-α stimulates ISG-15 transcription and production by human buccal epithelial cells. J Interferon Cytokine Res. 1999;19:923–928. doi: 10.1089/107999099313460. [DOI] [PubMed] [Google Scholar]
- 30.Yang XF, Weber GF, Cantor H. A novel Bcl-x isoform connected to the T cell receptor regulates apoptosis in T cells. Immunity. 1997;7:629–639. doi: 10.1016/s1074-7613(00)80384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streilein JW. Unraveling immune privilege. Science. 1995;270:1158–1159. doi: 10.1126/science.270.5239.1158. [DOI] [PubMed] [Google Scholar]
- 32.Chen YT. Cancer vaccine: identification of human tumor antigens by SEREX. Cancer J Sci Am. 2000;6(Suppl 3):S208–S217. [PubMed] [Google Scholar]
- 33.Hoeppner LH, Dubovsky JA, Dunphy EJ, McNeel DG. Humoral immune responses to testis antigens in sera from patients with prostate cancer. Cancer Immun. 2006;6:1. [PubMed] [Google Scholar]
- 34.Okada T, Akada M, Fujita T, Iwata T, Goto Y, Kido K, Okada T, Matsuzaki Y, Kobayashi K, Matsuno S, et al. A novel cancer testis antigen that is frequently expressed in pancreatic, lung, and endometrial cancers. Clin Cancer Res. 2006;12:191–197. doi: 10.1158/1078-0432.CCR-05-1206. [DOI] [PubMed] [Google Scholar]
- 35.Sivasubramanian N, Adhikary G, Sil PC, Sen S. Cardiac myotrophin exhibits rel/NF-κB interacting activity in vitro. J Biol Chem. 1996;271:2812–2816. doi: 10.1074/jbc.271.5.2812. [DOI] [PubMed] [Google Scholar]
- 36.Gupta S, Sen S. Myotrophin-κB DNA interaction in the initiation process of cardiac hypertrophy. Biochim Biophys Acta. 2002;1589:247–260. doi: 10.1016/s0167-4889(02)00178-7. [DOI] [PubMed] [Google Scholar]
- 37.Knuefermann P, Chen P, Misra A, Shi SP, Abdellatif M, Sivasubramanian N. Myotrophin/V-1, a protein up-regulated in the failing human heart and in postnatal cerebellum, converts NFκB p50–p65 heterodimers to p50–p50 and p65–p65 homodimers. J Biol Chem. 2002;277:23888–23897. doi: 10.1074/jbc.M202937200. [DOI] [PubMed] [Google Scholar]
- 38.Bullock TN, Patterson AE, Franlin LL, Notidis E, Eisenlohr LC. Initiation codon scanthrough versus termination codon readthrough demonstrates strong potential for major histocompatibility complex class I-restricted cryptic epitope expression. J Exp Med. 1997;186:1051–1058. doi: 10.1084/jem.186.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 41.Shastri N, Nguyen V, Gonzalez F. Major histocompatibility class I molecules can present cryptic translation products to T-cells. J Biol Chem. 1995;270:1088–1091. doi: 10.1074/jbc.270.3.1088. [DOI] [PubMed] [Google Scholar]
- 42.Aubsubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Screening of recombinant DNA libraries. In: Chanda V, editor. Current Protocols in Molecular Biology. Wiley; New York: 1999. p. A.1C.3. [Google Scholar]
- 43.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 44.Furuno M, Kasukawa T, Saito R, Adachi J, Suzuki H, Baldarelli R, Hayashizaki Y, Okazaki Y. CDS annotation in full-length cDNA sequence. Genome Res. 2003;13:1478–1487. doi: 10.1101/gr.1060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jameson BA, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci. 1988;4:181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- 46.Mahler M, Bluthner M, Pollard KM. Advances in B-cell epitope analysis of autoantigens in connective tissue diseases. Clin Immunol. 2003;107:65–79. doi: 10.1016/s1521-6616(03)00037-8. [DOI] [PubMed] [Google Scholar]
- 47.Preuss KD, Zwick C, Bormann C, Neumann F, Pfreundschuh M. Analysis of the B-cell repertoire against antigens expressed by human neoplasms. Immunol Rev. 2002;188:43–50. doi: 10.1034/j.1600-065x.2002.18805.x. [DOI] [PubMed] [Google Scholar]
- 48.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 49.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 50.Hennecke M, Kwissa M, Metzger K, Oumard A, Kroger A, Schirmbeck R, Reimann J, Hauser H. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29:3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraunus J, Schaumann DH, Meyer J, Modlich U, Fehse B, Brandenburg G, von Laer D, Klump H, Schambach A, Bohne J, Baum C. Self-inactivating retroviral vectors with improved RNA processing. Gene Ther. 2004;11:1568–1578. doi: 10.1038/sj.gt.3302309. [DOI] [PubMed] [Google Scholar]
- 52.Kralovics R, Stockton DW, Prchal JT. Clonal hematopoiesis in familial polycythemia vera suggests the involvement of multiple mutational events in the early pathogenesis of the disease. Blood. 2003;102:3793–3796. doi: 10.1182/blood-2003-03-0885. [DOI] [PubMed] [Google Scholar]
- 53.Bhattachary-Chatterjee M, Nath Baral R, Chatterjee SK, Das R, Zeytin H, Chakraborty M, Foon KA. Counterpoint: cancer vaccines: single-epitope anti-idiotype vaccine versus multiple-epitope antigen vaccine. Cancer Immunol Immunother. 2000;49:133–141. doi: 10.1007/s002620050612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 55.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 56.Zhao R, Xing S, Li Z, Fu X, Li Q, Krantz SB, Zhao ZJ. Identification of an acquired JAK2 mutation in polycythemia vera. J Biol Chem. 2005;280:22788–22792. doi: 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James C, Ugo V, Le Couedic PJ, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 58.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 59.Verma A, Kambhampati S, Parmar S, Platanias LC. Jak family of kinases in cancer. Cancer Metastasis Rev. 2003;22:423–434. doi: 10.1023/a:1023805715476. [DOI] [PubMed] [Google Scholar]
- 60.Jager D, Taverna C, Zippelius A, Knuth A. Identification of tumor antigens as potential target antigens for immunotherapy by serological expression cloning. Cancer Immunol Immunother. 2004;53:144–147. doi: 10.1007/s00262-003-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ehlken H, Schadendorf D, Eichmuller S. Humoral immune response against melanoma antigens induced by vaccination with cytokine gene-modified autologous tumor cells. Int J Cancer. 2004;108:307–313. doi: 10.1002/ijc.11537. [DOI] [PubMed] [Google Scholar]
- 62.Okada H, Attanucci J, Giezeman-Smits KM, Brissette-Storkus C, Fellows WK, Gambotto A, Pollack LF, Pogue-Geile K, Lotze MT, Bozik ME, Chambers WH. Immunization with an antigen identified by cytokine tumor vaccine-assisted SEREX (CAS) suppressed growth of the rat 9L glioma in vivo. Cancer Res. 2001;61:2625–2631. [PubMed] [Google Scholar]
- 63.Ono T, Sato S, Kimura N, Tanaka M, Shibuya A, Old LJ, Nakayama E. Serological analysis of BALB/c methylcholanthrene sarcoma meth A by SEREX: identification of a cancer/testis antigen. Int J Cancer. 2000;88:845–851. doi: 10.1002/1097-0215(20001215)88:6<845::aid-ijc1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 64.Schwab SR, Li KC, Kang C, Shastri N. Constitutive display of cryptic translation products by MHC class I molecules. Science. 2003;301:1367–1371. doi: 10.1126/science.1085650. [DOI] [PubMed] [Google Scholar]
- 65.Molldrem JJ, Lee PP, Wang C, Felio K, Kantarjian HM, Champlin RE, Davis MM. Evidence that specific T lymphocytes may participate in the elimination of chronic myelogenous leukemia. Nat Med. 2000;6:1018–1023. doi: 10.1038/79526. [DOI] [PubMed] [Google Scholar]
- 66.Gilbert HS. Current management in polycythemia vera. Semin Hematol. 2001;38:25–28. doi: 10.1016/s0037-1963(01)90137-4. [DOI] [PubMed] [Google Scholar]
- 67.Certa U, Wilhelm-Seiler M, Foser S, Broger C, Neeb M. Expression modes of interferon-α inducible genes in sensitive and resistant human melanoma cells stimulated with regular and pegylated interferon-α. Gene. 2003;315:79–86. doi: 10.1016/s0378-1119(03)00722-4. [DOI] [PubMed] [Google Scholar]
- 68.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ng B, Yang F, Huston DP, Yan Y, Yang Y, Xiong Z, Peterson LE, Wang H, Yang XF. Increased noncanonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes. J Allergy Clin Immunol. 2004;114:1463–1470. doi: 10.1016/j.jaci.2004.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ludewig B, Junt T, Hengartner H, Zinkernagel RM. Dendritic cells in autoimmune diseases. Curr Opin Immunol. 2001;13:657–662. doi: 10.1016/s0952-7915(01)00275-8. [DOI] [PubMed] [Google Scholar]
- 71.Zinkernagel RM, Hengartner H. Regulation of the immune response by antigen. Science. 2001;293:251–253. doi: 10.1126/science.1063005. [DOI] [PubMed] [Google Scholar]
- 72.Bauer C, Diesinger I, Brass N, Steinhart H, Iro H, Meese EU. Translation initiation factor eIF-4G is immunogenic, overexpressed, and amplified in patients with squamous cell lung carcinoma. Cancer. 2001;92:822–829. doi: 10.1002/1097-0142(20010815)92:4<822::aid-cncr1388>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 73.Nagata Y, Ono S, Matsuo M, Gnjatic S, Valmori D, Ritter G, Garrett W, Old LJ, Mellman I. Differential presentation of a soluble exogenous tumor antigen, NY-ESO-1, by distinct human dendritic cell populations. Proc Natl Acad Sci USA. 2002;99:10629–10634. doi: 10.1073/pnas.112331099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steinman RM, Inaba K, Turley S, Pierre P, Mellman I. Antigen capture, processing, and presentation by dendritic cells: recent cell biological studies. Hum Immunol. 1999;60:562–567. doi: 10.1016/s0198-8859(99)00030-0. [DOI] [PubMed] [Google Scholar]
- 75.Davidoff AM, Iglehart JD, Marks JR. Immune response to p53 is dependent upon p53/HSP70 complexes in breast cancers. Proc Natl Acad Sci USA. 1992;89:3439–3442. doi: 10.1073/pnas.89.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 77.Moudgil K, Sercarz EE. Antigenic determinants involved in induction and propagation of autoimmunity. In: Rose N, Mackay IR, editors. The Autoimmune Diseases. 3. Academic; San Diego: 1998. pp. 45–58. [Google Scholar]
- 78.Simpson E, Scott D, Chandler P. The male-specific histocompatibility antigen, H-Y: a history of transplantation, immune response genes, sex determination and expression cloning. Annu Rev Immunol. 1997;15:39–61. doi: 10.1146/annurev.immunol.15.1.39. [DOI] [PubMed] [Google Scholar]
- 79.Talpaz M, Ravandi F, Kurzrock R, Estrov Z, Kantarjian HM. Interferon-α and β: Clinical Application. Lippincott Williams & Wilkins; Philadelphia: 2000. [Google Scholar]
- 80.Bonnal S, Boutonnet C, Prado-Lourenco L, Vagner S. IRESdb: the Internal Ribosome Entry Site database. Nucleic Acids Res. 2003;31:427–428. doi: 10.1093/nar/gkg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kobayashi N, Saeki K, Yuo A. Granulocyte-macrophage colony-stimulating factor and interleukin-3 induce cell cycle progression through the synthesis of c-Myc protein by internal ribosome entry site-mediated translation via phosphatidylinositol 3-kinase pathway in human factor-dependent leukemic cells. Blood. 2003;102:3186–3195. doi: 10.1182/blood-2003-02-0567. [DOI] [PubMed] [Google Scholar]
- 83.Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469–473. doi: 10.1016/s0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- 84.Rivas-Estilla AM, Svitkin Y, Lopez Lastra M, Hatzoglou M, Sherker A, Koromilas AE. PKR-dependent mechanisms of gene expression from a subgenomic hepatitis C virus clone. J Virol. 2002;76:10637–10653. doi: 10.1128/JVI.76.21.10637-10653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stark GR, I, Kerr M, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]