Abstract
The spliceosome is the macromolecular machine responsible for pre-mRNA splicing, an essential step in eukaryotic gene expression. During splicing a myriad of subunits join and leave the spliceosome as it works on the pre-mRNA substrate. Strikingly, there are very few small molecules known to interact with the spliceosome. Splicing inhibitors are needed to capture transient spliceosome conformations and probe important functional components. Such compounds may also have chemotherapeutic applications, as links between splicing and cancer are increasingly uncovered. To identify new splicing inhibitors, we developed a high throughput assay for in vitro splicing using an RT-qPCR readout. In a pilot screen of 3,080 compounds we identified three small molecules that inhibit splicing in HeLa extract by interfering with different stages of human spliceosome assembly. Two of the compounds similarly impact spliceosomes in yeast extracts, suggesting selective targeting of conserved components. By examining related molecules, we identified chemical features required for the activity of two of the splicing inhibitors. In addition to verifying our assay procedure and paving the way to larger screens, these studies establish new compounds as chemical probes for investigating the splicing machinery.
Keywords: pre-mRNA splicing, spliceosome, RT-qPCR, inhibitor, high-throughput assay
Introduction
Pre-mRNA splicing is a critical process in eukaryotic gene expression. While the chemistry behind removing introns and ligating exons is well understood, the mechanisms by which the spliceosome recognizes its substrates and regulates splicing remain unclear. Spliceosomes assemble de novo at introns in a stepwise process from five uridine-rich small nuclear RNAs with associated proteins (U1, U2, U4, U5, and U6 snRNPs) and a large number of additional protein components1. In vitro studies using native gels have defined an ordered series of intermediate splicing complexes. In the first complex (E complex), U1 snRNP joins the pre-mRNA, followed by addition of U2 snRNP to create the pre-spliceosome or A complex. The U4, U5, and U6 tri-snRNP then join to create B complex, which is activated by release of U1 and U4 for splicing catalysis in C complex2. Complex rearrangements of protein-protein, protein-RNA and RNA-RNA interactions drive spliceosome assembly and progression. Given the complexity of the spliceosome, many additional complexes surely remain to be captured and characterized.
To make new intermediate spliceosome complexes available for biochemical and structural analysis, small molecule inhibitors that selectively target different components are needed to arrest spliceosome progression at discrete steps. With the large number of enzymatic activities and regulated rearrangements in spliceosomes, it is clear that a diverse set of compounds will be required. Some splicing inhibitors may also be useful as biological probes of spliceosome function in cells. With the recent finding of spliceosome mutations associated with progression of chronic lymphocytic leukemia and myelodysplastic syndrom3–6, such molecules may also hold promise for understanding and possibly treating human disease7.
High-throughput screening (HTS) with a sensitive and robust assay is an important strategy for identifying small molecule inhibitor candidates. An established human in vitro splicing system allows spliceosome function to be assessed in isolation from other cellular processes and provides a means to probe all of its ~one hundred components simultaneously8, 9. Here we describe HTS of ~3,000 compounds for splicing inhibitors using a new reverse transcription followed by quantitative PCR (RT-qPCR) assay system. We identified three structurally distinct small molecules that inhibit human in vitro splicing reactions in a dose-dependent manner. We characterized the effects of these compounds on splicing chemistry and spliceosome assembly using extracts and substrates in human and yeast to examine their selectivity. One compound, Tetrocarcin A (C1), an antibiotic with anti-tumor activity10, inhibits first step chemistry at an early stage of spliceosome assembly in extracts from both organisms. A family of naphthazarin compounds (C3) affects later stages of spliceosome assembly in human and yeast extracts, while a third indole derivative (C2) blocks the earliest stages of assembly in the human system only. With these results it is clear that we have an assay system that is robust in identifying new small molecule modulators of splicing. Furthermore, we can attribute effects of candidate inhibitors to discrete steps of splicing chemistry and spliceosome assembly.
Materials and Methods
In vitro splicing reactions
For the human splicing system, pre-mRNA substrate is derived from the adenovirus major late transcript. A G(5′)ppp(5′)G-capped substrate was generated by T7 run-off transcription followed by G50 gel filtration to remove unincorporated nucleoside triphosphates. Transcripts derived from a cDNA copy of spliced mRNA were used in some experiments as a control. For gel-based splicing assays, the substrate was body-labeled with 32P-UTP. Nuclear extract was prepared from HeLa cells grown in MEM/F12 1:1 and 5% (v/v) newborn calf serum11. For splicing reactions, we incubated substrate RNA at 10 nM concentration in 60 mM potassium glutamate, 2 mM magnesium acetate, 2 mM ATP, 5 mM creatine phosphate, 0.05 mg ml−1 tRNA, and 50% (v/v) HeLa nuclear extract at 30°C. For yeast splicing reactions, extracts were prepared according to Yan et al.12, and assayed using RP51A pre-mRNA at 4 nM as previously described13.
RT-qPCR reagents
RT-qPCR reactions were carried out using the TaqMan® One-Step RT-PCR kit (Applied Biosystems) with the following primers and TaqMan probe: 5′-TCTCTTCCGCATCGCTGTCT-3′ (forward primer) directed to the 5′ exon, 5′-GCGAAGAGTTTGTCCTCAACGT-3′ (reverse primer) directed to the 3′ exon, and 5′FAM-6-AGCTGTTGGGCTGCAG SPC3-BH13′ (TaqMan probe) directed to the exon junction. We determined the qPCR efficiency for these primers as (10(−1/slope)−1) where slope was derived from the linear regression analysis from a standard curve of values for cDNA containing spliced mRNA.
High-throughput splicing assay
In vitro splicing reactions were prepared in 384-well plates by dispensing 5 μL of nuclear extract by a liquid handling robot (Perkin Elmer Janus). A second robot equipped with a 384-pin tool (Perkin Elmer Janus MDT) transferred 200 nL of library or control compounds dissolved in DMSO into the nuclear extract, then 5 μL splicing mix containing pre-mRNA substrate and buffer were added for final concentrations of: 50% (v/v) nuclear extract, 200 μM library compound, 10 nM substrate RNA, 60 mM potassium glutamate, 2 mM magnesium acetate, 2 mM ATP, 5 mM creatine phosphate, 0.05 mg ml−1 yeast tRNA. Plates were sealed and incubated for 60 min at 30°C. After incubation, the splicing reaction was diluted 1:2 with water using a peristaltic dispenser (Matrix WellMate).
RT-qPCR analysis
15 nL of diluted splicing reaction were transferred by pin robot to a new 384-well plate containing 5 μl of RT-qPCR premix (1x TaqMan master mix, 0.8 μM reverse primer, 0.4 μM forward primer, 0.5 μM TaqMan probe). RT-qPCR plates were analyzed with an ABI PRISM 7900HT Sequence Detection System under the following conditions: RT-step: 30 min 48°C, 10 min 95°C; qPCR: 40 cycles of 30 sec 95°C, 50 sec 50°C, 50 sec 72°C followed by 7 min 72°C. Threshold cycle values (CT) for individual wells were normalized on a plate-to-plate basis to uninhibited control reactions.
Z′ calculation
We calculated a Z′ value for the assay using the following equation: Z′ = 1 − {(3σno splicing + 3σsplicing) / (μno splicing − μsplicing)}. ‘No splicing’ values were derived from splicing reactions containing a pre-mRNA substrate with a mutation that blocks the second step of splicing chemistry, whereas normal ‘splicing’ values were derived from splicing reactions with a pre-mRNA substrate competent for both steps of splicing chemistry.
Denaturing gel analysis
RNA was extracted from in vitro splicing reaction and separated on a 15% (v/v) denaturing polyacrylamide gel. 32P-labeled RNA species were visualized by phosphorimaging and quantified with ImageQuant software (Molecular Dynamics). Splicing efficiency is calculated as the amount of spliced mRNA relative to total RNA and normalized to a DMSO control reaction. IC50 is the concentration of inhibitor that causes 50% decrease of splicing efficiency estimated from plots of splicing efficiency vs. compound concentration.
Native gel analysis
For human splicing complexes, nuclear extract was pre-incubated at 30° C for 15 minutes to deplete endogenous ATP before setting up in vitro splicing reactions. Time point samples were kept on ice until all samples were ready for analysis. 10 μL of splicing reactions were mixed with 5 μL native gel loading buffer (20 mM Trizma base, 20 mM glycine, 25% (v/v) glycerol, 0.05% (w/v) cyan blue, 0.05% (w/v) bromophenol blue, 2.5 mg ml−1 heparin sulfate) and incubated at room temperature for five minutes before loading onto a 2.1% (w/v) agarose gel. Gels were run at 72 V for 3.5 hours, dried onto Whatman paper and exposed to phosphorimaging screens, which were digitized with a Typhoon Scanner (Molecular Dynamics).
For yeast splicing complexes, 5 μl of yeast splicing reactions were mixed with 5 μl complex buffer and yeast splicing complexes were separated using 0.5% (w/v) agarose; 3% 80:1 acrylamide:bis (v/v) gel run for 16 hours at 85V 13.
RESULTS
RT-qPCR assay to screen for inhibitors of in vitro splicing
To search for inhibitors of the human spliceosome, we used a synthetic pre-mRNA substrate consisting of two exons separated by an intron and HeLa nuclear “splicing” extract8. Spliceosomes assemble on the pre-mRNAs, which are then spliced in two chemical steps forming mRNA9, 14 (Figure 1a). Typically, the reaction is analyzed by denaturing gel electrophoresis to quantify the amount of splicing products, which is not conducive to scale up or automation. For HTS, we developed an RT-qPCR assay that employs a TaqMan® probe complementary to the unique splice junction sequence created by exon ligation (Figure 1b). This assay reports a threshold cycle (CT) that directly correlates to the amount of mRNA produced in the reaction by the spliceosome. A lower CT value indicates more mRNA and more splicing, while an increased CT value corresponds to reduced mRNA and less splicing. Using this system, we detect readily mRNA produced by in vitro splicing (Supplemental Figure 1a). We also verified that 2% DMSO, the compound in which most library molecules are dissolved, does not affect the assay (Supplemental Figure 1b). Finally, we do not detect mRNA in splicing reactions supplemented with the known splicing inhibitor SSA15 (Supplemental Figure 1b).
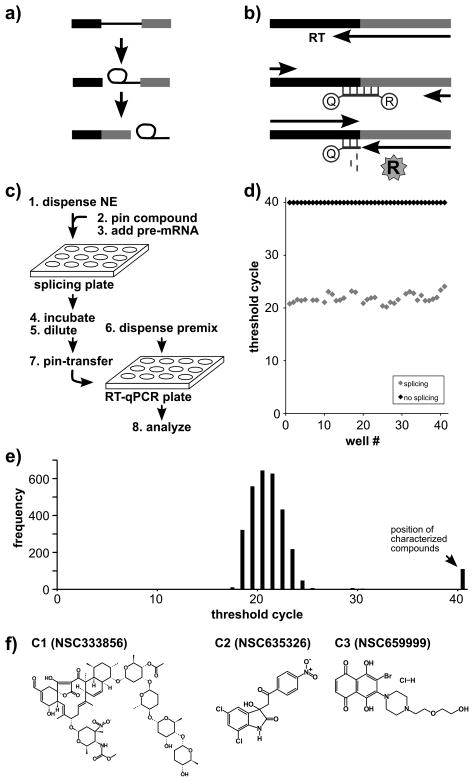
Figure 1.
High-throughput approach to identify splicing inhibitors. a) Two-step splicing reaction showing pre-mRNA, first step intermediates (5′ exon and lariat intron intermediates) and second step products (mRNA and intron lariat). b) Schematic of TaqMan® based RT-qPCR assay. mRNA is reverse transcribed (RT) and quantified via a dual-labeled oligo probe to the splice junction by release of a fluorophore reporter (R) from a quencher (Q). c) Schematic of automated splicing assay. d) Data used to calculate Z′ value for the assay using no-splicing control (black diamonds) and normal splicing (gray diamonds) reactions. For each reaction, CT is plotted vs. well number. e) Screening results of NCI library collection. The histogram plots the number of compounds screened vs. CT value, and the position of candidate inhibitors is indicated. f) The chemical structures of three verified hits (C1, C2 and C3).
To screen large chemical libraries for splicing inhibitors, we used liquid handling robots to set up the assay in 384-well plates. The steps of the protocol are schematized in Figure 1c: (1) 5 μl nuclear extract is dispensed to wells, (2) a pin robot transfers 0.2 μl of test compounds from a library plate into the nuclear extract, (3) 5 μl of pre-mRNA substrate in splicing buffer is dispensed into the nuclear extract, (4) splicing proceeds for sixty minutes, (5) the reactions are diluted with 10 μl of water, (6) 5 μl of the RT-qPCR reaction components are dispensed into a second 384-well plate, (7) a pin robot transfers 15 nl of the diluted splicing reactions as template to the RT-qPCR plate, and (8) RT-qPCR is carried out. To analyze the data, we compare the CT value for each well with control splicing wells into which either DMSO alone or SSA was added.
In scaling up for screening large compound libraries, some assay steps can be performed ahead of time. For example, we pre-fill plates with nuclear extract, which is labile to some extent, and store frozen to avoid extended waiting prior to setting up the splicing reaction. For convenience, we also pre-fill and freeze plates with the RT-qPCR reaction mix. Because the RT-qPCR analysis requires 3.5 hours for each plate, we seal and freeze the PCR plates after pinning from the diluted splicing reaction and perform the analysis over several days as needed. A second practical matter is generating large quantities of nuclear extract with reasonable splicing capacity. Our lab currently grows HeLa cells in 10L batches and prepares enough extract per month to screen 10,000 compounds. If desired, this capacity could be readily increased. Notably, we find that splicing efficiency varies with the source of the HeLa cell line (data not shown), so it is important to test extracts before using them for splicing reactions.
Before screening chemical libraries for splicing inhibitors, we characterized the splicing assay system by setting up a plate with alternating rows of reactions containing either wild-type pre-mRNA (normal splicing) or with pre-mRNA containing a splice site mutation that prevents the second step of splicing chemistry (no-splicing control). We measured mRNA amounts by RT-qPCR for each condition and found that normal splicing reactions cluster with an average CT value of 22±1.07, while the no-splicing control reactions never reach threshold (>40 CT), as expected for no spliced mRNA product (Figure 1d). From these CT distribution we calculate a Z′ value16 for the assay of 0.82 with good separation (~18 CT) between fully normal and no-splicing splicing reactions. We conclude that the automated splicing assay can measure loss of splicing allowing us to screen large chemical libraries for splicing inhibitors.
High-throughput screen for splicing inhibitors
Using the RT-qPCR splicing assay, we screened a 3,080 compounds library combining the ‘Structural Diversity Set’, ‘Challenge Set’, ‘Natural Products Set’, and ‘Mechanistic Diversity Set’ from the National Cancer Institute (NCI). The vast majority of compounds in the library did not have an effect on in vitro splicing and showed low CT values within a baseline range centered at a CT value of 21 (Figure 1e). About 3% or 100 compounds resulted in CT values in the fully inhibited splicing range (Figure 1e, CT >40). For these, we returned to the original splicing plate and manually repeated the RT-qPCR analysis of the candidate inhibitor wells in triplicate, which reconfirmed 40 (1.3% overall) as yielding high CT values. We ordered the compounds present in those wells and retested their effects on in vitro splicing by the same RT-qPCR assay. Six of these consistently interfered with in vitro splicing, but only three showed clear dose-dependent effects that are expected for a specific splicing inhibitor (Figure 1f).
Candidate compounds inhibit human and yeast pre-mRNA splicing
The three candidate splicing inhibitors that we identified from the pilot screen have very distinct structures (Figure 1f). Compound 1 (C1) is a large and complex natural compound known as Tetrocarcin A (NSC333856). It has been previously described as an antibiotic10 and as an antitumor compound that inhibits the anti-apoptotic gene Bcl210, 17, 18. The second compound (C2) is an indole derivative (NSC635326) with no known biological activity. The third compound (C3) is a naphthazarin derivative (NSC659999) and like several other naphthazarin compounds, has shown activity in a variety of biological contexts, including suppression of tumor growth18.
The RT-qPCR splicing assay detects inhibition of mRNA accumulation, but does not identify the molecular basis of this inhibition. To determine how the three compounds affect splicing chemistry, we performed in vitro splicing assays in HeLa nuclear extract using radiolabeled pre-mRNA substrate, followed by gel electrophoresis to visualize splicing intermediates and products. With all three compounds, unspliced pre-mRNA substrate levels did not change significantly, which indicates that RNA is generally stable in the reactions in the presence of compound. Therefore, RNA degradation induced by the compounds does not account for the absence of mRNA. Instead, the addition of all three compounds to splicing reactions results in a loss of splicing chemistry in a concentration-dependent manner (Figure 2a). The presence of C1 primarily resulted in loss of first step of splicing chemistry (and thereby second step chemistry as well). At 100 μM of C1 all splicing chemistry disappeared, and we determined an IC50 for the compound of ~25 μM. Second step chemistry appeared more sensitive to the C2 and C3 compounds, although first step chemistry was also lost at higher concentrations (Supplemental Figure 2). For C2, second step chemistry was completely lost at 200 μM, with IC50 of ~50 μM. C3 was the most potent splicing inhibitor. At 60 μM, its presence resulted in full loss of second step chemistry with an IC50 of ~20 μM. First step was lost at 200 μM, with an IC50 of ~50 μM (Supplemental Figure 2). The distinct sensitivities of 1st and 2nd splicing chemistry could mean that these compounds affects a spliceosome component differentially required for both steps. Alternatively, they may be selective for more than one target. Notably for all three compounds, splicing inhibition is reproducible with different preparations of nuclear extract and does not depend upon pre-incubation of the extract or pre-mRNA with the inhibitor.
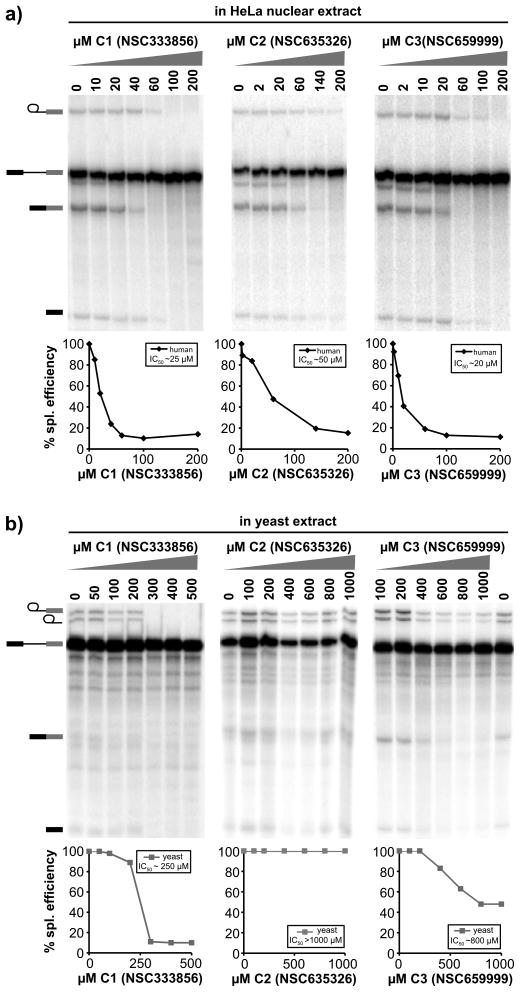
Figure 2.
Three compounds inhibit splicing chemistry in a dose-dependent manner. Denaturing gel analysis of RNA from splicing reactions with indicated concentration of C1, C2, and C3. Quantification of splicing efficiency vs. inhibitor concentration is plotted below each gel along with estimated IC50 values. a) Inhibition in HeLa nuclear extract. Identities of bands are schematized to the left as (from top to bottom) lariat intermediate, pre-mRNA, mRNA, 5′ exon intermediate. b) Inhibition in yeast extract. Identities of bands are schematized to the left as (from top to bottom) lariat intermediate, intron lariat, pre-mRNA, mRNA, 5′ exon intermediate.
We also examined the effects of these compounds on in vitro splicing in S. cerevisiae extracts with an RP51A splicing substrate19 (Figure 2b). Again, addition of C1 resulted in lost of first step chemistry, although higher concentrations were required to see an effect with an IC50 of ~250 μM. Similar to its effect in human extracts, addition of C3 primarily resulted in loss of second step, although much higher concentrations are required and splicing is never completely blocked. In contrast, C2 had no effect on yeast splicing. These results suggest that C1 and C3 interfere with a conserved mechanism of the splicing process, whereas C2 is selective for a factor specific to human splicing and does not simply inactivate all splicing extracts.
C1 stalls spliceosome assembly early in human and yeast extracts
The spliceosome assembles through a series of complex intermediates, and we expect that a splicing inhibitor selective for a component involved in complex formation will interfere with a specific assembly stage. We used native agarose gels to investigate the effect of inhibitor compounds on human spliceosome assembly20. In the absence of inhibitor (2% DMSO), these gels show the progression of spliceosome assembly through complex intermediates of E/H, A, B, and C complexes (Figure 3b- lanes 1–5). Both steps of splicing chemistry take place in C complex, which disassembles immediately after catalysis is completed. Direct inhibition of splicing chemistry typically causes C complex accumulation, while interference with a specific assembly step often results in accumulation of the preceding complex.
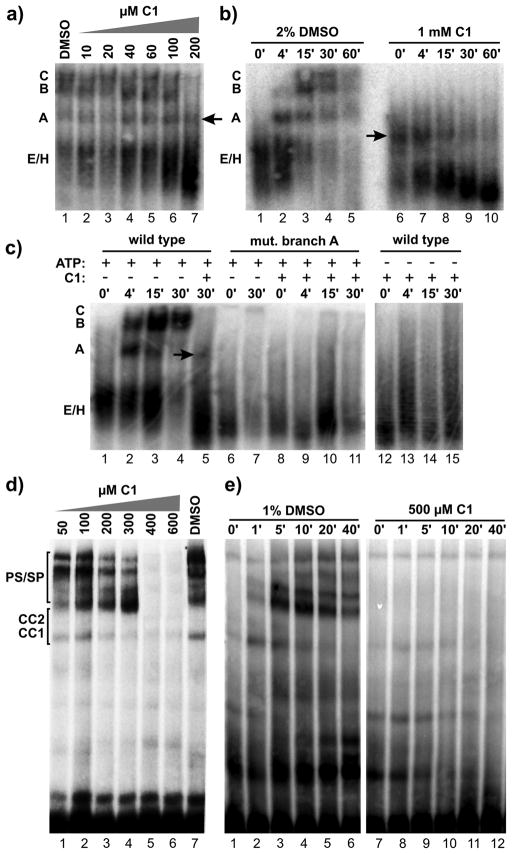
Figure 3.
C1 stalls human and yeast spliceosomes at an A-like complex. Native gel analysis of spliceosome assembly. a) Thirty minute time points of splicing reactions in HeLa nuclear extract supplemented with 2% DMSO or indicated concentration of C1. The identity of complexes is denoted with assembly occurring in the following order: H/E → A → B → C. The arrow indicates the A-like complex. b) Time course analysis of splicing reactions in HeLa nuclear extract in 2% DMSO or 1 mM C1. Time points are indicated in minutes. c) Time course analysis of splicing reactions in HeLa nuclear extract in the presence or absence of ATP and C1 as indicated using a pre-mRNA with wild type or mutant branch point. d) Twenty minute time points of splicing reactions in yeast extract supplemented with 1% DMSO or indicated concentration of C1. The identity of complexes is denoted with assembly occurring in the following order: CC1 → CC2 → PS/SP. e) Time course analysis of splicing reactions in yeast extract in 1% DMSO or 500 μM C1.
In the presence of increasing amounts of C1, spliceosome assembly is lost in parallel with the loss in splicing chemistry (Figures 3a and 2a). At a concentration that completely blocks splicing, a complex that migrates near the position of A complex (“Alike” complex) accumulates. Furthermore, B and C complexes do not appear over time, but instead an apparent conversion of the A-like complex to a stable faster migrating species becomes evident (Figure 3b- lanes 6–10).
Formation of A complex requires recognition of the branch point sequence and ATP hydrolysis2, 21. If the A-like complex is related to normal spliceosome assembly, it should have the same substrate and ATP requirements. Splicing reactions assembled in the presence of C1 with a pre-mRNA mutated at the branch point sequence no longer produce the A-like complex (Figure 3c- lanes 8–11). Like a branchpoint in the substrate, ATP is also required to form the A-like complex (Figure 3c- lanes 12–15). This result shows that compound C1 does not generally disturb all components of the extract, because specific dependencies of spliceosome assembly are maintained. It also suggests that C1 interferes with a component(s) involved in the stability of A complex or the transition to the next assembly stage.
We also looked at spliceosome assembly in yeast extracts treated with C1, and again see a dose dependent loss of spliceosome assembly compared to DMSO alone (Figure 3d). At lower concentrations the effect of C1 is most evident in the appearance of prespliceosomes/spliceosomes (PS/SP) complex bands, which are equivalent to A/B/C complexes in human extracts (Figure 3D- lanes 1–4), while at higher concentrations all PS/SP accumulation is lost (lanes 5–6). At 500 μM concentration C1 also dramatically reduces stable accumulation of commitment complexes CC1 and 2, which are formally equivalent to E/H complex in human extracts (Figure 3e- lanes 7–12).
From the yeast and human data, we conclude that C1 does not directly inhibit 1st splicing chemistry, but instead interferes with early stages of spliceosome assembly in both systems potentially by destabilizing complexes that form. These results also suggest that it targets a core component conserved in both human and yeast spliceosomes.
C3 causes accumulation of a B-like complex
C3 had a different effect on spliceosome assembly in human extracts. As the amount of C3 is increased in splicing reactions, C complex decreases with the same dose dependence as loss of second step chemistry (Figure 4a- lanes 2–7 and 2a). The compound does not inhibit or change the timing of A and B complex formation, even at concentrations that completely block both steps of splicing chemistry (Figure 4a- lanes 13–17). This result shows that C3 does not generally disrupt complex assembly. Instead, considering the different sensitivities of 1st and 2nd step chemistry to C3, there must be a factor(s) involved in late assembly that is selectively targeted by the presence of the compound.
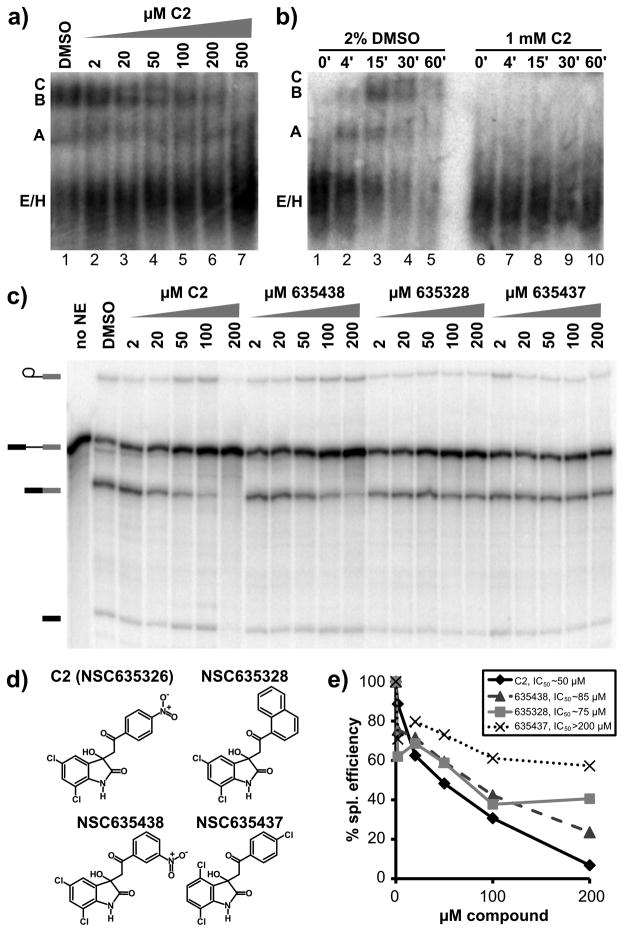
Figure 4.
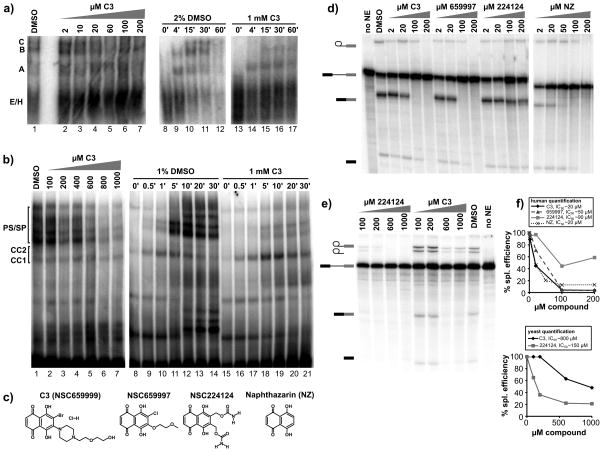
A B-like complex accumulates in the presence of the C3 compound. a) Native gel analysis of spliceosome assembly. Left panel shows thirty minute time points of splicing reaction in HeLa nuclear extract supplemented with 2% DMSO or indicated concentration of C3. Right panel shows time course analysis of splicing reactions in HeLa nuclear extract in 2% DMSO or 1 mM C3. Complexes are labeled as in Figure 3. b) Left panel shows twenty minute time points of splicing reactions in yeast extract supplemented with 1% DMSO or indicated concentration of C3. Right panel shows time course analysis of splicing reactions in yeast extract in 1% DMSO or 1 mM C3. Complexes are labeled as in Figure 3. c) Chemical structure of C3 and related compounds. d) Denaturing gel analysis of in vitro splicing reactions with increasing concentrations of C3 or indicated compounds in HeLa nuclear extract. Bands are schematized as in Figure 2. e) Denaturing gel analysis of in vitro splicing reactions with increasing concentrations of C3 or NSC224124 in yeast extract. f) Quantification of the splicing efficiency relative to compound concentration of the splicing reactions shown in (d) (top panel) and (e) (bottom panel). Estimated IC50 values are indicated.
In yeast extracts there is a similar dose dependent loss of spliceosome assembly (Figure 4b- lanes 2–7) with C3. In comparison to no inhibitor (1% DMSO- lanes 8–14), progression from commitment complexes (CC1/CC2) to prespliceosomes/spliceosomes (PS/SP) is decreased (Figure 4b- lanes15–21), which is formally equivalent to a decrease in A/B/C complex formation in human extracts.
C3 activity is related to redox potential in HeLa extract
To examine the structure activity relationships of the C3 inhibitor, which contains a naphthazarin backbone, we tested the effects of related compounds on in vitro splicing. In human extracts, another naphthazarin derivative (NSC659997) inhibits splicing primarily at second step, although with a slightly higher IC50 of ~50 μM (Figure 4c, 4d and 4f). Interestingly, NSC659997 also shows growth inhibition in a panel of tumor cell lines that is similar to the original C3 compound18. A second naphthazarin derivative (NSC224124) had a more limited effect on splicing (Figure 4c, 4d and 4f), and no effect on same panel of tumor cells. Finally, we tested the naphthazarin backbone alone (NZ), and find that it also inhibits splicing with an IC50 similar to C3 (Figure 4c, 4d and 4f). Together, these results suggest that the naphthazarin backbone alone mediates splicing inhibition, but that substitutions within the ring structure can affect the activity.
These results are in some contrast to what we observe with naphthazarin derivatives in yeast splicing. In this case, the naphthazarin backbone alone was not sufficient to inhibit splicing chemistry (data not shown). In contrast, NCS224124 is more potent in yeast than the C3 compound, but still has a relatively high IC50 of ~150μM (Figure 4e and 4f). This difference in activity points to the importance of substitutions on the naphthazarin ring in yeast splicing as well, but is it is not clear why the sensitivity to the substitutions is different for human splicing.
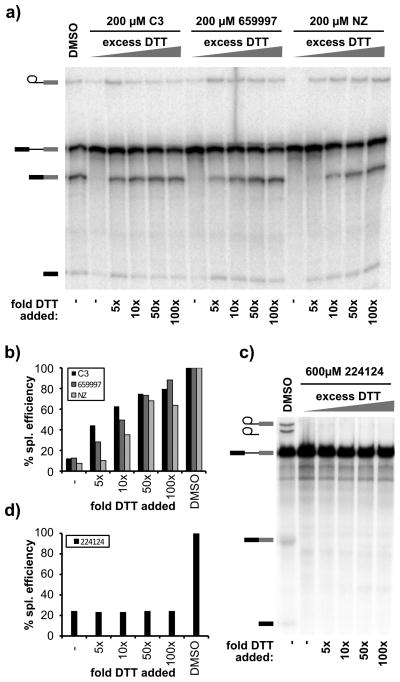
Naphthazarin and many of its derivatives are reactive compounds with well-known redox properties proposed to interfere with proteins by two mechanisms22. First, they can serve as electrophiles to covalently modify proteins, most commonly by thioether linkage with labile cysteine residues, which cannot by reverse by DTT. Alternatively, they also generate reactive oxygen species (ROS) that, among other effects, can oxidize thiol groups of cysteines, a process which can be blocked by addition of DTT. We tested the effect of DTT on HeLa splicing and found that excess DTT recovers about 70% of splicing in reactions inhibited by the naphthazarin compounds (Figure 5a and 5b). DTT alone does not affect splicing efficiency up to 20 mM concentration. Relief of splicing inhibition by DTT suggests that the compounds inhibit splicing through ROS generation that results in modification of a redox-sensitive cysteine(s). Because we still see assembly to B complex in the presence of the naphthazarin compounds at all concentrations, the redox-sensitive cysteine is specific and appears to be required for formation of a catalytically active spliceosome. Surprisingly, in yeast extracts, DTT did not reverse splicing inhibition by the most potent naphthazarin derivative (NCS224124) (Figure 5c and 5d), which suggests that the compound functions by a different mechanism in yeast splicing. Again, we cannot specify why different ring substitutions in the naphthazarin backbone affect the inhibitory properties of the different compounds. With human splicing, they may modulate the redox potential of the compounds, while splicing in yeast appears sensitive to their structure.
Figure 5.
Inhibition by naphthazarins is partially rescued by excess DTT. a) Denaturing gel analysis of in vitro splicing reactions in HeLa nuclear extract inhibited by C3 or related compounds supplemented with increasing concentrations of DTT. Bands are schematized as in Figure 2. b) Quantification of the splicing efficiency relative to compound concentration of the splicing reactions shown in (a). c) Same analysis as in (a) but with yeast splicing. d) Quantification of the splicing efficiency relative to compound concentration of the splicing reactions shown in (c). Estimated IC50 values are indicated.
C2 inhibits spliceosome assembly early in human extracts
The C2 compound only inhibits in the human splicing system and primarily affects very early spliceosome assembly. With increasing concentrations of C2, there is a loss of higher complexes that correlates with the loss of splicing chemistry (Figure 6a, 2a and Supplemental Figure 2b). At lower concentrations, where 2nd step is more sensitive to the compound, the spliceosome fully assembles to some extent indicating selectivity for a late-acting splicing factor (Figure 6a). In the presence of 1 mM C2, where all splicing chemistry is completely blocked, no complexes form beyond E/H (Figure 6b- lanes 6–10), which could indicate that the extract is generally inactivated. However, 1 mM C2 does not affect splicing in yeast extracts at all (Figure 2b and 2c), suggesting instead that C2 also has selectivity for a human splicing factor involved in the earliest assembly stages.
Figure 6.
Presence and orientation of a nitrophenyl ring are important for splicing inhibition by C2. a) Thirty minute time points of splicing reactions in HeLa nuclear extract supplemented with 2% DMSO or indicated concentration of C2. b) Time course analysis of splicing reactions in HeLa nuclear extract in 2% DMSO or 1 mM C2. c) Denaturing gel analysis of in vitro splicing reactions with increasing concentrations of C2 or indicated compounds in HeLa nuclear extract. d) Chemical structure of C2 and related compounds. e) Quantification of the splicing efficiency relative to compound concentration of the splicing reactions shown in (c). Estimated IC50 values are indicated.
We also examined structure activity relationships for C2. Two related compounds with different substitutions in place of the nitrophenyl group had very little effect on splicing chemistry (Figure 6c, 6d and 6e). A third compound, which differs only in the linkage position of the nitrophenyl group, inhibits splicing but with less potency than the original C2 compound (Figure 6c, 6c and 6e). We conclude that presence and orientation of a nitrophenyl ring from the oxoethyl group at the 3′ position of the indole ring structure are important for the mechanism of C2 inhibition of splicing. Possible explanations for the requirement include specific spliceosome component interaction with the nitrophenyl ring, increased stability of the compound in extracts or increased solubility.
DISCUSSION
Using a highly sensitive RT-qPCR splicing assay we identified three structurally diverse molecules (C1, C2 and C3) that inhibit pre-mRNA splicing at specific stages in spliceosome assembly in both HeLa and S. cerevisiae extracts. In HeLa extracts this inhibition has an IC50 in the 20–50 μM range. These three compounds are structurally distinct from previously identified splicing inhibitors23–25 and offer a unique opportunity to dissect important spliceosomal complexes not previously captured in either human or yeast splicing systems.
C1, also known as Tetrocarcin A, has demonstrated antibiotic activity, as well as cytotoxic effects in several cancer cell lines10. It has been shown to promote apoptosis by blocking BCL2 activity17, activating caspase-926 or inhibiting PI3K kinase activity27, depending on cell type, although its molecular mechanisms are not clear. One possibility for this wide range of effects is that Tetrocarcin A inhibits the splicing of one or more key gene products in the apoptosis pathway.
C2 impedes all stages of spliceosome assembly, eventually resulting in loss of all complexes beyond E/H. C2 has no effect on yeast in vitro splicing, therefore its target(s) are likely specific to higher eukaryotes. By comparing the activity of C2 to several structurally related compounds, we find that a nitrophenyl substituent is key to its inhibition of splicing. The lack of previous data demonstrating an effect of C2 on cell growth suggests cells may not readily take up the compound. However, it still may have in vitro utility by allowing concentration-dependent access to the earliest stages of spliceosome assembly.
The influence of C3 (NSC659999) on splicing is more complicated. At lower concentrations this compound primarily affects second step chemistry, which is mirrored by a clear loss of C complex accumulation. In yeast, C3 shows similar effects on splicing chemistry and complex assembly, although only at significantly higher concentration. In the human system, the effect of the C3 and related naphthazarin inhibitors can be reversed by the addition of excess DTT, which suggests that ROS generation by these compounds plays a role in their inhibition of splicing. It also indicates that a specific redox-sensitive cysteine in a splicing component functions in the transition from first to second step chemistry. Recently, the Dreyfuss lab identified a CDC25 phosphatase inhibitor in their screen for compounds that block in vitro splicing24. This compound is a naphthoquinone (NSC95397) with redox properties similar to the napthazarins. It also affects second step chemistry and C complex formation like the naphthazarins. It is notable that the phosphatase PP2A is also required for second step chemistry in human extracts and that the PP2A inhibitor Okadaic acid is also a splicing inhibitor28, 29. PP2A is sensitive to oxidation by H2O2, which can also be rescued by excess DTT30. We propose that at least some of the splicing inhibition observed with naphthoquinones, which includes the naphthazarins, is conferred by indirect inactivation of PP2A by oxidation. As of yet, there is no known role in splicing for the yeast PP2A ortholog. This may explain why most of the naphthazarin derivatives had no effect on yeast splicing and why the modest effect of NSC2241124 in yeast extracts cannot be rescued by DTT.
There are several directions of study that may be pursued with these new splicing inhibitors. Our results underscore their potential as tools in vitro to study splicing mechanisms. For those that can be synthesized and derivatized, their structure activity relationships can be further explored, and in some cases their targets potentially identified by addition of affinity tags. These studies may also open the door to their use as probes in studying the role of splicing in cells. Many splicing inhibitors have shown bioactivity in the growth of different tumor cell lines, including Tetrocarcin A and naphthazarins17, 22, 31, 32. An important next step will be to determine whether this activity is due to inhibition of spliceosomes and, if so, which splicing pathways are particularly affected.
Finally, with the extreme complexity of the spliceosome, the need is still great for a larger arsenal of compounds that modulate enzymatic functions and rearranging interactions involved in splicing. Fortunately, a great deal of chemical space remains unexplored. So far only relatively small libraries of bioactive molecules (<8000 compounds) and one larger library of synthetic small molecules (30,000 compounds) have been screened23–25. As we use the assay presented in this paper to screen more compound libraries, particularly those containing structurally diverse natural products, we will certainly increase the number of small molecule tools that will be useful for studying splicing mechanisms and cellular functions.
Supplementary Material
Acknowledgments
This work was funded by the University of California Cancer Research Coordinating Committee and National Institutes of Health grant R01CA136762 to M.S.J. We thank the UCSC Chemical Screening Center, supported by NIH 1S10RR022455 and grants from the California Institute for Quantitative Biosciences (QB3) and the US Department of State. We also thank J. Woo at the UCSF Genomics Core Facility for assistance with RT-qPCR analysis, M. Yoshida at the RIKEN Advanced Science Institute for providing SSA, B. Prichard for assistance in HeLa cultures and extract preparation and N. Weber for developing scripts to help analyze RT-qPCR results.
References
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Konarska MM, Sharp PA. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46(6):845–55. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 3.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, Vartanov A, Fernandes SM, Saksena G, Cibulskis K, Tesar B, Gabriel S, Hacohen N, Meyerson M, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, Mudie LJ, Stephens PJ, McLaren S, Massie CE, Tarpey PS, Varela I, Nik-Zainal S, Davies HR, Shlien A, Jones D, Raine K, Hinton J, Butler AP, Teague JW, Baxter EJ, Score J, Galli A, Della Porta MG, Travaglino E, Groves M, Tauro S, Munshi NC, Anderson KC, El-Naggar A, Fischer A, Mustonen V, Warren AJ, Cross NC, Green AR, Futreal PA, Stratton MR, Campbell PJ. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, Ramsay AJ, Bea S, Pinyol M, Martinez-Trillos A, Lopez-Guerra M, Colomer D, Navarro A, Baumann T, Aymerich M, Rozman M, Delgado J, Gine E, Hernandez JM, Gonzalez-Diaz M, Puente DA, Velasco G, Freije JM, Tubio JM, Royo R, Gelpi JL, Orozco M, Pisano DG, Zamora J, Vazquez M, Valencia A, Himmelbauer H, Bayes M, Heath S, Gut M, Gut I, Estivill X, Lopez-Guillermo A, Puente XS, Campo E, Lopez-Otin C. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 7.Webb TR, Joyner AS, Potter PM. The development and application of small molecule modulators of SF3b as therapeutic agents for cancer. Drug Discov Today. 2012 doi: 10.1016/j.drudis.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padgett RA, Hardy SF, Sharp PA. Splicing of adenovirus RNA in a cell-free transcription system. Proc Natl Acad Sci U S A. 1983;80(17):5230–4. doi: 10.1073/pnas.80.17.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruskin B, Krainer AR, Maniatis T, Green MR. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38(1):317–31. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 10.Tomita F, Tamaoki T, Shirahata K, Kasai M, Morimoto M, Ohkubo S, Mineura K, Ishii S. Novel antitumor antibiotics, tetrocarcins. J Antibiot (Tokyo) 1980;33(6):668–70. doi: 10.7164/antibiotics.33.668. [DOI] [PubMed] [Google Scholar]
- 11.Dignam JD, Lebovitz RM, Roeder RD. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18(9):5000–9. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perriman R, Ares M., Jr ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 2000;14(1):97–107. [PMC free article] [PubMed] [Google Scholar]
- 14.Padgett RA, Konarska MM, Grabowski PJ, Hardy SF, Sharp PA. Lariat RNA’s as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- 15.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9):576–83. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 16.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 17.Nakashima T, Miura M, Hara M. Tetrocarcin A inhibits mitochondrial functions of Bcl-2 and suppresses its anti-apoptotic activity. Cancer Res. 2000;60(5):1229–35. [PubMed] [Google Scholar]
- 18.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 19.Pikielny CW, Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell. 1986;45(6):869–77. doi: 10.1016/0092-8674(86)90561-1. [DOI] [PubMed] [Google Scholar]
- 20.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–8. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed R, Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988;2(10):1268–76. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- 22.You YJ, Zheng XG, Yong K, Ahn BZ. Naphthazarin derivatives: synthesis, cytotoxic mechanism and evaluation of antitumor activity. Arch Pharm Res. 1998;21(5):595–8. doi: 10.1007/BF02975381. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien K, Matlin AJ, Lowell AM, Moore MJ. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J Biol Chem. 2008;283(48):33147–54. doi: 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berg MG, Wan L, Younis I, Diem MD, Soo M, Wang C, Dreyfuss G. A quantitative high-throughput in vitro splicing assay identifies inhibitors of spliceosome catalysis. Mol Cell Biol. 2012;32(7):1271–83. doi: 10.1128/MCB.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samatov TR, Wolf A, Odenwalder P, Bessonov S, Deraeve C, Bon RS, Waldmann H, Luhrmann R. Psoromic acid derivatives: a new family of small-molecule pre-mRNA splicing inhibitors discovered by a stage-specific high-throughput in vitro splicing assay. Chembiochem. 2012;13(5):640–4. doi: 10.1002/cbic.201100790. [DOI] [PubMed] [Google Scholar]
- 26.Anether G, Tinhofer I, Senfter M, Greil R. Tetrocarcin-A--induced ER stress mediates apoptosis in B-CLL cells via a Bcl-2--independent pathway. Blood. 2003;101(11):4561–8. doi: 10.1182/blood-2002-08-2501. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima H, Sakaguchi K, Fujiwara I, Mizuta M, Tsuruga M, Magae J, Mizuta N. Apoptosis and inactivation of the PI3-kinase pathway by tetrocarcin A in breast cancers. Biochem Biophys Res Commun. 2007;356(1):260–5. doi: 10.1016/j.bbrc.2007.02.136. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Reddy B, Manley JL. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol Cell. 2006;23(6):819–29. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 29.Mermoud JE, Cohen P, Lamond AI. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20(20):5263–9. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foley TD, Armstrong JJ, Kupchak BR. Identification and H2O2 sensitivity of the major constitutive MAPK phosphatase from rat brain. Biochem Biophys Res Commun. 2004;315(3):568–74. doi: 10.1016/j.bbrc.2004.01.096. [DOI] [PubMed] [Google Scholar]
- 31.Nakajima H, Hori Y, Terano H, Okuhara M, Manda T, Matsumoto S, Shimomura K. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J Antibiot (Tokyo) 1996;49(12):1204–11. doi: 10.7164/antibiotics.49.1204. [DOI] [PubMed] [Google Scholar]
- 32.Mizui Y, Sakai T, Iwata M, Uenaka T, Okamoto K, Shimizu H, Yamori T, Yoshimatsu K, Asada M. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J Antibiot (Tokyo) 2004;57(3):188–96. doi: 10.7164/antibiotics.57.188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.