Abstract
Normal biological tissues harbour different populations of cells with intricate spacial distribution patterns resulting in heterogeneity of their overall cellular composition. Laser microdissection involving direct viewing and expertise by a pathologist, enables access to defined cell populations or specific region on any type of tissue sample, thus selecting near-pure populations of targeted cells. It opens the way for molecular methods directed towards well-defined populations, and provides also a powerful tool in studies focused on a limited number of cells. Laser microdissection has wide applications in oncology (diagnosis and research), cellular and molecular biology, biochemistry and forensics for tissue selection, but other areas have been gradually opened up to these new methodological approaches, such as cell cultures and cytogenetics. In clinical oncology trials, molecular profiling of microdissected samples can yield global “omics” information which, together, with the morphological analysis of cells, can provide the basis for diagnosis, prognosis and patient-tailored treatments. This remarkable technology has brought new insights in the understanding of DNA, RNA, and the biological functions and regulation of proteins to identify molecular disease signatures. We review herein the different applications of laser microdissection in a variety of fields, and we particularly focus attention on the pre-analytical steps that are crucial to successfully perform molecular-level investigations.
Keywords: Laser microdissection, histopathology, quality control, snap-freezing, DNA, RNA, proteomics, in situ cellular and molecular analyses
Introduction
Tumoral tissues often contain a wide array of tumor cells disseminated in an abundant connective tissue called the stroma. Stromal tissue is composed of fibrous tissue, vessels and inflammatory cells and possibly necrotic or haemorrhagic areas. Thus, when the aim is to focus an analysis on a homogeneous cell population, containing only tumor cells, or conversely only stromal cells, this heterogeneity needs to be dealt with. A differential approach to molecular mechanisms in heterogeneous tissues may thus require the procurement of pure populations of cells. Although flow cytometry has long been used to enrich a particular cell type, it cannot easily purify cells from solid tissue samples, such as biopsies. So, the major disadvantage of flow cytometry for solid tissue analysis is that it requires a suspension of individual cells and specific fluorescent marker for identification. This also means that the tissue architecture and any information about the spatial relationship between different cells are lost. For the in situ analysis of tissue sections, laser microdissection provides the same technological scope as flow cytometry for cell suspensions and, in contrast to flow cytometry, can be applied to all sample types, including plant tissues.
Laser microdissection overcomes the problem of cellular heterogeneity that characterizes tissues. It aims to recover a target cluster of cells, or a single cell precisely selected under microscope guidance, from a complex tissue section (frozen or fixed by prior paraffin-embedding) for subsequent molecular analysis [1,2]. Developed in 1996, this technique for isolating specific cells from a sample responds to the need for miniaturization of analytical techniques applicable to very small cell numbers.
Cells isolated by microdissection have been widely used as a genomic DNA isolation tool for library generation. Although a large number of studies have described analyses of DNA and RNA extracted from laser-captured tissue fragments, protein or lipid-based studies are still scarce. In addition, biopsies performed nowadays are often radiologically guided needle biopsies yielding smaller amounts of tissue than former surgical procedures [3]. This powerful tool has reduced the amount of material collected while improving its quality, enabling the performance of more sophisticated and reproducible analyses.
The identification of genomic abnormalities, genetic or epigenetic tumors can now establish prognosis for individual patients and predict susceptibility and resistance to many anticancer drugs, including new targeted therapies. In the near future, increasing numbers of research laboratories will develop high-level platforms for such studies on patients entering clinical trials. Ultimately, validation of this technique will enable individualized therapeutic approaches and thus better management of patients. However, it should be noted that whatever the laser-assisted microdissector system used, subsequent molecular analyses can be significantly hampered by flaws in the initial preparation of the tissues. Any tissue preparation method must be a compromise between obtaining good morphological contrast and the ability to preserve the biological material for further molecular analyses. Processing-induced artifacts can occur at each step of tissue treatment. In the area of laser microdissection techniques, we intend here to delineate a “yellow brick road” that should be followed for the successful characterization of biological samples.
Principle of tissue microdissection
Originally, tissue microdissection was performed manually under control of a light microscope, with or without a micromanipulator. Specific histological zones of interest in the tissue section were selected and scraped from the section using a modified Pasteur pipette with an ultrafine pointed tip [4]. The development of this approach using a laser beam has greatly increased the precision and effectiveness of biological material collection (Figure 1).
Figure 1.
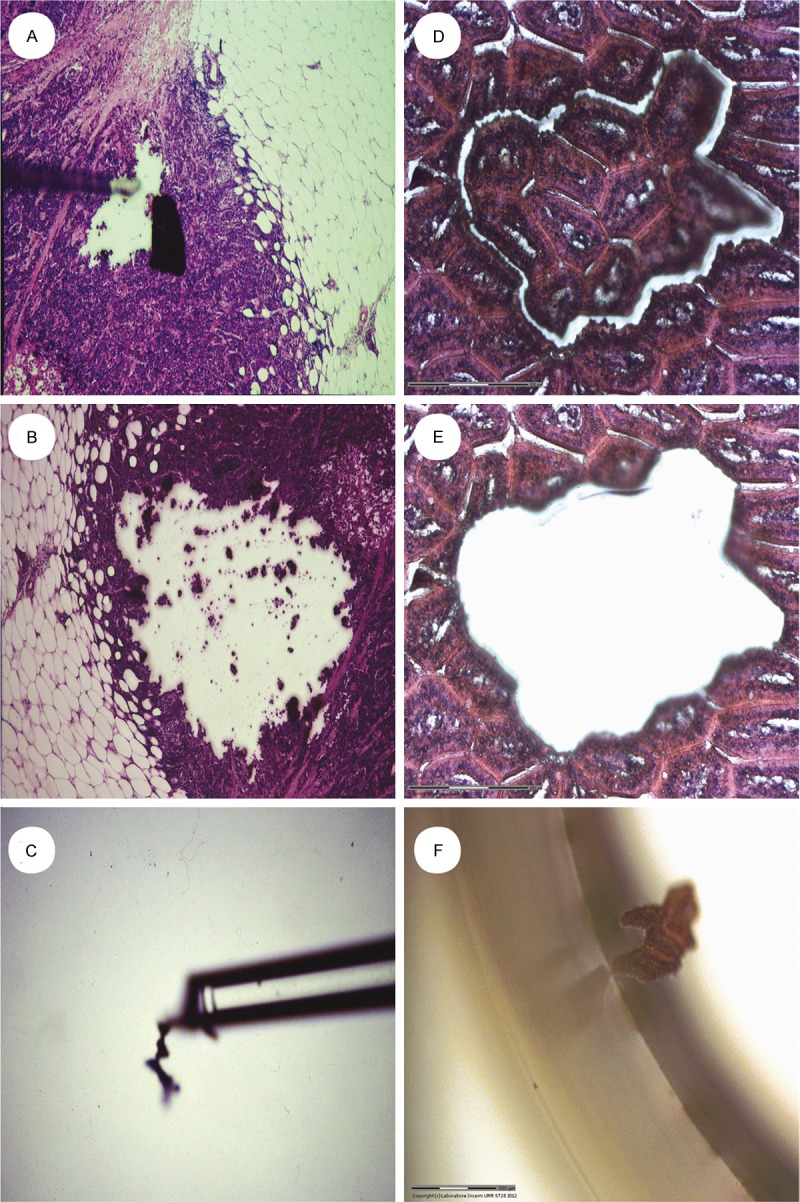
Comparing two techniques of microdissection, one on the left panel where the tissue is manually scraped with a sharp point (A-C) and another one on the right panel with the assistance of a laser beam, which enables to cut and to recover an area of tissue with much greater precision (D-F).
Sample selection is still performed under the microscope by a trained pathologist. Automation of a large part of the process has greatly facilitated its development. Microdissection of tissue from archival histological material can be used when the target analyte is DNA [5], but the use of frozen tissue can also be recommended when the objective is to study RNA, proteins or even lipids. When structure identification from morphology alone seems difficult, it is also possible to use immuno-histochemical staining or immuno-fluorescence labelling, which provides valuable information concerning the localization of antigenic epitopes for targeted detection of specific cells [6]. Importantly, microdissection should always involve rapid techniques with short steps, to prevent degradation of the molecular components. The task is thus to obtain a sufficient amount of tissue material and to preserve tissue quality throughout the entire process.
Different microdissection systems
Membrane-based microdissection, first described in 1992 after selective ultraviolet radiation fractionation [7], has emerged as a valuable tool for the selection of homogeneous cell populations from solid tumors. Over the past 15 years, laser-assisted microdissection devices have gradually become more user-friendly. A laser microdissector is composed of a microscope (upright or inverted) coupled to a laser beam (wavelength in the IR-infrared and/or UV-ultraviolet range) which operates under the control of a computer program. The two main features of the UV laser are that it combines high photon density for cold laser ablation and that it works at short wavelengths. The heat generated during microdissection is thus limited and the impact on cellular biomolecules minimal [8] At the appropriate magnification, one or more cells can be selected on the computer screen using a graphical wizard (freehand drawing or predefined geometric shapes), and according to the manufacturer they can then be ablated by laser beam and collected in a conventional tube (Eppendorf tube®) or on a specific support (capsule or adhesive-cap tube). The recovered microdissected tissue samples can then be assessed in the tube at lower magnification.
Four laser-assisted microdissection systems are currently available, based on different operating principles:
The LCM system (for Laser Capture Microdissection, PIXCEL II®, and more recently the VERITAS™ system, both from Arcturus Engineering and now under the label of Life Technologies), was originally conceived and developed in 1996 as a research tool at the National Institutes of Health [9] (Figure 2). From an inverted microscope, the collection of tissue fragments is performed via adherence to a thermoplastic film made of an ethylene vinyl acetate-EVA membrane, following tissue exposure to low-energy infrared (IR) laser pulses (wave length 980 nm). Capsules coated with this film (CapSure® LCM Caps, Arcturus Engineering, Inc., USA) are placed in contact with tissue sections, maintaining the active capture area twelve microns off the surface of the sample, which is deposited on a regular histological glass slide. Pulsing the laser through this capsule causes the thermoplastic film to melt onto the cells of interest by a range of impacts within a zone of pre-established diameter (7.5, 15 or 30 micrometers). Lifting off the cap removes the target cell(s) attached to the cap. Using an appropriate buffer, tissue lysis can be performed for the analysis of extracted products. Successful LCM requires tissue sections to be absolutely dry. If any moisture is present in the tissue, the IR laser will react with the water molecules, causing the tissue to heat, thus degrading RNA. Microdissection using VERITAS™ provides a new standard, for the first time combining IR and UV lasers in a single instrument, thus enabling the UV-destruction of unwanted cells.
Figure 2.
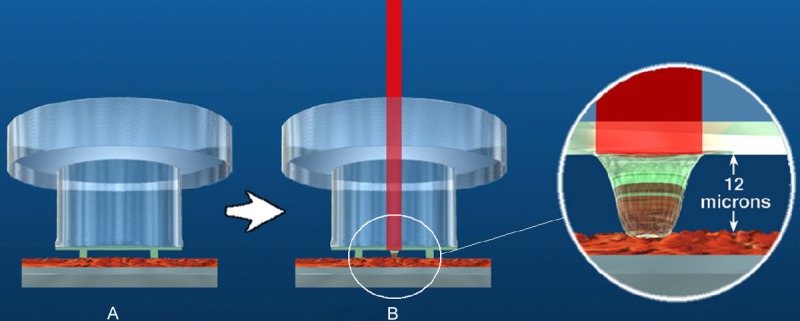
A: Laying of the capsule in contact with the histological section. B: IR laser firing and heat-sensitive adhesion of the membrane under the laser pulse (http://www.excilone.com/).
The LMPC system (for Laser Microdissection and Pressure Catapulting) (3D-PALM Microlaser Technologies, Carl Zeiss MicroImaging Technologies, Munich, Germany) associates an inverted microscope with an ultraviolet laser (UV-A pulsed, wave length 335 nm) (Figure 3). The use of specially designed glass slides (MembraneSlides®) enables efficient collection of microdissected fragments. These slides are coated with a thin polyethylene napthalate (PEN) membrane over their entire surface except for a large central rectangle where a space of 1 μm remains between the glass and the membrane on which the tissue is laid. The membranes are mounted either on regular glass slides (for PEN membrane), or on steel frames PET (Polyethylene Terephthalate) or POL (Polyester) membrane. Thus, depending on tissue type and thickness, and the methods of analysis (i.e. with or without immunofluorescent detection), one slide type may work better than another for a defined application. Therefore it is advisable to test different combinations of membranes and slides in order to optimize results. To overcome the hydrophobic nature of the membrane it is advisable to irradiate with UV light at 254 nm for 30 minutes. When the membrane becomes more hydrophilic, therefore the cryo- and paraffin sections adhere better. Positive side effects are sterilization and destruction of potentially contaminating nucleic acids. According to parameters governing energy and focus, the laser beam will first enable the isolation of the targeted tissue region by photo-ablation. The separated specimen is extracted from the tissue section using a software-controlled laser pulse, the energy being increased with a de-focused laser pulse to generate a photonic force. Thus, the material is catapulted off the slide into the cap of a microcentrifuge tube containing 15 to 25 μl of a buffer, positioned a few millimeters above the tissue section [10]. It remains possible, although more difficult and less efficient, to cut and/or directly catapult from regular cleaned and degreased glass slides.
Figure 3.
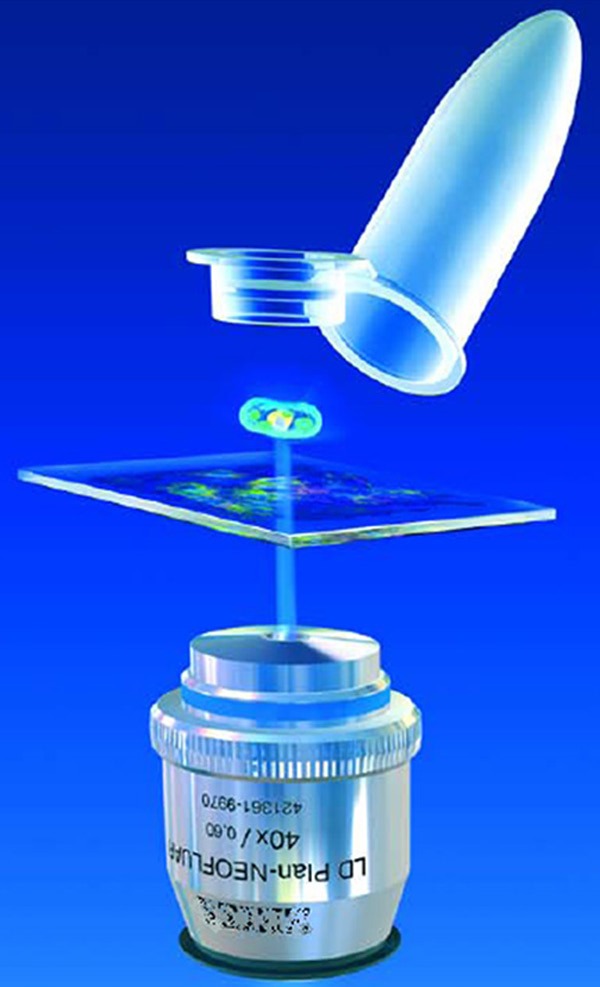
Catapulting of a selected tissue region into the cap of a conventional tube containing a few microliters of extraction buffer (Copyright by http://www.zeiss.com).
The LMD system (Leica Microsystems) is closed to the LMPC system (Figure 4). The main difference between the two systems is the way to recover the biological material. Dissection is performed using an upright microscope and the UV laser-cut area drops by gravity into the cap of a tube placed just below. In this system, unlike the others, the tissue section remains stationary on the microscope and the laser beam moves over it when cutting [11].
Figure 4.
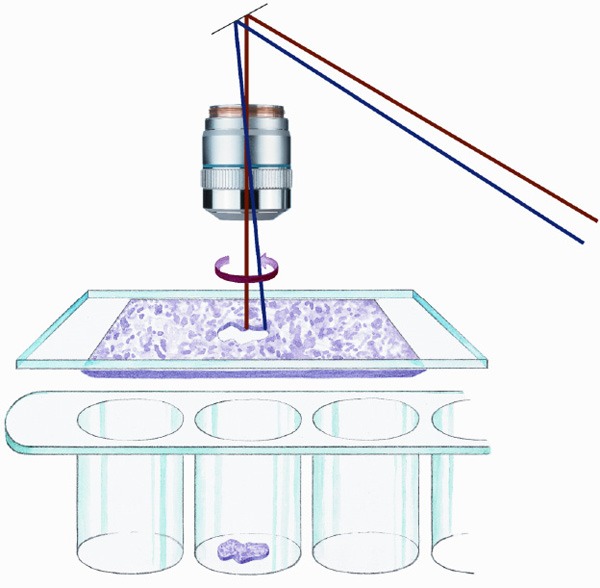
Cutting a tissue region of interest, and recovery by gravity into the cap of a conventional tube positioned below the tissue section (Copyright by http://www.leica-microsystems.com).
The MMI Caplift technology, called MMi Cellcut® operates with a UV laser (wave length 335 nm) on an inverted microscope (Figure 5). The principle of this system is the use of specific recovery tubes (Insulation Cap®) with an adhesive part inside the cap. The histological section is deposited on the thin membrane in the center of a mounted metal frame slide. Then the MMI MembraneSlide is inverted and placed on a glass slide for protection against contamination. The sample is then sandwiched between the membrane and the glass. The adhesive part of the recovery tubes will contact one face of this membrane on the underside of the histological section. The laser beam cuts the selected area or cells, which are mechanically detached from the tube: the cells remain attached to the cap and can be analyzed after solubilization in a suitable buffer [12]. During the microdissection procedure, the fragments of the tissue are positioned on the adhesive surface of the cap. Difficulty in removing the fragments from the cap can occur, and it is recommended to add the extraction buffer to the tube, then to close the lid and leave the tube upside down in order to let it soak the desired amount of time, followed by spinning.
Figure 5.
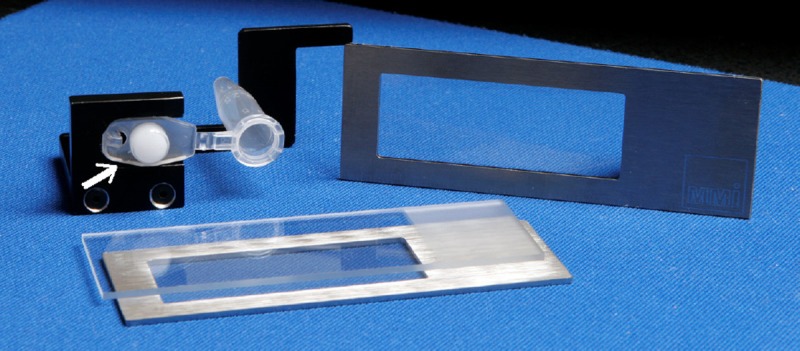
Adhesive side of the cap of a tube (white arrow) for recovering a tissue region of interest that has been preselected and then cut by the laser beam before separation of the histological slide and the cap (Copyright by http://www.molecular-machines.com).
Main applications
For patient diagnosis and experimental studies, biological tissue can be either analyzed under a microscope after immuno-histostaining or crushed for further molecular analysis. Laser microdissection provides a valuable link between these two approaches. It gives new insights into cellular mechanisms, genetic disorders, tumor biomarker identification patient-tailored therapy [13], and even extensive knowledge of the lipid composition which contributes to the pathogenesis of Alzheimer’s disease [14] (Figure 6).
Figure 6.
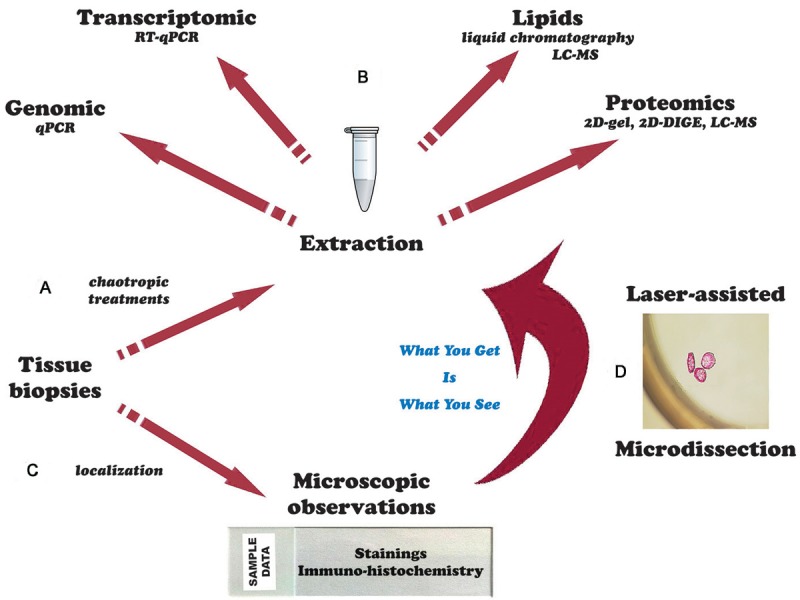
(A) The entire tissue sample is crushed to extract the derivatives for (B) molecular analyses. (C) The tissue sample is observed under the microscope and can be analyzed by immunohistochemistry performed on histological section. (D) The technique of laser microdissection enables simultaneous observation and sampling of the cells of interest on histological sections that will then be studied by molecular analysis techniques.
Tissue microdissection is used to obtain phenotypically identical cells, while other cells can be ignored or destroyed, therefore increasing the sensitivity and specificity of subsequent molecular analyses. Combination of microdissection and microarray analysis can for example enable comparison of gene expression in different breast cancer areas, such as the intra-ductal component or the infiltrating component, with greater accuracy than global gene expression analyses [15], as it is also demonstrated in different cell populations from the same sample, such as tumor cells and normal cells in peritumoral tissue in colon cancers [16].
Neoplasia is the main field of application of this technique for selecting tissue, but other fields have gradually opened up to these new methodological approaches, for instance for microdissecting living cells from a cell culture, for which special equipment has been developed, with the possibility of re-culturing the isolated cells [8]. De Spiegelaere W et al. used this technique for the isolation of small tissue fractions from developing embryos [17]. Sethi et al. have described its usefulness in unique case of renal amyloidosis in native and transplant kidney biopsies [18].
Genomics
The tumor genome is the main target of studies using tissue microdissection techniques. From cell fragments, one of the most common applications has been the search for loss of heterozygosity (LOH) in malignant tumors. This type of study is based on individual DNA polymorphisms. Thus, tissue microdissection has been widely used in lung [19], colon [20] or prostate cancer [21]. In our laboratory, we have shown that tissue microdissection greatly increased the sensitivity of LOH detection in inflammatory breast cancer [22]. However, there is a possible pitfall in these analyses if too few cells are used for allelic amplification: allelic drop-out can occur, due to sectioning of tissue and artificially-induced allelic imbalance, especially if tissue sections are microdissected [23].
It has also been possible to search for changes in microsatellite sequences [24], to highlight point mutations [25] and to demonstrate the clonal nature of a tumor population, or possible rearrangements of genes [26].
Microarray technology
Microarray technology evolved from Southern Blotting where fragmented DNA is attached to a substrate and then probed with known DNA sequences to simultaneous measurement of the expression levels of large numbers of genes. DNA microarrays, such as the CGH-array technique (Comparative Genomic Hybridization), can be used to detect small genomic rearrangements through the use of DNA chips, and it enables highly resolutive analysis of a large number of genomic DNA fragments [27]. This method is rapid and sensitive and enables comparison of the genomes of two populations of cells (one healthy and one tumoral for example), or listing of expressed genes in a given tissue. In association with laser microdissection, it enables the detection of SNPs (Single Nucleotide Polymorphism), a type of DNA polymorphism in which two chromosomes differ by a single base pair [28].
Tissue microarray (TMA) consists of paraffin blocks in which up to 1000 separate tissue cores are assembled in array fashion to allow multiplex histological analysis on a single standard glass slide [29]. An important advantage of this technique is that all the samples are evaluated under the same technical conditions with multiple stages of tumor progression analyzed in one experiment. However, the major concern is the lack of representation of the original tumor from which the sample is derived. We can imagine difficulties arising during microarray analysis of tissue when there are varying proportions of tumor cells versus normal and stromal cells. In addition, the large number of genes that will be identified as being differentially expressed will require the development of databases that will enable categorization of results. TMA is performed using different paraffin blocks [30], so that considerable time is required to prepare the block containing the right targeted areas, and there is a risk of increased tension within the paraffin wax causing considerable damage to the donor and receiving block [31].
Cytogenetics
Cytogenetic studies, enabling analysis of the complex chromosome rearrangements that have been frequently detected in many malignancies and congenital diseases are also greatly facilitated by microdissection [32]. The characterization of chromosomal fragments or bands combines the microdissection technique with micro-FISH (Fluorescent In Situ Hybridization) technology [33]. This technique is mainly used on sex-specific X and Y chromosome identification, and requires cell fixation on the slide. Some authors have developed a S-FISH and reported the use of cells in suspension in forensic research generating full DNA profiles from as little as ten buccal cells [34].
A recently developed technology also needs to be mentioned. The AmpliGrid (Advalitix AG, Germany), a chip with hydrophilic and hydrophobic spots on which the reaction takes place has been developed as an amplification platform for ultra-low-volume applications in the 1 μl range from material isolated by different laser microdissection devices [35].
Transcriptomic studies
While genomic approaches can be performed at room temperature because of the extreme resistance of the DNA molecule, transcriptome studies should be performed on frozen microdissected samples [36]. To obtain the necessary RNA amount for whole genome cDNA microarrays, the isolated total RNAs can be amplified by T7-based RNA-polymerase in vitro transcription [37,38].
The quality and integrity of RNA extracts are assessed by measuring the A260/A280 ratio and by studying the electrophoretic pattern on agarose gel. This step constitutes the main drawback of this technique because of the amount of RNA required. A computer program has been developed by Agilent Technologies (http://www.agilent.com/chem/RIN) to assign an index of quality to RNA extracts from very small amounts of tissue according to the 18S and 28S ribosomal RNA electrophoretic migration profiles. This index, called RNA Integrity Number (RIN), ranges from 1 (for completely degraded RNA) to 10 (RNA of excellent quality). Usually, only indexes over 8 with a A260/A280 ratio above 1.8 are considered sufficient for a study of RNA using the cDNA microarray technique [39,40]. Quantification of total RNA extracts is now increasingly performed using spectrophotometric assay assisted by the Nanodrop® ND-1000 Spectrophotomer (Nano-Drop Technologies, Labtech, France). A major advantage of this system is the very low sample consumption (1 to 2 μl), which is especially important for precious specimens such as human biopsies or laser microdissected samples [41].
The method of RNA extraction can also be crucial. Different RNA isolation kits have been tested with significant differences in RNA quantity and quality. The recovery rate of RNA samples of known concentration should always be tested before using a new method or kit in the RNA extraction process. The reference procedure is acid-phenol-chloroform extraction (e.g. TRIzol), followed by alcohol precipitation as initially described by Chomczynski and Sacchi [42].
RNA-Seq
Sequencing-based approaches (RNA-Seq) are ultra-high-throughput technologies emerging as attractive alternatives for measuring mRNA expression. They can overcome some limitations of microarrays (high background levels, lack of sensitivity, reliance upon existing knowledge about the genome sequence and cost of the experiment) [43]. Although as yet not completely finalized, combining laser microdissection, linear amplification starting from 1 ng of total RNA, with RNA-Seq provides a powerful tool for transcriptome analysis which can be used at the single cell level, where the expression of 75% more genes than in microarray techniques has been reported [44,45].
Using miRNA
To partially overcome the problem of RNA degradation, it has been recommended that microRNA (miRNA) expression should be monitored. Several years ago, mi-RNAs were invoked in tissue-specific gene regulation (see the microRNA database at http://www.mirbase.org/, and the miRWalk database at http://www.ma.uni-heidelberg.de/apps/zmf/mirwalk/), and their signatures could provide a good tool for studying the transcriptome in cancer [46-48]. MiRNAs are small, highly conserved regulatory molecules (19-24 nucleotides) of non-coding single-stranded RNA, which down-regulate gene expression by binding to messenger RNAs and preventing them from being translated into proteins. In oesophageal squamous cell carcinoma, Zhu et al. analyzed tumor-associated miRNA levels in normal squamous epithelial cells and related tumors, and identified several dysregulated microRNAs [49]. Due to their small size, they are more resistant to fixatives, and will therefore be less degraded than mRNA after pre-analytical treatments. For instance, Nonn et al. compared miRNA expression from formalin-fixed and paraffin-embedded (FFPE) and frozen prostate biopsies from the same patient [50]. In agreement with other studies [51,52], they showed close PCR amplification results from either material, suggesting the possibility of studying miRNA profiles on FFPE tissue samples archived in pathology departments [53]. Schuster et al. have also demonstrated that miRNA expression could be reliably quantified by real-time PCR on archived FFPE tissue samples, even after different immunohistochemical staining procedures with high temperature antigen retrieval, while total RNA was degraded [54].
Proteomic
Proteomic analysis in tissue samples imposes restrictions on experimental protocols used for tissue processing. This is partly due to the diversity of the physico-chemical properties of proteins, and also to the lack of suitable method for amplifying proteins, unlike nucleic acids [55]. Depending on its amino acid sequence and associated post-translational modifications, a protein has a tri-dimensional structure and specific reactive sites. It has a given mass and density, a certain degree of hydrophilicity or hydrophobicity, and exhibits specific behaviors at different pH. Proteins may also be localized in a given region or organelle of the cell, thus requiring more sophisticated extraction protocol. In the initial phase of a study it is very important to define and characterize the process by which the proteins from a given tissue are to be extracted. Hence, the lysis of tissues or cells by sonication or osmotic shock, the optimal solubilization of all proteins using proteomic analysis compatible detergents and the breaking of covalent bonds, all need to be considered. The physicochemical properties of the target proteins have to be known to ensure effective protection against the activity of proteases. It is usually recommended to work at low temperature and in the presence of proteases inhibitors. At higher SDS concentrations (i.e. 10%) protease inhibitors are not necessary in the lysis buffer, because SDS itself acts as a strong protein denaturant [56]. However, SDS is not compatible with downstream analytical techniques such as mass spectrometry, unless preceded by gel separation.
Gel electrophoresis
Polyacrylamide gel electrophoresis (PAGE) can separate proteins according to their molecular weight and can be followed by Western Blot analysis for highly specific protein characterization [57,58]. However, this approach is limited by the availability of appropriate antibodies and by the amount of proteins necessary to obtain a signal. A sensitive Western Blot protocol has recently been developed for the detection of phosphorylated proteins in small amounts of tissue [59]. Alternatively the proteome can be studied by using two-dimensional gel electrophoresis (2D-PAGE), identifying proteins from both isoelectric point and molecular weight, followed by staining with silver nitrate [60,61]. This rather laborious strategy has shown its limits for the analysis of proteins of high molecular weight or with an extreme isoelectric point (pI > 11), and for very hydrophobic proteins. In these cases, there is poor reproducibility and poor detection of low-abundance proteins without prior pre-treatment [62]. The linear dynamic range of silver staining techniques is also reported to be lower than that of other protein detection methods, making the silver staining technique essentially non-quantitative [63]. The use of 2D-PAGE with digestion, extraction, and mass spectrometric analysis of individual protein spots is a time-consuming process with numerous technical hurdles [64]. On the whole, as demonstrated by De Souza et al. who spent 11 days, totalling 70 h of cutting time for two cell populations (blood vessels and myocytes), the association of laser microdissection and 2D-PAGE analysis is mainly limited by the analytical throughput and the low protein content of microdissected samples [65].
In terms of visualization of proteins separated by 2D electrophoresis, the use of fluorescent probes has significantly improved protein quantification with a greater range of linearity. Reactive cyanine dyes (propyl-C3, green, and methyl-C5, red), that exhibit fluorescence at different wavelengths, are more sensitive than Coomassie or silver staining and have been used for high-sensitivity protein visualization [66]. With these stains, comparative studies can be performed using a technique known as 2D-DIGE (2D-DIfferential Gel Electrophoresis) which is based on differential labeling of two samples with two different fluorophores, and their subsequent separation on the same 2D gel. This process yields two superimposed images, one at each wavelength, which can be processed to assess the differences in protein content between the two samples. Such staining is also compatible with protein identification by mass spectrometry, with similar performances to lysine reactive dyes [67].
Mass spectrometry
Many of the limitations of proteome analysis using 2D-PAGE have been overcome by the development of chromatographic separation of proteolytic digests combined with tandem mass spectrometry (LC-MS/MS) [68]. Thus, it has become possible to detect hundreds to thousands of proteins directly from a complex protein mixture with a sensitivity approaching atomolar levels. The power of shotgun LC-MS/MS-based proteomics technology has been documented by successful proteomic analysis of subcellular structures in mammalian cells, such as the nucleolus [69] or the mitochondria [70].
LC-MS/MS is typically used after trypsin digestion of the proteins of interest. Trypsin cleaves specific positions in the protein sequence (i.e. at the C-terminal side of the basic amino acids lysine and arginine) and yields a mixture of peptides (small protein fragments) for each protein in the sample. To manage the complexity of such digests, peptide mixtures are separated by high performance nanoscale liquid chromatography before being ionized using the application of a high voltage (few kV) to a needle coupled to the LC column in a process known as electrospray ionization [71]. The mass of each ionized peptide is determined by its interaction with different electric or magnetic fields. Individual peptides can be fragmented by collision with a background gas (e.g. helium or nitrogen). Upon collision, peptides dissociate into sequence-specific fragment ions whose masses are also measured. An experiment of this type, called tandem mass spectrometry (or MS/MS) measurement, yields two kinds of information: the mass of the analyzed peptide, and a list of sequence-specific fragments. The data collected are subjected to computer analysis using dedicated search engines (e.g. Mascot, Sequest, X-Tandem, Phenyx or OMSSA) that match the experimental data to sequences found in specific protein databases (e.g. ExPASy Proteomics: http://www.expasy.org/proteomics). This process enables the identification of the proteins in the sample from their amino acid sequence, and discriminates between background noise and true identifications [72].
A simple and robust alternative method for mass analysis of less complex peptides mixtures is the so-called Matrix-Assisted Laser for Desorption/Ionization-Time of Flight (MALDI-ToF) [73]. In MALDI-ToF, the peptides to be analyzed are deposited on a surface and dried in the presence of an organic matrix that absorbs at a particular wavelength. After crystallization, a laser is used to desorb the intact analyte from the matrix and ionize the molecules [74]. The ions formed are then accelerated by a high electric field, and their mass is deduced from the time they require to “fly” through an electric field-free region, hence the term time-of-flight. Derived from the previous method, the Surface-Enhanced Laser Desorption/Ionization-ToF (SELDI-ToF originally proposed by Ciphergen Biotechnologies Inc., and continued by Bio-Rad: http://www3.bio-rad.com, as the ProteinChip Seldi System) is based on the use of “chips” or strips that contain sites of specific affinity. The sites consist of surface chemical (covalent, ionic, hydrophobic, hydrophilic, metal) or biochemical (antibody, receptor, DNA) components for the specific retention of certain proteins/peptides of interest [75]. A laser is then used to desorb and ionize the retained components which yield a pattern of masses in a low resolution time of flight mass spectrometer. The downside of this technology is that protein identification has to be performed separately by MALDI-ToF or LC-MS/MS. In addition, prior knowledge of the associated specific properties of the targeted protein families is highly desirable.
Other approaches
An innovative strategy, called “Accurate Mass and Time Tags” or “AMT tags”, has recently shown great potential in proteomics research, especially due to its increased throughput, enabling the management of quantitative proteomic analyses of large numbers of samples with high sensitivity [76]. This strategy enables the identification of peptides on the basis of their chromatographic retention times and their mass, accurately determined using high resolution mass spectrometers such as ion cyclotron resonance [Fourier transform instruments] [77]. For example, from a sample of 3000 microdissected tumor cells of breast cancer, the equivalent of 300 ng of total protein, this technology has enabled the identification of thousand unique proteins involved in a wide variety of biological functions and cellular compartments [78]. Some of these proteins were subsequently highlighted as a signature associated with therapy resistance in breast cancer recurrence [79]. Dos Santos et al. used a similar strategy to determine the differential protein profiles between tumoral and non-tumoral microdissected cholangiocytes. Defining specific proteome alterations, they detected a panel of proteins that could be therapeutic targets and/or tumor biomarkers [80].
Reverse Phase Protein (micro) Array (RPMA) is another way to analyze proteins from microdissected tissues. It relies on simultaneous deposition of a small amount of total protein (from the picogram to femtogram value) retained in the form of a spot by adsorption on a nitrocellulose membrane, with the equivalent of 200 cells deposited per spot on a slide. Different spots correspond to protein samples reflecting different stages or physiological conditions in which a specific antibody is tested [81].
The study of proteomics is booming and laser microdissection is clearly becoming an essential tool that can overcome the constraints resulting from the cellular heterogeneity of tissues [82], Laser microdissection, which challenges conventional proteomics techniques in terms of sensitivity and quantification, is a suitable tool for experiments to discover protein biomarkers, provided that an appropriate analytical strategy is selected. Successful investigations have thus been reported for prostate cancer [83], breast cancer [79] and cholangiocarcinoma [80].
Technical factors hampering analyses
Major technical advances have automated many functions of laser microdissection systems making them relatively easy and friendly to use. However the quality and reproducibility of results obtained by microdissection are not only based on the quality of biological tissues considered but also some technical issues that should not be overlooked during the processing steps before microdissection. Thus, some products may interfere with the analysis and distort the results that will be difficult to interpret. The quality of the samples is a crucial parameter, but also the way in which this quality is evaluated. And finally it is important to take into account the physiological state of the cells.
Many factors in samples, such as the presence of haemoglobin [84], fat, glycogen or cell constituents, have been shown to inhibit the retro-transcription process (RT) as well as the Polymerase Chain Reaction (PCR) [85,86]. To ensure quality of the measures for qPCR and RT-qPCR assays, Bustin SA et al. have provided a checklist, named the MIQE guidelines, for authors preparing reports, enabling reviewers to evaluate their work and other investigators to reproduce it [87].
Depending on its composition, the very small volume of buffer contained in the cap of the microcentrifuge tubes can either be evaporated (i.e. lysis buffer) or crystallized (i.e. guanidinium isothiocyanate or urea) during microdissection. It is therefore advocated to work in an air-conditioned room where the temperature can be regulated according to external conditions (in summer or winter) (Figure 7).
Figure 7.
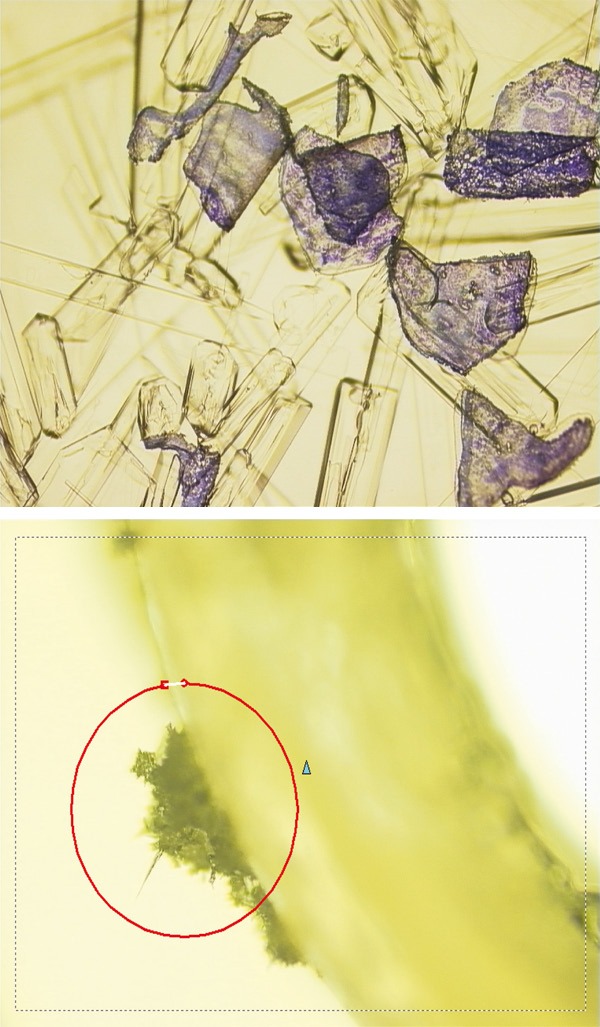
Crystal formation in the buffer contained in the cap tube during recovery of microdissected areas.
RNA quality control
RNA degradation remains one of the main concerns, depending on the tissue [for review, see Fleige et al [88]. To assess tissue RNA quality, RNAs left after laser removal from the remaining tissue section can be extracted by pipetting buffer onto the slide and then gently scraping off for analysis. Simultaneously, cryosections of tissue can be immediately treated together to extract RNA using the same protocol as that for section scrapes after microdissection [89].
The time required to recover enough biological material is often a limiting factor when dealing with a totally isolated cell located in a complex tissue. In some complicated case, more than 20 min can often be required by technicians to search for and identify cells of interest in the section. Clément-Ziza et al. have applied a method that stabilizes RNA, whereby sections are exposed to an argon flux during the microdissection time for up to 90 min. Less than 5% of RNA degradation was observed when slides were kept for up to 90 min under the microscope, whereas 20% degradation was reached after 30 min without argon [90].
Storage of sections can also result in oxidation of RNA on the exposed surface and storage at different temperatures influences the extent of RNA fragmentation [91]. If the sample surface has been exposed to air it can be altered, therefore the first 2-3 tissue sections from the block should be discarded. Any prolonged aqueous phase steps can also enhance RNase activity.
The laser microdissection technique is not an end in itself and before any protocol is embarked upon it is imperative to ensure the integrity of the RNA contained in the blocks that will be analyzed. Finally, when performing microdissection the use of an adhesive cap (dry capturing) instead of liquid recovery could potentially avoid aqueous conditions, and hence nuclease activity.
Housekeeping normalization
When comparative analysis of mRNA expression levels of genes studied is conducted in different cells and tissues, methods for accurate normalization are required [92]. Thus, housekeeping genes, which do not vary markedly through the cell cycle or in response to different experimental conditions, should be carefully employed as internal standards. Several housekeeping genes should be included, because some are more susceptible to degradation than others, resulting for example in lower normalized expression values in the thawed samples after 30 minutes [93]. Experimental studies have shown significant variations in expression levels of some of these genes between tissues, and unstable expression in relation to experimental conditions. For example, Bas A. et al. have reported that the 18s rRNA is more suitable for normalization of mRNA expression levels in resting and activated T lymphocytes [94]. Some authors also indicate that GAPDH mRNA levels may vary among cancer cell lines, and different cancers and normal tissues [95,96], or even during epithelial differentiation [97].
To determine the most stable reference genes from a set of tested candidate housekeeping genes in a given panel, some authors use algorithm applications such as a VBA applet for Microsoft Excel. The geNorm © method [93], BestKeeper © [98], NormFinder © software [99] or recently RefGenes © which enables users to search for the most stable genes in microarray samples [100] have been employed. A gene expression normalization factor can be calculated for each sample on the basis of the geometric mean of a user-defined number of reference genes. Use of the three most stable internal control genes for calculation of an RT-PCR normalization factor is recommended.
The right ratio
Tissue sample study will be more difficult for the more complex cellular systems, and in case of variations in size in a given tissue area. It is thus best to compare samples quantitatively in terms of surface areas rather than in terms of numbers of cells. Furthermore, as the protein fraction is essentially cytoplasmic, it is also important to consider the nucleus-cytoplasm ratio of the cells [101] which is increased in tumor cells [102].
Sampling and conditioning
At all stages of sample processing, rigorous procedures should be employed to reduce sample contamination. It is essential to work with scrubs and gloves to avoid introducing RNases or keratins in the reaction media. Only RNase-free filter pipette tips should be used and other instruments (razor-blade, tweezers...) should be properly cleaned and sterilized to keep away contamination, or replaced by single-use sterile equipment. A new razor-blade should be employed for each new sample to avoid contamination from one tissue sample to another. Fingerprints, hair, dead skin flakes and dust are the usual sources of contaminating keratins which derive mainly from the operator [103]. If they are present in concentrations greater than that of the protein studied, their abundance can lead to false results. Precautions should be taken, including carefully washing the surfaces contacting the samples or gels with acetonitrile, performing procedures under a laminar flow or still air hood, passing solutions through 0.22 μm filters, and wearing lab coat and latex gloves at all times when samples or gels are handled [104,105]. For protein studies in tissues rich in proteases, e.g. pancreas, spleen and lung, it may be advisable to add a mixture of protease inhibitors to the dye solution and to work at low temperature.
The samples for study should meet the requirements of quality and quantity, and be properly marked for efficient tracking. Tissue sections, frozen or fixed and embedded in paraffin, should be neither too thick (which would interfere with microscope observation), nor too thin (thus reducing the amount of the material obtained). In practice, most laboratories use sections from 7 to 8 micrometers thick.
The Superfrost® glass slide (Menzel Gläser, Braunschweig, Germany) have a permanent positive charge which electrostatically attracts frozen tissue sections and cytology preparations, binding them to it with covalent bonds. This prevents their use for microdissection.
DEPC treatment
The need for diethyl pyrocarbonate (DEPC)-treated autoclaved solutions in RNA molecular biological procedures on a variety of tissues and cultured tissue cells is a prerequisite, although purified water obtained by ultrafiltration has been found as efficient as DEPC treatment for suppressing RNase activity [106]. The histological slides should be cleaned, lipid-free and pretreated with a 0.1% DEPC solution in water prior to sterilization to limit RNA degradation. All DEPC-treated solutions should be autoclaved before use to remove DEPC which is a modifying agent for single-stranded molecules like RNA; DEPC reacts with purine residues and weakly with cytosine [107]. DEPC, which is already known to modify histidine and tyrosine residues [108], can also act on serine and threonine residues [109] and it also alters proteins. In contrast, DEPC and potassium permanganate have been recognized as useful reagents for detecting DNA distortions, especially melted regions [110].
The merits of RNALater®
Well-preserved quality of RNA is observed using RNALater® to protect tissue against RNases attack. However, laser microdissection should not be recommended on tissues containing high concentrations of chaotropic salts from this compound. Therefore, when using this procedure, RNALater® concentration should be reduced in the tissue to a level which did not interfere with laser microdissection [111]. In one observation, cryosections from RNALater®-fixed tissue never achieved the histomorphological quality of cryosections from snap-frozen tissue [112]. Samples in RNALater® will not freeze in the cryostat without previous wash steps in PBS for 5 min at 4°C, otherwise the cell outlines appear blurred, with tissue fragmentation occurrences observed in some specimens, making pathological interpretation difficult [113]. Conversely, it has been shown that RNALater®, normally devoted to RNA preservation, could be useful in the preparation and extraction of protein contents [114].
Tissue component preservation
A protocol should be established first, taking into account each step of the sampling procedure from the surgical removal to the Pathology Department, ensuring both a quick transport in sterile conditions and a microscope analysis to ascertain that the sample includes the entire lesion to be studied. Whatever the nature of the disease, the tissue should preferably be preserved by snap freezing directly in liquid nitrogen at 196°C for 10-15 minutes after resection during surgery or needle biopsy. Samples are then stored at 80°C until use.
The shorter the time of the freezing process immediately after biopsy the better the tissue preservation. The time elapsing between vascular clamping and excision, called warm ischemia, leads to rapid cell necrosis [115]. Organ protection against damage is based on blast-freezing. The influence of warm ischemia time on the degradation of RNA has been studied in the rat by Lazarus [116]. At 37°C for 60 min this showed that warm ischemia time exceeding 30 minutes resulted in a degradation of part of the RNA in the tissues of the surgical specimen. A 50% drop in RNA molecular weight in the kidneys was demonstrated after one hour in warm conditions.
The results of molecular analysis after microdissection will depend not only on the tissue type itself but also on the quality and preparation of the different samples. In practice, the reliability of results should enable molecular biology techniques to be applied after biological samples have been collected by microdissection, while allowing for good morphological observation of tissues under the microscope. Frozen biopsy provides better preservation and better quality of the tissue-derived products, thus increasing the sensitivity of molecular analysis techniques. Although it is possible to study and analyze samples from fixed tissue samples prior to paraffin embedding [117,118], all treatments applied to the tissue can alter the biological molecules and destabilize in situ a large number of interactions and molecular structures.
Finally, staining solutions could contain a complete protease inhibitor “cocktail”, but the need for protease inhibitors needs to be explored further, including malignant tissues where endogenous proteolytic activity may be expected to be more prevalent, as also observed with denaturing buffer [119]. Inclusion of RNAase inhibitor during the staining steps preparatory to laser capture microdissection appears to be important in protecting from RNA damage that can occur, with a consequent loss of representation of certain genes in particular in microarray hybridization analyses [120].
Frozen tissues
The freezing process
Freezing a tissue sample is intended to provide a hardened matrix for quick sectioning, and to preserve the biochemical or immunological properties of a specimen. An extremely rapid cooling speed should be used to produce vitreous ice without crystallisation damage in the cells. The best procedure is snap-freezing using liquid nitrogen, where tissue samples are placed in a cryotube under sterile RNase-free conditions. Tissues will be then stored at temperatures below -75°C in chest freezers with the necessary security measures to prevent thawing and/or exposure to large changes in temperature. The tubes should be stored in specially designed boxes until their use. Fatty and other soft tissues often remain too soft to be easily cut. It is possible to spray the surface with a special cooling spray, or to work at an even lower temperature to harden it prior to cutting. Frozen tissues should be stored close to the room where the cryotome sectioning will be performed in order to minimize warming events during transfer. Although storage should be at -70 or -80°C for nucleic acids, -20°C may however be sufficient for many antigens.
An aqueous solution of glycols and resins, which provides an inert matrix for sectioning, Optimal Cutting Temperature (OCT) Tissue-Tek embedding medium (Sakura Finetek, Inc., Torrance, CA, USA) is ordinarily used as an embedding medium for sectioning frozen tissue samples on a cryostat. Unfortunately, the storage of pathological specimens in OCT compound has been found to affect results, for example to inhibit DNA amplification in PCR [121]. A chilled mixture such as isopentane and dry ice mixture (-79°C) is sometimes provided for a rapid freezing process. A freezing step in liquid nitrogen remains preferable, to minimize any contact of the tissues with chemical substances that could interfere with downstream molecular analyses. High adipose content can make the process of obtaining thin, flat, intact cryosections suitable for LCM a difficult challenge, requiring colder cutting temperatures [122].
Special tips
For storage, frozen tissue should never be wrapped in aluminium foil to avoid warming of the sample when recovering the tissue for analysis, nor should it be put directly into the cryotube, to prevent adhesion to the tube. In our experience, for accurate tissue recovery, as well as for morphology, sterility and RNAse-free content, we first lay down the fresh samples on a sterile plastic strip to which it will then stick (Figure 8), and then we place the strip in a 2 mL cryotube for preservation by instantaneous snap freezing in liquid nitrogen. We then only have to take the tissue-carrying strip out of the tube with a sterile clamp in the cryotome and apply the tissue fragment directly to the specimen stub trimmed by as small as possible amount of OCT. The production of a frozen section is greatly assisted by having a well-orientated specimen with a flat cutting surface within a small rim of embedding medium. After cutting, the sections are immediately fixed in 70% ethanol and then dehydrated through a graded series of ethanol and xylene applications and briefly air-dried because over-dried RNA cannot be perfectly dissolved again.
Figure 8.
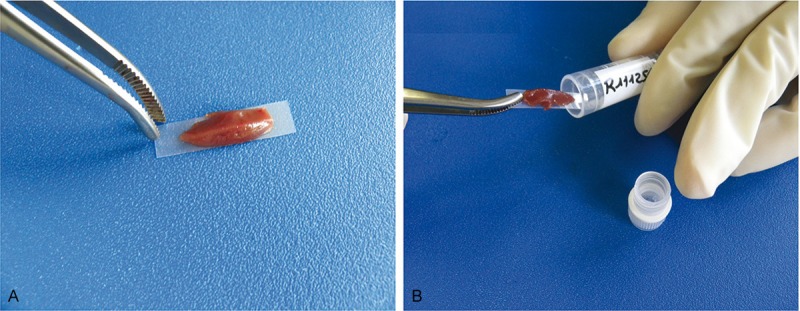
A: The fresh tissue sample is laid down with a sterile pinch on a RNAse-free plastic strip. B: The strip and the sample are then introduced into a classical cryotube for preservation by instantaneous snap freezing in liquid nitrogen.
Only one section is thawed and the time-lapse for the microdissection step is no longer than 15 min to minimise degradation processes.
The impact of thaws
Time to freezing and storage markedly affects RNA integrity [123-125]. Moreover, repetitive thawing leads to molecular degradations and should be banned. The process of thawing initiates RNA degradation after 30 minutes in unfixed tissues [126]. However, some authors report that repetitive thawing and freezing appears not detrimental to RNA as long as the total thawing time is short [127]. Others have shown that microdissected cells can be subjected to freeze-thaw cycles and the cell lysate then is collected into a solution designed to minimize RNA degradation. Subsequent RT-PCR reactions have been performed from the lysate without the need for any additional processing. With this technique the authors [128] demonstrated that just a couple of hundred microdissected cells in frozen breast carcinoma sections can provide adequate RNA template for 80-100 RT-PCR reactions.
Fixation of tissues and paraffin-embedding processes
The objective of fixation is to preserve cells and tissue morphology before hardening biological tissue by inclusion in paraffin blocks for serial cutting.
Fixation processes
The most fixative substances are formalin or formalin derivatives, and fixed- and embedded-samples are referred to as FFPE (Formaldehyde Fixed Paraffin Embedded) samples. Formalin, like other aldehyde fixatives, forms cross-linked methylene bridges and Schiff bases between basic amino acid (lysine) residues of proteins [129]. This cross-linking stabilizes the proteins in situ, which is the basis of fixation. The changes in immunoreactivity observed after formaldehyde fixation are explained by a reduced availability for the secondary antibody which is masked by cross-linking of globular proteins [130].
The speed of fixation with formaldehyde is highly pH-dependent [131] and immunoreactivity is irreversibly reduced by formaldehyde fixation [132] which also degrades genomic DNA and creates artefactual mutations [133]. This unstable product can be converted into formic acid; therefore its pH should be buffered before any use [134]. Formaldehyde modified by acidification and/or addition of alcoholic solution has given better results than formaldehyde alone as a fixative [135]. In experimental protocols, immunohistochemical analysis is improved after intravascular formalin injection [136]. Some investigators have achieved acceptable RNA recovery by using precipitative fixatives, such as ethanol and acetone. For example, in the study by Su et al., the RNA yield from ethanol-fixed brain tissues was 70% of the yield from fresh frozen specimens, and suitable for expression profiling of brain tissues by LCM and cDNA microarray [137].
The Just-In-Time concept
Formaldehyde also cross-links proteins to DNA depending on the duration of fixation [138,139]. It must be performed fast to avoid alteration of nucleic acids and proteins, in no more than 2 hours for small fragments. A surfix may decrease the efficiency of extraction of nucleic acids [140,141]. As the fixation time is extended, alterations to DNA will limit the available range of restriction enzyme/probe combinations and gel electrophoresis migration [142]. Excessively extended time periods of formaldehyde fixation result in increased RNA degradation [143]. While it is possible to use samples processed by formaldehyde fixation for Western Blot analysis [144], prolonged fixation time is known to reduce protein immunoreactivity [141,145].
The variable outcome of PCR [146], which is most probably the result of DNA fragmentation, has been shown by Williams et al. in tissues fixed for a long time in neutral buffered formaldehyde [133]. A zinc-based fixative developed by Lykidis et al. efficiently preserves DNA and RNA integrity, and facilitates protein analysis using 2D gel electrophoresis, in comparison with neutral buffered formalin fixation procedures, whilst maintaining optimal tissue morphology even over a 6-14-month period of storage [147].
To avoid excessively long fixation time prior to paraffin-embedding, the fixative should be removed and can be replaced with a freshly-prepared PBS storage solution before quick embedding.
Formaldehyde, or what else?
Although used extensively in pathology laboratories, formaldehyde and its aldehyde variants have another major drawback, their high toxicity. Formaldehyde (all physical forms) was nominated by NIEHS for reclassification in the 12th RoC, based on the 2004 review by the International Agency for Research on Cancer (IARC 2006), which concluded that there was sufficient evidence for the carcinogenicity of formaldehyde in humans [148]. Classified Class I carcinogen since the decree of July 13th 2006 in the list of substances, preparations and carcinogenic processes in section A 231-56 in the Labor Code, based on IARC epidemiological studies, it had been demonstrated to cause respiratory disease and respiratory cancers [149], and is therefore banned from use without certain precautions. In fact, this obligation has oriented laboratories towards the use of other types of fixatives, mostly alcoholic, some of which are still under evaluation, excluding formaldehyde-based products. Among these less toxic fixatives recently available on the market under the names of FineFIX® [150]; Glyofix [151]; UMFIX® [152], and protein and HOPE [153], One, known as RCL2®, developed since 1999, seems to be more successful in maintaining both cellular and tissue structures and remaining compatible with the requirements of molecular biology [154]. Nevertheless, there are probably other toxicities underlying the current use of these new fixatives, but not yet proven due to lack of use in daily practice.
To avoid shrinkage of tissues and optimize the penetration rate of the fixatives, we use a mix of ethanol-formalin-acetic acid, called AFA (Labonord SAS, Templemars, France) as a fixative and this provides a good compromise for satisfactory paraffin-embedded tissue preservation and good genomic analysis [22,36,155-157]. Ethanol fixation prior to paraffin-embedding of tissues also yields excellent histomorphology and good preservation of derivative products [158,159]. It has also been successfully used for proteomic analyses by mass spectrometry [160], even if authors have reported that sectioning ethanol-fixed tissues was difficult [161].
Finally, Bouin solution [162], which enables excellent morphological analysis, is not at all recommended for molecular biology techniques [163]. Reversible cross-linkers, such as dithiobis (succinimidyl) propionate (DSP), have been applied successfully with downstream extraction of sufficient quantities of nucleic acids [164] but they produce peptides and protein fragments rather than intact proteins [165]. Although still described in the literature for some applications, mainly for FFPE-treated archival samples, the study of RNA from fixed tissue is generally avoided.
Paraffin embedding processes
When several pieces of a sample are embedded in a single block, the paraffin coating process must be carefully carried out to align the pieces appropriately so that on the histological slide each sample included will be spread evenly. In addition, embedding of some tissue fragments (brain, kidney, skin or oesophageal biopsies for example) should be properly oriented in the block of paraffin so that sectioning can be performed in the optimal axis. Serial paraffin sections 7 μm thick on average can be cut to provide good histological detail for microdissection. One paraffin formulation (Paraplast® embedding media, Electron Microscopy Sciences, Hatfield, PA, USA) greatly improves small biopsy specimen observation with minimal cell shrinkage and distortion, and generates much lower cross-linking of proteins. While it is quite possible to extract DNA from tissue samples embedded in paraffin, it is also possible to study proteins, but the treatments greatly alter the protein structures. However, authors showed the presence of extra bands development in PCR due to the harshness of the methodology used to isolate nucleic acids from formalin-fixed and paraffin embedded tissue samples or the nature of the fixation procedure, or because of the time passed during storage in which alteration in the chromosomal DNA would take place [166]. The fixation and inclusion steps, involving especially the use of paraffin heated to 56°C over a period of ten hours to enable a proper infiltration of tissues, may alter the immunohistochemical detection of some antigens on tissue by storage of paraffin unstained slides as short a time as three months [167].
Tissue staining
Staining of tissue sections facilitates the analysis of cell populations laid out on glass slides. However it is clear that identification of cells or tissue areas without staining is preferable [168].
Since no coverslip is used in laser-assisted microdissection, the reduction in refractive index means that most light passing through the tissue is scattered, which can obscure cellular detail at high magnifications [169,170]. Thus, morphology sometimes appears quite different when compared with coverslipped tissue sections. In order to prevent this difficulty we add 10-15 μl of ethanol directly on the tissue section prior to the microdissection step. This enables better morphological analysis before evaporation. The PALM company synthetized a fluid cover medium in order to improve morphology without affecting downstream molecular applications, and particularly RNA integrity [171]. Therefore, it should be noted that morphological identification may take longer if there is no coverslip and mounting medium on the tissue section.
Standard staining substances
The widely used haematoxylin/eosin stain can result in DNA degradation [172] requiring more PCR cycles [173]. Haematoxylin/eosin staining [65] and other stains such as methyl green [174] or toluidine blue [175,176] are compatible with global protein profiling and 2D-PAGE proteomic analysis after laser capture microdissection. Nuclear Fast Red [177] or cresyl violet [89] [are best in terms of preserving RNA integrity. Although HistoGene has been reported to yield high-quality RNA in frozen brain LCM [178], it has also been found to cause significant RNA degradation in microdissected epithelia from other tissues, e.g. mammary [122], prostate [179] and cervix [180], and thus great care is required in its use. The haematoxylin and eosin combination is the most common staining technique used in histology. The diagnosis of most malignancies is based largely on this procedure. Haematoxylin, however, does not stain by itself but must be oxidized to hematein (usually at an acid pH) to act as a dye. The overall coloration of the stained specimen is the result of the balance of the concentration of the alum-haematoxylin and eosin. The most frequently-used form of eosin is eosin Y which is a tetrabrominated derivative of fluorescein. Thus, eosin provides autofluorescence and on its own constitutes a useful tool for morphological survey [181]. However, eosin staining complicates data interpretation using capillary electrophoresis in the analysis of fluorescent labelled PCR amplification products [182] and has a detrimental effect on protein separation [160]. Tangrea et al. have used immuno-guided microdissection known as expression microdissection (xMD) using immunohistochemically stained tissue to guide the cell selection process [183]. Through the EVA film the light energy is absorbed only where there is a large deposit of highly absorbent immunostain.
Immuno-staining procedure
One-hour immunohistochemical staining protocols can lead to significant degradation of RNA by RNases activated in aqueous environments [184]. In 1999, Fend et al. proposed a rapid immuno-staining procedure (total procedure time from 12 to 25 minutes) for frozen sections, followed by laser-capture microdissection and RNA extraction, which enables a targeted mRNA analysis of immuno-phenotypically defined cell populations [6]. Together with short-term formalin fixation, a reduction of antibody incubation times and digestion with proteinase K, Fink et al proposed the use of immunofluorescence applied to microdissection [185]. Burbach et al. in 2004 also described a rapid immunofluorescence staining approach combined with laser microdissection on frozen sections that does not interfere with RNA recovery and integrity for quantitative RT-PCR [186]. It thus appears that immunostaining in microdissection requires a significant adaptation from conventional immunostaining protocols, to obtain more accurate qualitative and quantitative laser microdissection. Shortened protocols are therefore needed, such as those that allow double fluorescence labelling to be performed in one incubation of only 5 minutes [187].
Staining or not, that is the question
It is sometimes advisable, when enough tissue is available, to prepare a subsequent tissue section that should be classically stained for comparison with the unstained microdissection slide. To this end, some manufacturers have invented a computerized tracking system whereby, from serial histological sections, a section is stained on a slide and used as a reference for other unstained sections to be microdissected, for example the software integrated into PALM. There is also the similar image analysis software, such as the Leica LMD AutoVision Control system. The AutoScanXT Software Module from Life Technologies found on the Arcturus system is an image analysis program that automatically identifies immuno-stained regions of interest with minimal supervision by the investigator [188].
The fluorescent background
The PEN membrane is well-known to present a stronger fluorescent background, mainly with the DAPI and Hoechst dye. Thus, if weak fluorescence needs to be detected it could be advis able to work with a polyethylene teraphthalate (PET) MembraneSlide. The PET-membrane does not absorb the laser as much as the PEN membrane does. Therefore you need to increase the energy for cutting compared to PEN membrane slides. And, to get rid of the background immunofluorescence it might help to proceed to a non-specific binding which is performed by covering the section with a stronger protein-blocking solution.
Cell count analysis before laser-assisted microdissection can also be performed on a PC using the ImageJ program developed in 1997 by Wayne Rasband at the Research Services Branch of the National Institute of Mental Health [189]. This is a Java-based image processing programme, in open-access on the Internet at the following link: http://rsb.info.nih.gov/ij/.
Concluding remarks
Many factors need to be taken into account in the process of molecular analysis of microdissected tissue. Each step increasing the purity of the samples may also cause loss of significant amounts of material. It is therefore recommended to reduce the number of steps and the time required for each of them.
In tissue samples, we need to cope with a limiting factor that is especially crucial for proteomics, that is the small quantities of material obtained by microdissection. This implies spending much more time on the microdissection of specific tissues, entailing a risk of impaired quality of the material recovered. This is even more drastic for the analysis of RNA if no information is known about the number of copies and the half-life of the transcript studied.
As already underlined by others [190], providing high quality tissue at the molecular level is dependent on the protocol for tissue procurement and for processing. There is no single way to prepare samples. Hence, each protocol will differ depending on the type of sample and the type of experiment. Today we need to think about each step upstream and downstream of the microdissection step. It is fundamental to develop a method that preserves the original quality of the sample, considering that the technical approach will depend crucially on the tissue studied, its texture and particularly on the presence or absence of nucleases or proteases, more abundant in the pancreas, spleen or even lung. Finally, the preservation of this tissue (frozen or paraffin-embedded after fixation) and the proposed study, which may include DNA, RNA and proteins, are also to be considered.
In line with Bova et al. [191], we invite investigators not to consider laser microdissection as a tool that is sufficient in itself. On a general basis, the development of an optimal protocol for any tissue procurement is therefore a prerequisite, providing guidelines for clinicians and researchers for satisfactory preparation of tissues and an optimisation of the classic techniques for molecular analysis. These analyzes can be used clinically as aids in cancer diagnosis, clinical management, genomic profile studies, and targeted therapy.
The technique of laser microdissection is of great importance in the field of molecular analysis at the level of the cell. It helped to bring greater precision in the selection of biological tissues that can be seen under a microscope according to their physiological or pathological state or not. This precision in the recovery of biological samples is primarily a function of expertise in the recognition of biological tissues. The ability to use different markers as specific dyes or not, conjugated-antibody or not, has extended greatly the scope for future research from frozen or even paraffin-embedded tissue after fixation. Then, the very fine cutting laser beam and the ability to avoid or destroy any unwanted cells or cell areas only strengthen the precision, quality and reproducibility of the analyzes made on these samples later. The laser-capture microdissection process does not alter or damage the morphology or chemistry of the sample collected, nor the surrounding cells. This represents a major step forward in many areas of application to obtain better results from very small sample. Taken together, the technique of laser-assisted tissue microdissection has obvious advantages in the molecular analysis of very small samples corroborated by numerous publications showing high rates of accuracy. However, this technique does not make sense if the quality of the sample is not strictly controlled from the beginning throughout the various stages of treatment. If the DNA molecule is relatively strong, it is not the same for RNA which we know the extreme fragility depending on their environment and drastic precautions that should be considered to preserve their integrity. This also applies, but to a lesser extent for the study of proteins with which a specific protocol must be defined in terms of the family of proteins to which it belongs, if it is known. In addition, the inability to amplify recovered products, as it is the case for nucleic acids, makes longer and more complicated the protein studies.
Nevertheless, while the cost of the investment for acquiring such a system remains high, significant costs should be added with respect to consumables (special histological Membraneslides®, capsules and recovery tubes) and to updating software. It is important to note that tissue microdissection cannot yet be implemented in the context of daily diagnosis, but it is perfectly suited to research protocols defined as part of a multi-partnership. Long-term methodological back-up is required both for the technical maintenance of the system and for the protocols developed by researchers, technicians, engineers and students.
Acknowledgements
Authors are gratefully indebted to Dr Philippe Ratajczak for critical reading and helpful discussions, to Nathalie Martin and Irmine Ferreira for technical assistance. We also thank Angela Swaine Verdier for kindly editing the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Bertheau P, Meignin V, Janin A. [Microdissection on histologic and cytologic preparation: an approach to tissue heterogeneity] . Ann Pathol. 1998;18:110–119. [PubMed] [Google Scholar]
- 2.Simone NL, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14:272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 3.de Kerviler E, Guermazi A, Zagdanski AM, Meignin V, Gossot D, Oksenhendler E, Mariette X, Brice P, Frija J. Image-guided core-needle biopsy in patients with suspected or recurrent lymphomas. Cancer. 2000;89:647–652. [PubMed] [Google Scholar]
- 4.Zhuang Z, Bertheau P, Emmert-Buck MR, Liotta LA, Gnarra J, Linehan WM, Lubensky IA. A microdissection technique for archival DNA analysis of specific cell populations in lesions < 1 mm in size. Am J Pathol. 1995;146:620–625. [PMC free article] [PubMed] [Google Scholar]
- 5.Going JJ. Extraction of DNA from microdissected archival tissues. Methods Mol Med. 2001;39:291–298. doi: 10.1385/1-59259-071-3:291. [DOI] [PubMed] [Google Scholar]
- 6.Fend F, Emmert-Buck MR, Chuaqui R, Cole K, Lee J, Liotta LA, Raffeld M. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibata D, Hawes D, Li ZH, Hernandez AM, Spruck CH, Nichols PW. Specific genetic analysis of microscopic tissue after selective ultraviolet radiation fractionation and the polymerase chain reaction. Am J Pathol. 1992;141:539–543. [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel A, Horneffer V, Lorenz K, Linz N, Huttmann G, Gebert A. Principles of laser microdissection and catapulting of histologic specimens and live cells. Methods Cell Biol. 2007;82:153–205. doi: 10.1016/S0091-679X(06)82005-4. [DOI] [PubMed] [Google Scholar]
- 9.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 10.Bohm M, Wieland I, Schutze K, Rubben H. Microbeam MOMeNT: non-contact laser microdissection of membrane-mounted native tissue. Am J Pathol. 1997;151:63–67. [PMC free article] [PubMed] [Google Scholar]
- 11.Kolble K. The LEICA microdissection system: design and applications. J Mol Med (Berl) 2000;78:B24–25. [PubMed] [Google Scholar]
- 12.Böhm M, Schmidt C, Wieland I, Leclerc N. Membrane-Based Laser Microdissection in Molecular Oncology. Onkologie. 1999;22:296–301. [Google Scholar]
- 13.Domazet B, Maclennan GT, Lopez-Beltran A, Montironi R, Cheng L. Laser capture microdissection in the genomic and proteomic era: targeting the genetic basis of cancer. Int J Clin Exp Pathol. 2008;1:475–488. [PMC free article] [PubMed] [Google Scholar]
- 14.Panchal M, Loeper J, Cossec JC, Perruchini C, Lazar A, Pompon D, Duyckaerts C. Enrichment of cholesterol in microdissected Alzheimer’s disease senile plaques as assessed by mass spectrometry. J Lipid Res. 2010;51:598–605. doi: 10.1194/jlr.M001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu G, Reynolds L, Crnogorac-Jurcevic T, Gillett CE, Dublin EA, Marshall JF, Barnes D, D’Arrigo C, Van Trappen PO, Lemoine NR, Hart IR. Combination of microdissection and microarray analysis to identify gene expression changes between differentially located tumour cells in breast cancer. Oncogene. 2003;22:3742–3748. doi: 10.1038/sj.onc.1206428. [DOI] [PubMed] [Google Scholar]
- 16.Sugiyama Y, Farrow B, Murillo C, Li J, Watanabe H, Sugiyama K, Evers BM. Analysis of differential gene expression patterns in colon cancer and cancer stroma using microdissected tissues. Gastroenterology. 2005;128:480–486. doi: 10.1053/j.gastro.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 17.De Spiegelaere W, Filliers M, Van Soom A. Laser capture microdissection for gene expression analysis of specific cell populations in single blastocysts. Methods Mol Biol. 2012;853:29–37. doi: 10.1007/978-1-61779-567-1_4. [DOI] [PubMed] [Google Scholar]
- 18.Sethi S, Vrana JA, Theis JD, Leung N, Sethi A, Nasr SH, Fervenza FC, Cornell LD, Fidler ME, Dogan A. Laser microdissection and mass spectrometry-based proteomics aids the diagnosis and typing of renal amyloidosis. Kidney Int. 2012;82:226–234. doi: 10.1038/ki.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mead LJ, Gillespie MT, Hung JY, Rane US, Rayeroux KC, Irving LB, Campbell LJ. Frequent loss of heterozygosity in early non-small cell lung cancers at chromosome 9p21 proximal to the CDKN2a gene. Int J Cancer. 1997;71:213–217. doi: 10.1002/(sici)1097-0215(19970410)71:2<213::aid-ijc15>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang Z, Emmert-Buck MR, Roth MJ, Gnarra J, Linehan WM, Liotta LA, Lubensky IA. von Hippel-Lindau disease gene deletion detected in microdissected sporadic human colon carcinoma specimens. Hum Pathol. 1996;27:152–156. doi: 10.1016/s0046-8177(96)90368-8. [DOI] [PubMed] [Google Scholar]
- 21.Vocke CD, Pozzatti RO, Bostwick DG, Florence CD, Jennings SB, Strup SE, Duray PH, Liotta LA, Emmert-Buck MR, Linehan WM. Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12-21. Cancer Res. 1996;56:2411–2416. [PubMed] [Google Scholar]
- 22.Bertheau P, Plassa LF, Lerebours F, de Roquancourt A, Turpin E, Lidereau R, de The H, Janin A. Allelic loss detection in inflammatory breast cancer: improvement with laser microdissection. Lab Invest. 2001;81:1397–1402. doi: 10.1038/labinvest.3780353. [DOI] [PubMed] [Google Scholar]
- 23.Heinmoller E, Dietmaier W, Zirngibl H, Heinmoller P, Scaringe W, Jauch KW, Hofstadter F, Ruschoff J. Molecular analysis of microdissected tumors and preneoplastic intraductal lesions in pancreatic carcinoma. Am J Pathol. 2000;157:83–92. doi: 10.1016/S0002-9440(10)64520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giuffre G, Muller A, Brodegger T, Bocker-Edmonston T, Gebert J, Kloor M, Dietmaier W, Kullmann F, Buttner R, Tuccari G, Ruschoff J. Microsatellite analysis of hereditary nonpolyposis colorectal cancer-associated colorectal adenomas by laser-assisted microdissection: correlation with mismatch repair protein expression provides new insights in early steps of tumorigenesis. J Mol Diagn. 2005;7:160–170. doi: 10.1016/S1525-1578(10)60542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coulbault L, Herlicoviez D, Chapon F, Read MH, Penniello MJ, Reynier P, Fayet G, Lombes A, Jauzac P, Allouche S. A novel mutation in the mitochondrial tRNA Asn gene associated with a lethal disease. Biochem Biophys Res Commun. 2005;329:1152–1154. doi: 10.1016/j.bbrc.2005.02.083. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy RP, Zhang S, Bostwick DG, Qian J, Eble JN, Wang M, Lin H, Cheng L. Molecular genetic evidence for different clonal origins of epithelial and stromal components of phyllodes tumor of the prostate. Am J Pathol. 2004;165:1395–1400. doi: 10.1016/S0002-9440(10)63397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morandi L, Marucci G, Foschini MP, Cattani MG, Pession A, Riva C, Eusebi V. Genetic similarities and differences between lobular in situ neoplasia (LN) and invasive lobular carcinoma of the breast. Virchows Arch. 2006;449:14–23. doi: 10.1007/s00428-006-0192-7. [DOI] [PubMed] [Google Scholar]
- 28.Harada T, Chelala C, Bhakta V, Chaplin T, Caulee K, Baril P, Young BD, Lemoine NR. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27:1951–1960. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 30.Coudry RA, Meireles SI, Stoyanova R, Cooper HS, Carpino A, Wang X, Engstrom PF, Clapper ML. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2007;9:70–79. doi: 10.2353/jmoldx.2007.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubin MA. Use of laser capture microdissection, cDNA microarrays, and tissue microarrays in advancing our understanding of prostate cancer. J Pathol. 2001;195:80–86. doi: 10.1002/path.892. [DOI] [PubMed] [Google Scholar]
- 32.Hu L, Sham JS, Tjia WM, Tan YQ, Lu GX, Guan XY. Generation of a complete set of human telomeric band painting probes by chromosome microdissection. Genomics. 2004;83:298–302. doi: 10.1016/j.ygeno.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 33.Meltzer PS, Guan XY, Burgess A, Trent JM. Rapid generation of region specific probes by chromosome microdissection and their application. Nat Genet. 1992;1:24–28. doi: 10.1038/ng0492-24. [DOI] [PubMed] [Google Scholar]
- 34.Vandewoestyne M, Van Hoofstat D, Van Nieuwerburgh F, Deforce D. Suspension fluorescence in situ hybridization (S-FISH) combined with automatic detection and laser microdissection for STR profiling of male cells in male/female mixtures. Int J Legal Med. 2009;123:441–447. doi: 10.1007/s00414-009-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroneis T, Gutstein-Abo L, Kofler K, Hartmann M, Hartmann P, Alunni-Fabbroni M, Walcher W, Dohr G, Petek E, Guetta E, Sedlmayr P. Automatic retrieval of single microchimeric cells and verification of identity by on-chip multiplex PCR. J Cell Mol Med. 2009;14:954–969. doi: 10.1111/j.1582-4934.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao WL, Mourah S, Mounier N, Leboeuf C, Daneshpouy ME, Legres L, Meignin V, Oksenhendler E, Maignin CL, Calvo F, Briere J, Gisselbrecht C, Janin A. Vascular endothelial growth factor-A is expressed both on lymphoma cells and endothelial cells in angioimmunoblastic T-cell lymphoma and related to lymphoma progression. Lab Invest. 2004;84:1512–1519. doi: 10.1038/labinvest.3700145. [DOI] [PubMed] [Google Scholar]
- 37.Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 38.Schlomm T, Luebke AM, Sultmann H, Hellwinkel OJ, Sauer U, Poustka A, David KA, Chun FK, Haese A, Graefen M, Erbersdobler A, Huland H. Extraction and processing of high quality RNA from impalpable and macroscopically invisible prostate cancer for microarray gene expression analysis. Int J Oncol. 2005;27:713–720. [PubMed] [Google Scholar]
- 39.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Spiegelaere W, Cornillie P, Van Poucke M, Peelman L, Burvenich C, Van den Broeck W. Quantitative mRNA expression analysis in kidney glomeruli using microdissection techniques. Histol Histopathol. 2011;26:267–275. doi: 10.14670/HH-26.267. [DOI] [PubMed] [Google Scholar]
- 42.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- 43.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid MW, Schmidt A, Klostermeier UC, Barann M, Rosenstiel P, Grossniklaus U. A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS One. 2012;7:e29685. doi: 10.1371/journal.pone.0029685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 46.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 47.Medina PP, Slack FJ. MicroRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 48.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu L, Yan W, Rodriguez-Canales J, Rosenberg AM, Hu N, Goldstein AM, Taylor PR, Erickson HS, Emmert-Buck MR, Tangrea MA. MicroRNA analysis of microdissected normal squamous esophageal epithelium and tumor cells. Am J Cancer Res. 2011;1:574–584. [PMC free article] [PubMed] [Google Scholar]
- 50.Nonn L, Vaishnav A, Gallagher L, Gann PH. mRNA and micro-RNA expression analysis in laser-capture microdissected prostate biopsies: valuable tool for risk assessment and prevention trials. Exp Mol Pathol. 2010;88:45–51. doi: 10.1016/j.yexmp.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, Guenther SM, O’Leary JJ, Sheils O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szafranska AE, Davison TS, Shingara J, Doleshal M, Riggenbach JA, Morrison CD, Jewell S, Labourier E. Accurate molecular characterization of formalin-fixed, paraffin-embedded tissues by microRNA expression profiling. J Mol Diagn. 2008;10:415–423. doi: 10.2353/jmoldx.2008.080018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siebolts U, Varnholt H, Drebber U, Dienes HP, Wickenhauser C, Odenthal M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2009;62:84–88. doi: 10.1136/jcp.2008.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuster C, Budczies J, Faber C, Kirchner T, Hlubek F. MicroRNA expression profiling of specific cells in complex archival tissue stained by immunohistochemistry. Lab Invest. 2011;91:157–165. doi: 10.1038/labinvest.2010.134. [DOI] [PubMed] [Google Scholar]
- 55.von Eggeling F, Melle C, Ernst G. Microdissecting the proteome. Proteomics. 2007;7:2729–2737. doi: 10.1002/pmic.200700079. [DOI] [PubMed] [Google Scholar]
- 56.Jones GL, Khawar SL, Watson K. A polymorphic variation in the sodium dodecyl sulfate-polyacrylamide gel electrophores separation of carboxymethylated human hair proteins. Electrophoresis. 1999;20:3347–3348. doi: 10.1002/(SICI)1522-2683(19991101)20:17<3347::AID-ELPS3347>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 57.Natkunam Y, Rouse RV, Zhu S, Fisher C, van De Rijn M. Immunoblot analysis of CD34 expression in histologically diverse neoplasms. Am J Pathol. 2000;156:21–27. doi: 10.1016/S0002-9440(10)64701-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang DH, Tai LK, Wong LL, Sethi SK, Koay ES. Proteomics of breast cancer: enhanced expression of cytokeratin19 in human epidermal growth factor receptor type 2 positive breast tumors. Proteomics. 2005;5:1797–1805. doi: 10.1002/pmic.200401069. [DOI] [PubMed] [Google Scholar]
- 59.Maitre M, Roullot-Lacarriere V, Piazza PV, Revest JM. Western blot detection of brain phosphoproteins after performing Laser Microdissection and Pressure Catapulting (LMPC) J Neurosci Methods. 2011;198:204–212. doi: 10.1016/j.jneumeth.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Craven RA, Totty N, Harnden P, Selby PJ, Banks RE. Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: evaluation of tissue preparation and sample limitations. Am J Pathol. 2002;160:815–822. doi: 10.1016/S0002-9440(10)64904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ornstein DK, Gillespie JW, Paweletz CP, Duray PH, Herring J, Vocke CD, Topalian SL, Bostwick DG, Linehan WM, Petricoin EF 3rd, Emmert-Buck MR. Proteomic analysis of laser capture microdissected human prostate cancer and in vitro prostate cell lines. Electrophoresis. 2000;21:2235–2242. doi: 10.1002/1522-2683(20000601)21:11<2235::AID-ELPS2235>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 62.Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci U S A. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishihara JC, Champion KM. Quantitative evaluation of proteins in one- and two-dimensional polyacrylamide gels using a fluorescent stain. Electrophoresis. 2002;23:2203–2215. doi: 10.1002/1522-2683(200207)23:14<2203::AID-ELPS2203>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 64.Gorg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 65.De Souza AI, McGregor E, Dunn MJ, Rose ML. Preparation of human heart for laser microdissection and proteomics. Proteomics. 2004;4:578–586. doi: 10.1002/pmic.200300660. [DOI] [PubMed] [Google Scholar]
- 66.Zhou G, Li H, DeCamp D, Chen S, Shu H, Gong Y, Flaig M, Gillespie JW, Hu N, Taylor PR, Emmert-Buck MR, Liotta LA, Petricoin EF 3rd, Zhao Y. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol Cell Proteomics. 2002;1:117–124. doi: 10.1074/mcp.m100015-mcp200. [DOI] [PubMed] [Google Scholar]
- 67.Greengauz-Roberts O, Stoppler H, Nomura S, Yamaguchi H, Goldenring JR, Podolsky RH, Lee JR, Dynan WS. Saturation labeling with cysteine-reactive cyanine fluorescent dyes provides increased sensitivity for protein expression profiling of laser-microdissected clinical specimens. Proteomics. 2005;5:1746–1757. doi: 10.1002/pmic.200401068. [DOI] [PubMed] [Google Scholar]
- 68.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 69.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 70.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 71.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 72.Anderson DC, Li W, Payan DG, Noble WS. A new algorithm for the evaluation of shotgun peptide sequencing in proteomics: support vector machine classification of peptide MS/MS spectra and SEQUEST scores. J Proteome Res. 2003;2:137–146. doi: 10.1021/pr0255654. [DOI] [PubMed] [Google Scholar]
- 73.Palmer-Toy DE, Sarracino DA, Sgroi D, LeVangie R, Leopold PE. Direct acquisition of matrix-assisted laser Desorption/Ionization time-of-flight mass spectra from laser capture microdissected tissues. Clin Chem. 2000;46:1513–1516. [PubMed] [Google Scholar]
- 74.Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int J Mass Spectrom Ion Proc. 1987;78:53–68. [Google Scholar]
- 75.von Eggeling F, Davies H, Lomas L, Fiedler W, Junker K, Claussen U, Ernst G. Tissue-specific microdissection coupled with ProteinChip array technologies: applications in cancer research. Biotechniques. 2000;29:1066–1070. doi: 10.2144/00295rr02. [DOI] [PubMed] [Google Scholar]
- 76.Shen Y, Tolic N, Masselon C, Pasa-Tolic L, Camp DG 2nd, Lipton MS, Anderson GA, Smith RD. Nanoscale proteomics. Anal Bioanal Chem. 2004;378:1037–1045. doi: 10.1007/s00216-003-2329-8. [DOI] [PubMed] [Google Scholar]
- 77.Page JS, Masselon CD, Smith RD. FTICR mass spectrometry for qualitative and quantitative bioanalyses. Curr Opin Biotechnol. 2004;15:3–11. doi: 10.1016/j.copbio.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 78.Umar A, Luider TM, Foekens JA, Pasa-Tolic L. NanoLC-FT-ICR MS improves proteome coverage attainable for approximately 3000 laser-microdissected breast carcinoma cells. Proteomics. 2007;7:323–329. doi: 10.1002/pmic.200600293. [DOI] [PubMed] [Google Scholar]
- 79.Umar A, Kang H, Timmermans AM, Look MP, Meijer-van Gelder ME, den Bakker MA, Jaitly N, Martens JW, Luider TM, Foekens JA, Pasa-Tolic L. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol Cell Proteomics. 2009;8:1278–1294. doi: 10.1074/mcp.M800493-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dos Santos A, Court M, Thiers V, Sar S, Guettier C, Samuel D, Brechot C, Garin J, Demaugre F, Masselon CD. Identification of cellular targets in human intrahepatic cholangiocarcinoma using laser microdissection and accurate mass and time tag proteomics. Mol Cell Proteomics. 2010;9:1991–2004. doi: 10.1074/mcp.M110.000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 82.Emmert-Buck MR, Strausberg RL, Krizman DB, Bonaldo MF, Bonner RF, Bostwick DG, Brown MR, Buetow KH, Chuaqui RF, Cole KA, Duray PH, Englert CR, Gillespie JW, Greenhut S, Grouse L, Hillier LW, Katz KS, Klausner RD, Kuznetzov V, Lash AE, Lennon G, Linehan WM, Liotta LA, Marra MA, Munson PJ, Ornstein DK, Prabhu VV, Prange C, Schuler GD, Soares MB, Tolstoshev CM, Vocke CD, Waterston RH. Molecular profiling of clinical tissue specimens: feasibility and applications. Am J Pathol. 2000;156:1109–1115. doi: 10.1016/S0002-9440(10)64979-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wellmann A, Wollscheid V, Lu H, Ma ZL, Albers P, Schutze K, Rohde V, Behrens P, Dreschers S, Ko Y, Wernert N. Analysis of microdissected prostate tissue with ProteinChip arrays--a way to new insights into carcinogenesis and to diagnostic tools. Int J Mol Med. 2002;9:341–347. [PubMed] [Google Scholar]
- 84.Radstrom P, Knutsson R, Wolffs P, Lovenklev M, Lofstrom C. Pre-PCR processing: strategies to generate PCR-compatible samples. Mol Biotechnol. 2004;26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 85.Rossen L, Norskov P, Holmstrom K, Rasmussen OF. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol. 1992;17:37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 86.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 88.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 89.Bevilacqua C, Makhzami S, Helbling JC, Defrenaix P, Martin P. Maintaining RNA integrity in a homogeneous population of mammary epithelial cells isolated by Laser Capture Microdissection. BMC Cell Biol. 2011;11:95. doi: 10.1186/1471-2121-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clement-Ziza M, Munnich A, Lyonnet S, Jaubert F, Besmond C. Stabilization of RNA during laser capture microdissection by performing experiments under argon atmosphere or using ethanol as a solvent in staining solutions. Rna. 2008;14:2698–2704. doi: 10.1261/rna.1261708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS One. 2007;2:e1261. doi: 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 93.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bas A, Forsberg G, Hammarstrom S, Hammarstrom ML. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol. 2004;59:566–573. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- 95.Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem. 2001;295:17–21. doi: 10.1006/abio.2001.5171. [DOI] [PubMed] [Google Scholar]
- 96.Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 97.Steele BK, Meyers C, Ozbun MA. Variable expression of some “housekeeping” genes during human keratinocyte differentiation. Anal Biochem. 2002;307:341–347. doi: 10.1016/s0003-2697(02)00045-3. [DOI] [PubMed] [Google Scholar]
- 98.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 99.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 100.Hruz T, Wyss M, Docquier M, Pfaffl MW, Masanetz S, Borghi L, Verbrugghe P, Kalaydjieva L, Bleuler S, Laule O, Descombes P, Gruissem W, Zimmermann P. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics. 2011;12:156. doi: 10.1186/1471-2164-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rudzki Z, Szczudrawa J, Stachura J. Morphometry of normal, regenerating and cancerous hepatocytes. Folia Histochem Cytobiol. 1989;27:141–148. [PubMed] [Google Scholar]
- 102.Serio G, Giardina C, Valente T, Pennella A, Lettini T, Dalena AM, D’Eredita G, Ricco R, Resta L. Quantitative morphometrical investigation of basal cell layer in laryngeal premalignant lesions. J Exp Clin Cancer Res. 2002;21:495–502. [PubMed] [Google Scholar]
- 103.Shapiro SZ. Elimination of the detection of an artefactual 65 kDa keratin band from immunoblots. J Immunol Methods. 1987;102:143–146. doi: 10.1016/s0022-1759(87)80019-4. [DOI] [PubMed] [Google Scholar]
- 104.Ochs D. Protein contaminants of sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1983;135:470–474. doi: 10.1016/0003-2697(83)90714-5. [DOI] [PubMed] [Google Scholar]
- 105.Sinha P, Poland J, Schnolzer M, Rabilloud T. A new silver staining apparatus and procedure for matrix-assisted laser desorption/ionization-time of flight analysis of proteins after two-dimensional electrophoresis. Proteomics. 2001;1:835–840. doi: 10.1002/1615-9861(200107)1:7<835::AID-PROT835>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 106.Mabic S, Kano I. Impact of purified water quality on molecular biology experiments. Clin Chem Lab Med. 2003;41:486–491. doi: 10.1515/CCLM.2003.073. [DOI] [PubMed] [Google Scholar]
- 107.Okamoto T, Okabe S. Ultraviolet absorbance at 260 and 280 nm in RNA measurement is dependent on measurement solution. Int J Mol Med. 2000;5:657–659. doi: 10.3892/ijmm.5.6.657. [DOI] [PubMed] [Google Scholar]
- 108.Hnizda A, Santrucek J, Sanda M, Strohalm M, Kodicek M. Reactivity of histidine and lysine side-chains with diethylpyrocarbonate -- a method to identify surface exposed residues in proteins. J Biochem Biophys Methods. 2008;70:1091–1097. doi: 10.1016/j.jbbm.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 109.Mendoza VL, Vachet RW. Protein surface mapping using diethylpyrocarbonate with mass spectrometric detection. Anal Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kahl BF, Paule MR. The use of diethyl pyrocarbonate and potassium permanganate as probes for strand separation and structural distortions in DNA. Methods Mol Biol. 2009;543:73–85. doi: 10.1007/978-1-60327-015-1_6. [DOI] [PubMed] [Google Scholar]
- 111.Thelen P, Burfeind P, Grzmil M, Voigt S, Ringert RH, Hemmerlein B. cDNA microarray analysis with amplified RNA after isolation of intact cellular RNA from neoplastic and non-neoplastic prostate tissue separated by laser microdissections. Int J Oncol. 2004;24:1085–1092. [PubMed] [Google Scholar]
- 112.Stemmer K, Ellinger-Ziegelbauer H, Lotz K, Ahr HJ, Dietrich DR. Establishment of a protocol for the gene expression analysis of laser microdissected rat kidney samples with affymetrix genechips. Toxicol Appl Pharmacol. 2006;217:134–142. doi: 10.1016/j.taap.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 113.Ellis M, Davis N, Coop A, Liu M, Schumaker L, Lee RY, Srikanchana R, Russell CG, Singh B, Miller WR, Stearns V, Pennanen M, Tsangaris T, Gallagher A, Liu A, Zwart A, Hayes DF, Lippman ME, Wang Y, Clarke R. Development and validation of a method for using breast core needle biopsies for gene expression microarray analyses. Clin Cancer Res. 2002;8:1155–1166. [PubMed] [Google Scholar]
- 114.Rodrigo MC, Martin DS, Redetzke RA, Eyster KM. A method for the extraction of high-quality RNA and protein from single small samples of arteries and veins preserved in RNAlater. J Pharmacol Toxicol Methods. 2002;47:87–92. doi: 10.1016/s1056-8719(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 115.Huang J, Qi R, Quackenbush J, Dauway E, Lazaridis E, Yeatman T. Effects of ischemia on gene expression. J Surg Res. 2001;99:222–227. doi: 10.1006/jsre.2001.6195. [DOI] [PubMed] [Google Scholar]
- 116.Lazarus HM. Alterations of high molecular weight nuclear RNA following kidney storage. Exp Mol Pathol. 1974;21:155–165. doi: 10.1016/0014-4800(74)90086-0. [DOI] [PubMed] [Google Scholar]
- 117.Burgemeister R. Nucleic acids extraction from laser microdissected FFPE tissue sections. Methods Mol Biol. 2011;724:117–129. doi: 10.1007/978-1-61779-055-3_8. [DOI] [PubMed] [Google Scholar]
- 118.Scicchitano MS, Dalmas DA, Bertiaux MA, Anderson SM, Turner LR, Thomas RA, Mirable R, Boyce RW. Preliminary comparison of quantity, quality, and microarray performance of RNA extracted from formalin-fixed, paraffin-embedded, and unfixed frozen tissue samples. J Histochem Cytochem. 2006;54:1229–1237. doi: 10.1369/jhc.6A6999.2006. [DOI] [PubMed] [Google Scholar]
- 119.Rabilloud T. Solubilization of proteins for electrophoretic analyses. Electrophoresis. 1996;17:813–829. doi: 10.1002/elps.1150170503. [DOI] [PubMed] [Google Scholar]
- 120.Wang H, Owens JD, Shih JH, Li MC, Bonner RF, Mushinski JF. Histological staining methods preparatory to laser capture microdissection significantly affect the integrity of the cellular RNA. BMC Genomics. 2006;7:97. doi: 10.1186/1471-2164-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Turbett GR, Sellner LN. The use of optimal cutting temperature compound can inhibit amplification by polymerase chain reaction. Diagn Mol Pathol. 1997;6:298–303. doi: 10.1097/00019606-199710000-00009. [DOI] [PubMed] [Google Scholar]
- 122.McGinley JN, Zhu Z, Jiang W, Thompson HJ. Collection of epithelial cells from rodent mammary gland via laser capture microdissection yielding high-quality RNA suitable for microarray analysis. Biol Proced Online. 2010;12:9026. doi: 10.1007/s12575-010-9026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.De Cecco L, Musella V, Veneroni S, Cappelletti V, Bongarzone I, Callari M, Valeri B, Pierotti MA, Daidone MG. Impact of biospecimens handling on biomarker research in breast cancer. BMC Cancer. 2009;9:409. doi: 10.1186/1471-2407-9-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jewell SD, Srinivasan M, McCart LM, Williams N, Grizzle WH, LiVolsi V, MacLennan G, Sedmak DD. Analysis of the molecular quality of human tissues: an experience from the Cooperative Human Tissue Network. Am J Clin Pathol. 2002;118:733–741. doi: 10.1309/VPQL-RT21-X7YH-XDXK. [DOI] [PubMed] [Google Scholar]
- 125.Medeiros F, Rigl CT, Anderson GG, Becker SH, Halling KC. Tissue handling for genome-wide expression analysis: a review of the issues, evidence, and opportunities. Arch Pathol Lab Med. 2007;131:1805–1816. doi: 10.5858/2007-131-1805-THFGEA. [DOI] [PubMed] [Google Scholar]
- 126.Botling J, Edlund K, Segersten U, Tahmasebpoor S, Engstrom M, Sundstrom M, Malmstrom PU, Micke P. Impact of thawing on RNA integrity and gene expression analysis in fresh frozen tissue. Diagn Mol Pathol. 2009;18:44–52. doi: 10.1097/PDM.0b013e3181857e92. [DOI] [PubMed] [Google Scholar]
- 127.Rupp C, Dolznig H, Puri C, Schweifer N, Sommergruber W, Kraut N, Rettig WJ, Kerjaschki D, Garin-Chesa P. Laser capture microdissection of epithelial cancers guided by antibodies against fibroblast activation protein and endosialin. Diagn Mol Pathol. 2006;15:35–42. doi: 10.1097/00019606-200603000-00006. [DOI] [PubMed] [Google Scholar]
- 128.To MD, Done SJ, Redston M, Andrulis IL. Analysis of mRNA from microdissected frozen tissue sections without RNA isolation. Am J Pathol. 1998;153:47–51. doi: 10.1016/S0002-9440(10)65544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Metz B, Kersten GF, Hoogerhout P, Brugghe HF, Timmermans HA, de Jong A, Meiring H, ten Hove J, Hennink WE, Crommelin DJ, Jiskoot W. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- 130.Mason JT, O’Leary TJ. Effects of formaldehyde fixation on protein secondary structure: a calorimetric and infrared spectroscopic investigation. J Histochem Cytochem. 1991;39:225–229. doi: 10.1177/39.2.1987266. [DOI] [PubMed] [Google Scholar]
- 131.Berod A, Hartman BK, Pujol JF. Importance of fixation in immunohistochemistry: use of formaldehyde solutions at variable pH for the localization of tyrosine hydroxylase. J Histochem Cytochem. 1981;29:844–850. doi: 10.1177/29.7.6167611. [DOI] [PubMed] [Google Scholar]
- 132.Hoetelmans RW, van Slooten HJ, Keijzer R, van de Velde CJ, van Dierendonck JH. Routine formaldehyde fixation irreversibly reduces immunoreactivity of Bcl-2 in the nuclear compartment of breast cancer cells, but not in the cytoplasm. Appl Immunohistochem Mol Morphol. 2001;9:74–80. [PubMed] [Google Scholar]
- 133.Williams C, Ponten F, Moberg C, Soderkvist P, Uhlen M, Ponten J, Sitbon G, Lundeberg J. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Plenat F, Montagne K, Weinbreck N, Corby S, Champigneulle J, Antunes L, Bonnet C, Maire C, Monhoven N. [Molecular consequences of fixation and tissue processing: the examples of nucleic acids and proteins] . Ann Pathol. 2006;26:8–21. doi: 10.1016/s0242-6498(06)70655-1. [DOI] [PubMed] [Google Scholar]
- 135.Bartkova J, Bartek J, Lukas Z, Vojtesek B, Staskova Z, Bursova H, Pavlovska R, Rejthar A, Kovarik J. Effects of tissue fixation conditions and protease pretreatment on immunohistochemical performance of a large series of new anti-keratin monoclonal antibodies: value in oncopathology. Neoplasma. 1991;38:439–446. [PubMed] [Google Scholar]
- 136.De Marzo AM, Fedor HH, Gage WR, Rubin MA. Inadequate formalin fixation decreases reliability of p27 immunohistochemical staining: probing optimal fixation time using high-density tissue microarrays. Hum Pathol. 2002;33:756–760. doi: 10.1053/hupa.2002.126187. [DOI] [PubMed] [Google Scholar]
- 137.Su JM, Perlaky L, Li XN, Leung HC, Antalffy B, Armstrong D, Lau CC. Comparison of ethanol versus formalin fixation on preservation of histology and RNA in laser capture microdissected brain tissues. Brain Pathol. 2004;14:175–182. doi: 10.1111/j.1750-3639.2004.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Crisan D, Mattson JC. Retrospective DNA analysis using fixed tissue specimens. DNA Cell Biol. 1993;12:455–464. doi: 10.1089/dna.1993.12.455. [DOI] [PubMed] [Google Scholar]
- 139.Warford A, Pringle JH, Hay J, Henderson SD, Lauder I. Southern blot analysis of DNA extracted from formal-saline fixed and paraffin wax embedded tissue. J Pathol. 1988;154:313–320. doi: 10.1002/path.1711540406. [DOI] [PubMed] [Google Scholar]
- 140.Foss RD, Guha-Thakurta N, Conran RM, Gutman P. Effects of fixative and fixation time on the extraction and polymerase chain reaction amplification of RNA from paraffin-embedded tissue. Comparison of two housekeeping gene mRNA controls. Diagn Mol Pathol. 1994;3:148–155. doi: 10.1097/00019606-199409000-00003. [DOI] [PubMed] [Google Scholar]
- 141.Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, Wong DT, Jordan RC. Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod Pathol. 2002;15:979–987. doi: 10.1097/01.MP.0000026054.62220.FC. [DOI] [PubMed] [Google Scholar]
- 142.Dubeau L, Chandler LA, Gralow JR, Nichols PW, Jones PA. Southern blot analysis of DNA extracted from formalin-fixed pathology specimens. Cancer Res. 1986;46:2964–2969. [PubMed] [Google Scholar]
- 143.Krafft AE, Duncan BW, Bijwaard KE, Taubenberger JK, Lichy JH. Optimization of the Isolation and Amplification of RNA From Formalin-fixed, Paraffin-embedded Tissue: The Armed Forces Institute of Pathology Experience and Literature Review. Mol Diagn. 1997;2:217–230. doi: 10.1054/MODI00200217. [DOI] [PubMed] [Google Scholar]
- 144.Ikeda K, Monden T, Kanoh T, Tsujie M, Izawa H, Haba A, Ohnishi T, Sekimoto M, Tomita N, Shiozaki H, Monden M. Extraction and analysis of diagnostically useful proteins from formalin-fixed, paraffin-embedded tissue sections. J Histochem Cytochem. 1998;46:397–403. doi: 10.1177/002215549804600314. [DOI] [PubMed] [Google Scholar]
- 145.Casasco A, Casasco M, Cornaglia AI, Danova M, Giordano M, Calligaro A. Tissue fixation for immunohistochemical detection of proliferating cell nuclear antigen with PC10 monoclonal antibody. Biotech Histochem. 1994;69:112–117. doi: 10.3109/10520299409106270. [DOI] [PubMed] [Google Scholar]
- 146.Greer CE, Wheeler CM, Manos MM. Sample preparation and PCR amplification from paraffin-embedded tissues. PCR Methods Appl. 1994;3:S113–122. doi: 10.1101/gr.3.6.s113. [DOI] [PubMed] [Google Scholar]
- 147.Lykidis D, Van Noorden S, Armstrong A, Spencer-Dene B, Li J, Zhuang Z, Stamp GW. Novel zinc-based fixative for high quality DNA, RNA and protein analysis. Nucleic Acids Res. 2007;35:e85. doi: 10.1093/nar/gkm433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.National Toxicology Program. Final report on carcinogens background document for formaldehyde. Rep Carcinog Backgr Doc. 2010:i–512. [PubMed] [Google Scholar]
- 149.Speit G, Schmid O, Neuss S, Schutz P. Genotoxic effects of formaldehyde in the human lung cell line A549 and in primary human nasal epithelial cells. Environ Mol Mutagen. 2008;49:300–307. doi: 10.1002/em.20386. [DOI] [PubMed] [Google Scholar]
- 150.Stanta G, Mucelli SP, Petrera F, Bonin S, Bussolati G. A novel fixative improves opportunities of nucleic acids and proteomic analysis in human archive’s tissues. Diagn Mol Pathol. 2006;15:115–123. doi: 10.1097/00019606-200606000-00009. [DOI] [PubMed] [Google Scholar]
- 151.Umlas J, Tulecke M. The effects of glyoxal fixation on the histological evaluation of breast specimens. Hum Pathol. 2004;35:1058–1062. doi: 10.1016/j.humpath.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 152.Vincek V, Nassiri M, Nadji M, Morales AR. A tissue fixative that protects macromolecules (DNA, RNA, and protein) and histomorphology in clinical samples. Lab Invest. 2003;83:1427–1435. doi: 10.1097/01.lab.0000090154.55436.d1. [DOI] [PubMed] [Google Scholar]
- 153.Olert J, Wiedorn KH, Goldmann T, Kuhl H, Mehraein Y, Scherthan H, Niketeghad F, Vollmer E, Muller AM, Muller-Navia J. HOPE fixation: a novel fixing method and paraffin-embedding technique for human soft tissues. Pathol Res Pract. 2001;197:823–826. doi: 10.1078/0344-0338-00166. [DOI] [PubMed] [Google Scholar]
- 154.Delfour C, Roger P, Bret C, Berthe ML, Rochaix P, Kalfa N, Raynaud P, Bibeau F, Maudelonde T, Boulle N. RCL2, a new fixative, preserves morphology and nucleic acid integrity in paraffin-embedded breast carcinoma and microdissected breast tumor cells. J Mol Diagn. 2006;8:157–169. doi: 10.2353/jmoldx.2006.050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Janin A, Murata H, Leboeuf C, Cayuela JM, Gluckman E, Legres L, Desveaux A, Varna M, Ratajczak P, Soulier J, de The H, Bertheau P, Socie G. Donor-derived oral squamous cell carcinoma after allogeneic bone marrow transplantation. Blood. 2009;113:1834–1840. doi: 10.1182/blood-2008-07-171702. [DOI] [PubMed] [Google Scholar]
- 156.Rivet J, Mourah S, Murata H, Mounier N, Pisonero H, Mongiat-Artus P, Teillac P, Calvo F, Janin A, Dosquet C. VEGF and VEGFR-1 are coexpressed by epithelial and stromal cells of renal cell carcinoma. Cancer. 2008;112:433–442. doi: 10.1002/cncr.23186. [DOI] [PubMed] [Google Scholar]
- 157.Varna M, Soliman H, Feugeas JP, Turpin E, Chapelin D, Legres L, Plassa LF, de Roquancourt A, Espie M, Misset JL, Janin A, de The H, Bertheau P. Changes in allelic imbalances in locally advanced breast cancers after chemotherapy. Br J Cancer. 2007;97:1157–1164. doi: 10.1038/sj.bjc.6603937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 1999;25:86–91. [PubMed] [Google Scholar]
- 159.Perlmutter MA, Best CJ, Gillespie JW, Gathright Y, Gonzalez S, Velasco A, Linehan WM, Emmert-Buck MR, Chuaqui RF. Comparison of snap freezing versus ethanol fixation for gene expression profiling of tissue specimens. J Mol Diagn. 2004;6:371–377. doi: 10.1016/S1525-1578(10)60534-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ahram M, Flaig MJ, Gillespie JW, Duray PH, Linehan WM, Ornstein DK, Niu S, Zhao Y, Petricoin EF 3rd, Emmert-Buck MR. Evaluation of ethanol-fixed, paraffin-embedded tissues for proteomic applications. Proteomics. 2003;3:413–421. doi: 10.1002/pmic.200390056. [DOI] [PubMed] [Google Scholar]
- 161.Bostwick DG, al Annouf N, Choi C. Establishment of the formalin-free surgical pathology laboratory. Utility of an alcohol-based fixative. Arch Pathol Lab Med. 1994;118:298–302. [PubMed] [Google Scholar]
- 162.Sanders CT, Sanchez N, Ballantyne J, Peterson DA. Laser microdissection separation of pure spermatozoa from epithelial cells for short tandem repeat analysis. J Forensic Sci. 2006;51:748–757. doi: 10.1111/j.1556-4029.2006.00180.x. [DOI] [PubMed] [Google Scholar]
- 163.Ben-Ezra J, Johnson DA, Rossi J, Cook N, Wu A. Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J Histochem Cytochem. 1991;39:351–354. doi: 10.1177/39.3.1704393. [DOI] [PubMed] [Google Scholar]
- 164.Xiang CC, Mezey E, Chen M, Key S, Ma L, Brownstein MJ. Using DSP, a reversible cross-linker, to fix tissue sections for immunostaining, microdissection and expression profiling. Nucleic Acids Res. 2004;32:e185. doi: 10.1093/nar/gnh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Namimatsu S, Ghazizadeh M, Sugisaki Y. Reversing the effects of formalin fixation with citraconic anhydride and heat: a universal antigen retrieval method. J Histochem Cytochem. 2005;53:3–11. doi: 10.1177/002215540505300102. [DOI] [PubMed] [Google Scholar]
- 166.Cataloluk O, Cakmak EA, Buyukberber N, Barlas O. Formalin fixing and paraffin embedding may lead to extra band development in PCR. New Microbiol. 2003;26:193–198. [PubMed] [Google Scholar]
- 167.Bertheau P, Cazals-Hatem D, Meignin V, de Roquancourt A, Verola O, Lesourd A, Sene C, Brocheriou C, Janin A. Variability of immunohistochemical reactivity on stored paraffin slides. J Clin Pathol. 1998;51:370–374. doi: 10.1136/jcp.51.5.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Becker KF, Schott C, Becker I, Hofler H. Guided protein extraction from formalin-fixed tissues for quantitative multiplex analysis avoids detrimental effects of histological stains. Proteomics Clin Appl. 2008;2:737–743. doi: 10.1002/prca.200780106. [DOI] [PubMed] [Google Scholar]
- 169.Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, Geho DH, Petricoin EF 3rd, Liotta LA. Laser-capture microdissection. Nat Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 170.Gjerdrum LM, Hamilton-Dutoit S. Laser-assisted microdissection of membrane-mounted sections following immunohistochemistry and in situ hybridization. Methods Mol Biol. 2005;293:139–149. doi: 10.1385/1-59259-853-6:139. [DOI] [PubMed] [Google Scholar]
- 171.Micke P, Bjornsen T, Scheidl S, Stromberg S, Demoulin JB, Ponten F, Ostman A, Lindahl P, Busch C. A fluid cover medium provides superior morphology and preserves RNA integrity in tissue sections for laser microdissection and pressure catapulting. J Pathol. 2004;202:130–138. doi: 10.1002/path.1496. [DOI] [PubMed] [Google Scholar]
- 172.Murase T, Inagaki H, Eimoto T. Influence of histochemical and immunohistochemical stains on polymerase chain reaction. Mod Pathol. 2000;13:147–151. doi: 10.1038/modpathol.3880028. [DOI] [PubMed] [Google Scholar]
- 173.Burton MP, Schneider BG, Brown R, Escamilla-Ponce N, Gulley ML. Comparison of histologic stains for use in PCR analysis of microdissected, paraffin-embedded tissues. Biotechniques. 1998;24:86–92. doi: 10.2144/98241st01. [DOI] [PubMed] [Google Scholar]
- 174.Okuducu AF, Janzen V, Hahne JC, Ko Y, Wernert N. Influence of histochemical stains on quantitative gene expression analysis after laser-assisted microdissection. Int J Mol Med. 2003;11:449–453. [PubMed] [Google Scholar]
- 175.Kirana C, Ward T, Jordan TW, Rawson P, Royds J, Shi HJ, Stubbs R, Hood K. Compatibility of toluidine blue with laser microdissection and saturation labeling DIGE. Proteomics. 2009;9:485–490. doi: 10.1002/pmic.200800197. [DOI] [PubMed] [Google Scholar]
- 176.Li C, Hong Y, Tan YX, Zhou H, Ai JH, Li SJ, Zhang L, Xia QC, Wu JR, Wang HY, Zeng R. Accurate qualitative and quantitative proteomic analysis of clinical hepatocellular carcinoma using laser capture microdissection coupled with isotope-coded affinity tag and two-dimensional liquid chromatography mass spectrometry. Mol Cell Proteomics. 2004;3:399–409. doi: 10.1074/mcp.M300133-MCP200. [DOI] [PubMed] [Google Scholar]
- 177.Burgemeister R, Gangnus R, Haar B, Schutze K, Sauer U. High quality RNA retrieved from samples obtained by using LMPC (laser microdissection and pressure catapulting) technology. Pathol Res Pract. 2003;199:431–436. doi: 10.1078/0344-0338-00442. [DOI] [PubMed] [Google Scholar]
- 178.Ordway GA, Szebeni A, Duffourc MM, Dessus-Babus S, Szebeni K. Gene expression analyses of neurons, astrocytes, and oligodendrocytes isolated by laser capture microdissection from human brain: detrimental effects of laboratory humidity. J Neurosci Res. 2009;87:2430–2438. doi: 10.1002/jnr.22078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kube DM, Savci-Heijink CD, Lamblin AF, Kosari F, Vasmatzis G, Cheville JC, Connelly DP, Klee GG. Optimization of laser capture microdissection and RNA amplification for gene expression profiling of prostate cancer. BMC Mol Biol. 2007;8:25. doi: 10.1186/1471-2199-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.DeCarlo CA, Escott NG, Werner J, Robinson K, Lambert PF, Law RD, Zehbe I. Gene expression analysis of interferon kappa in laser capture microdissected cervical epithelium. Anal Biochem. 2008;381:59–66. doi: 10.1016/j.ab.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bonsib SM, Reznicek MJ. Renal biopsy frozen section: a fluorescent study of hematoxylin and eosin-stained sections. Mod Pathol. 1990;3:204–210. [PubMed] [Google Scholar]
- 182.Murphy KM, Berg KD, Geiger T, Hafez M, Flickinger KA, Cooper L, Pearson P, Eshleman JR. Capillary electrophoresis artifact due to eosin: implications for the interpretation of molecular diagnostic assays. J Mol Diagn. 2005;7:143–148. doi: 10.1016/S1525-1578(10)60021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tangrea MA, Chuaqui RF, Gillespie JW, Ahram M, Gannot G, Wallis BS, Best CJ, Linehan WM, Liotta LA, Pohida TJ, Bonner RF, Emmert-Buck MR. Expression microdissection: operator-independent retrieval of cells for molecular profiling. Diagn Mol Pathol. 2004;13:207–212. doi: 10.1097/01.pdm.0000135964.31459.bb. [DOI] [PubMed] [Google Scholar]
- 184.Gjerdrum LM, Lielpetere I, Rasmussen LM, Bendix K, Hamilton-Dutoit S. Laser-assisted microdissection of membrane-mounted paraffin sections for polymerase chain reaction analysis: identification of cell populations using immunohistochemistry and in situ hybridization. J Mol Diagn. 2001;3:105–110. doi: 10.1016/S1525-1578(10)60659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fink L, Kinfe T, Seeger W, Ermert L, Kummer W, Bohle RM. Immunostaining for cell picking and real-time mRNA quantitation. Am J Pathol. 2000;157:1459–1466. doi: 10.1016/S0002-9440(10)64784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Burbach GJ, Dehn D, Nagel B, Del Turco D, Deller T. Laser microdissection of immunolabeled astrocytes allows quantification of astrocytic gene expression. J Neurosci Methods. 2004;138:141–148. doi: 10.1016/j.jneumeth.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 187.von Smolinski D, Blessenohl M, Neubauer C, Kalies K, Gebert A. Validation of a novel ultra-short immunolabeling method for high-quality mRNA preservation in laser microdissection and real-time reverse transcriptase-polymerase chain reaction. J Mol Diagn. 2006;8:246–253. doi: 10.2353/jmoldx.2006.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tangrea MA, Hanson JC, Bonner RF, Pohida TJ, Rodriguez-Canales J, Emmert-Buck MR. Immunoguided microdissection techniques. Methods Mol Biol. 2011;755:57–66. doi: 10.1007/978-1-61779-163-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 190.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bova GS, Eltoum IA, Kiernan JA, Siegal GP, Frost AR, Best CJ, Gillespie JW, Su GH, Emmert-Buck MR. Optimal molecular profiling of tissue and tissue components: defining the best processing and microdissection methods for biomedical applications. Mol Biotechnol. 2005;29:119–152. doi: 10.1385/MB:29:2:119. [DOI] [PubMed] [Google Scholar]


