Abstract
Background: MicroRNAs (miRNAs) are small, non-coding RNAs (18-25 nucleotides) that post-transcriptionally modulate gene expression by negatively regulating the stability or translational efficiency of their target mRNAs. The aim of this study was to investigate the expression pattern of microRNA-107 (miR-107) in human breast cancer, and its potential role in disease pathogenesis. Methods: Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to determine the expression level of miR-107 in 30 breast cancer specimens and adjacent normal breast tissues. MTT and colony formation assays, transwell and wound healing test, cell cycle assays were conducted to explore the potential function of miR-107 in human MDA-MB-231 breast cancer cells. Luciferase reporter assays were employed to validate regulation of a putative target of miR-107. The effect of modulating miR-107 on endogenous levels of this target were subsequently confirmed via Western blotting. Results: miR-107 expression was relatively decreased in breast cancer specimens compared with adjacent normal tissues (P<0.01). Overexpression of miR-107 suppressed MDA-MB-231 cell proliferation and migration, meanwhile the cells were arrested at G0/G1 phase. Luciferase assays using a reporter carrying a putative miR-107 target site in the 3’, untranslated region (3’-UTR) of CDK8 revealed that miR-107 directly targets CDK8. Overexpression of miR-107 led to downregulation of CDK8 at the mRNA and protein level, as assessed by Western blotting. Conclusions: miR-107 may play an important role in breast cancer progression, which might negatively regulate the expression of CDK8 and inhibit the proliferation and migration of MDA-MB-231 cell line.
Keywords: Breast cancer, miR-107, CDK8
Introduction
Breast cancer is a common highly heterogeneous malignancy, and is one of the main gynecological cancers in the world [1]. miRNAs are a class of 18 to 25 nucleotides single-stranded non-coding RNA, and can regulate gene expression at post-transcriptional level through inhibiting protein translation or degrading mRNA of target gene [2]. There has been significant evidence showing that miRNAs regulate as many as 30% of the human protein coding genes [3]. Current molecular cancer research has mostly focused on miRNA function in the regulation of tumor initiation, development and molecular targeted therapy. About half of the miRNA upstream genes locate in tumor-associated region on chromosome, and miRNA are abnormally expressed in a variety of tumors, suggesting that miRNAs may function as a tumor suppressor gene or oncogene. In the past few decades, it has been shown that miR-107 acts as a tumor suppressor gene in several tumor development process, but so far the role and mechanism of miR-107 in breast cancer growth, invasion and metastasis are still unclear [4-6].
CDK8 (cyclin dependent kinase 8) locating on chromosome13q12 has five transcripts and only one transcript encodes protein product containing 464 amino acid residues (molecular weight 53.2 kD). CDK8 has important function on the regulation of gene transcription [7]. Recent studies suggest that CDK8 is important in the process of tumor development [8]. CDK8 plays a key role in the regulation of cell cycle and cell growth on post-transcriptional level, and promotes the development and progression of colorectal cancer [9]. But the role of CDK8 in breast cancer has not been reported yet.
This study explored the function of miRNA-107 in human breast cancer cell MDA-MB-231 and underlying mechanism. Firstly, total RNAs of collected 30 cases of breast invasive ductal carcinoma were extracted and used to detect miR-107 expression in breast cancer tissues by qRT-PCR. Then the effects of miR-107 on proliferation activity, migration and cell cycle of MDA-MB-231 were verified by in vitro cell culture experiments, which further clarified the function of miRNA-107 in breast cancer development. Finally, miR-107 downstream target gene CDK8 was figured out through online prediction software, and specific binding between miR-107 and CDK8 was confirmed by luciferase reporter gene assay. Besides, Western blotting further validated that miR-107 negatively regulated CDK8 expression.
Material and methods
Specimens
30 cases of breast cancer specimens and adjacent normal breast tissue (>5 cm apart from cancer tissue) were obtained from department of Thyroid and Breast Surgery, Shanghai Tenth People’s Hospital. Removed specimens were immediately stored in liquid nitrogen. All specimens were pathologically confirmed as invasive ductal carcinoma, and all patients did not receive chemotherapy and radiotherapy.
Cell line
Human breast cancer cell line MDA-MB-231 was purchased from Chinese Academy of Sciences in Shanghai, human embryonic kidney HEK293 cells were purchased from American Type Culture Collection (ATCC).
Reagents
DMEM/high glucose medium and fetal bovine serum were purchased from Hyclone. miR-107 mimics and negative control (NC) were purchased from Shanghai GenePharma Ltd. Lipofectamine 2000 reagents were purchased from Invitrogen. CDK8 3’-UTR plasmid was constructed by Shanghai Sbo-bio Company. Dual luciferase reporter gene assay kit was purchased from Promega. BCA protein concentration assay kit was purchased from Beyotime Biotechnology Research Institute. CDK8 monoclonal antibody was purchased from Cell Signaling Company. MTT cell proliferation assay reagents were purchased from Sigma. Transwell chambers were purchased from Corning. The remaining chemical reagents were all analytical grade products.
Cells culture
Human breast cancer cells MDA-MB-231 were cultured in DMEM/high glucose medium containing 10% fetal bovine serum at 37°C, 5% CO2. Cell were passaged every other day using 25 cm2 cell culture flasks and maintained in good condition.
Total RNA extraction and qRT-PCR
Breast cancer and normal tissues removed from liquid nitrogen were grounded in liquid nitrogen and processed according to instructions of miRcute microRNA Extraction Kit from TIANGEN Company. Extracted total RNA was placed at -20°C. Reverse transcription reaction reagents were prepared according to instructions of KaPa kit. RT conditions: 37°C, 15 min; 85°C, 5 sec. Synthesized cDNA was used as template for qPCR amplification. Premix, miRNA-107 primer and ROX1 were added into 10 μl reaction system according TaKaRa kit instruction. qPCR conditions: 95°C, 5 min; 95°C, 5 s for 40 cycles; 60°C, 30 s. Fluorescence was analyzed. 10 μl reaction system. GAPDH was used as an internal control.
MTT assay for proliferation activity
MDA-MB-231 cells was cultured in 25 cm2 culture flask to approximately 80%-90% density collected by digestion and centrifugation, and then seeded into 96 well plates at 1,000 cells/well. 96-well plate was placed in cell culture incubator until cell monolayer reaches 40%-50% density. 50 nM or 100 nM mimics was transfected into cells according to Invitrogen Lipofectamine reagent instructions, with small RNA fragments transfected group (NC) as the negative control. 20 μl MTT solution (5 mg/ml) was added daily for 4 days. Supernatant was discarded and 150 μl DMSO (dimethyl sulfoxide) was added to each well. Then plates were placed on low-speed shaker for 10 min to fully dissolve the crystals. Absorbance at 490 nm was measured by the multi-plate reader. Experiment was performed with sextuplicates and repeated for 3 times.
Colony-forming unit assay
miR-107 mimics or negative control was transfected into MDA-MB-231 cells at 100 nM concentrations. 48 h later transfected cells were seeded into 6-well plates as 500 cell/well with triplicate and incubated at 37°C, 5% CO2 for 7 days until visible cloning were observed in the dish. Culture medium was discarded. Each well was washed with PBS twice carefully. Then cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet. After washed with running water three times and dried at room temperature, each well was observed and photographed. Cell colonies with more than 50 cells were counted under the microscope. Clone formation rate was calculated as following formula: Clone formation rate = number of formed colony / number of seeded cells × 100%.
Transwell experiment
In accordance with Transwell chamber instructions, DMEM/high glucose medium containing 10% FBS was added to the lower chamber while MDA-MB-231 cell suspension transfected with miR-107 mimics or NC for 48 h was added to the upper chamber. After incubation at 37°C, 5% CO2 for 12-18 h, the lower chamber was observed using inverted microscope. Incubation was terminated when cells passed into lower chamber. Inside of the upper chamber was cleaned with a cotton swab. Lower chamber was immersed and washed with PBS, fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, washed for three times with running water, and then photographed. Membrane-binding crystal violet was dissolved with 300 μl 33% glacial acetic acid, and then absorbance at 573 nm was measured using microplate reader.
Cell scratch assay
MDA-MB-231 cells was cultured in 25 cm2 culture flask to approximately 80%-90% confluency, collected by digestion and centrifugation, and then seeded into 6-well plates at 2.5×105 cells/well. 6-well plate was placed in cell culture incubator until cell monolayer reaches 40%-50% density. 100 nM miR-107 mimics was transfected into MDA-MB-231 cells. The transfected cells was incubated until the cell monolayer reached to 100% density. The bottom of 6-well plates was scratched with a P200 pipette tip and washed 3 times with PBS to wash off unattached cells. The width of scratch was observed at 0 h, 24 h and 48 h using inverted microscope (50-fold). Each experiment was repeated three times.
Cell cycle analysis
Transfection was performed as mentioned above. 48 h post transfection, cells were digested, resuspended, centrifuged at 4°C, 3200 r/min for 10 min and washed with PBS for three times. Then cells was stained with 250 μl 0.05 g/L propidium iodide (PI) in dark at room temperature for 30 min, and analyzed by flow cytometry (FACSCanto ™ II, BD Biosciences).
miR-107 target gene prediction and 3’-UTR plasmid vectors construction
miRNA target genes were predicted using online prediction software miRanda, TargetScan and PicTar. 3’-UTR region of CDK8 including miR-107 targeting sequence was amplified using PCR amplification. Upstream primer: 5’-GCCGGCATAGACGCGTGCTGCATCGGAATCTTGTC-3’, downstream primer: 5’-ATCCTTTATTAAGCTTACCACATACAAAGACAAATGCTT-3’ (Mlu I, Hind III restriction sites were underlined). Target sequence after T-A clone was sub-cloned into the vector pMIR and inserted into the downstream of firefly luciferase gene. This recombinant vector was named as pMIR-REPORT. All constructed plasmids were validated by restriction analysis and DNA sequencing.
Luciferase report gene detection
HEK293 cells in logarithmic growth phase were seeded into 96-well culture plate, incubated at 37°C, 5% CO2 for 24 h, and co-transfected with pMIR-REPORT and miR-107 mimics (or NC) using Lipofectamine 2000. Experiment was performed with sextuplicates and repeated for 3 times. Detection was performed following instruction of dual luciferase reporter gene assay kit. Cell culture medium was discarded after 24 h transfection. Wells were washed three times with PBS and treated by 20 μl cell lysis buffer for 15 min at room temperature. 100 μl firefly luciferase detection solution was added to detect firefly luciferase activity, then 100 μl renilla luciferase dection reagent was added to measure renilla luciferase activity. Luciferase activity (C) = firefly luciferase / renilla luciferase activity. Relative luciferase activity was calculated in each group.
Western blotting
Three groups of monolayer cells were washed with ice-cold PBS for 2 times, added RIPA lysis buffer [1% Triton X-100, 50 mmol/L (Tris pH 7.4), 150 mmol/L NaCl, 20 mmol/L iodoacetamide, 1 mmol/L PMSF, 1% aprotinin], lysed on ice for 30 min, scrapped off, transferred into EP tube and centrifuged at 4°C, 12000xg for 30 min. The supernatants were collected and protein concentration was determined using BCA protein quantitation kit. Each sample with 60 μg protein was added 1X SDS sample buffer [100 mmol/L Tri-HCl (pH 6.8), 4% SDS, 0.2% bromophenol blue, 20% glycerol, 200 mmol/L β-mercaptoethanol], and denatured at 95°C for 5 min. Proteins were separated by 10% SDS-PAGE electrophoresis and transferred to 0.45 μm NC membrane. Membrane was blocked with 5% skim milk for 1 h, and incubated with 1:1000 diluted CDK8 monoclonal antibodies at 4°C overnight. The membrane was then incubated with 1:1000 diluted β-actin antibody at room temperature for 1 h, washed with PBST three times, and detected with Odyssey system.
Statistical methods
Experimental data was showed as mean ± SD. Two groups were compared using t-test comparison and multi-groups were compared using variance analysis by SPSS17.0 statistical software. P<0.05 indicates significant difference.
Results
miR-107 in breast cancer tissues was reduced
To analyze miR-107 expression in breast cancer tissues, total RNA of 30 cases of invasive breast cancer and adjacent normal tissues were extracted and diluted to 10 ng/μl as template. qRT-PCR was performed to assess miR-107 expression with GAPDH as an internal control. The results showed that miR-107 in breast cancer tissues (2.645 ± 0.6832) significantly decreased compared with normal tissues (8.487 ± 0.8821, P<0.01) (Figure 1).
Figure 1.
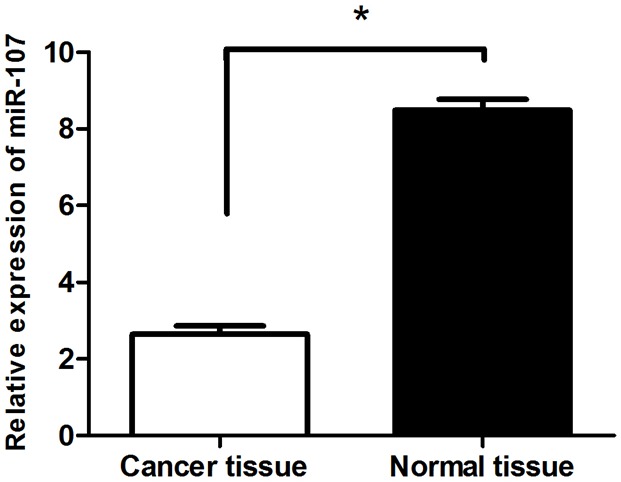
miR-107 was significantly decreased in breast cancer tissues. The mRNA expression of miR-107 was measured by qRT-PCR in breast cancer tissues (Cancer tissue) and adjacent normal breast tissue (Normal tissue). Graph represents the 2-ΔΔCt values + SD, p<0.01.
miR-107 inhibited proliferation of breast cancer cell MDA-MB-231
Breast cancer cell MDA-MB-231 were treated with 50 nM, 100 nM miR-107 mimics for 24 h, 48 h, 72 h or 96 h and measured the absorbance at 490 nm. Inhibition rate was calculated as following: inhibition rate (%) = (OD value of the control group - OD value of experimental group) / OD value of control group × 100%. Compared with the control group, miR-107 treated group was inhibited in a dose and time dependent manner. Cell proliferation was strongest inhibited when cells were treated with 100 nM miR-107 mimics for 72 h with the inhibition rate of 47.4% (P<0.01) (Figure 2).
Figure 2.
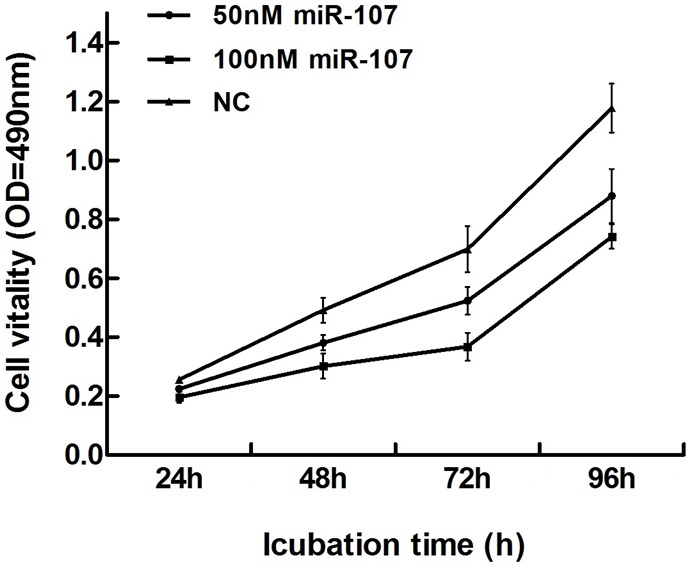
miR-107 inhibited cell proliferation. The MTT assay was performed to measure the proliferation level of MDA-MB-231 cells transfected with negative control (NC) or miR-107 mimics at the indicated concentrations. miR-107 had strongest inhibitory effect on cell proliferation (inhibition ratio up to 47.4%) at concentration of 100 nM for 72 h. Graph represented OD at 490 nm ± SD, p<0.01.
miR-107 decreased MDA-MB-231 clone formation rate
Clone formation rate of miR-107 mimics transfected group (9.667 ± 0.0213) was significantly lower than that of NC group (20.333 ± 0.0768), demonstrating that miR-107 mimics significantly inhibited MDA-MB-231 colony formation (P<0.05) (Figure 3).
Figure 3.
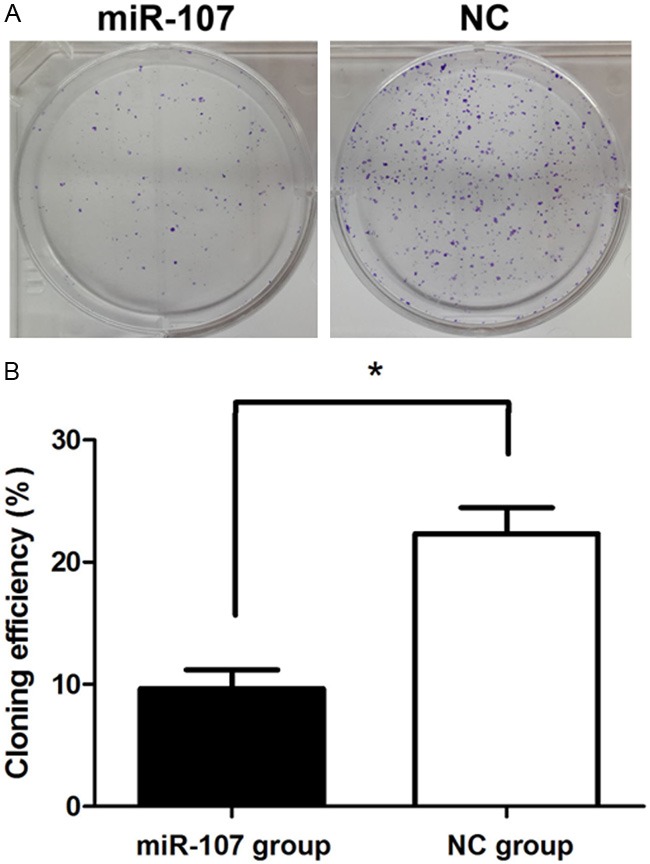
miR-107 inhibited cell colony formation. A: Representative images of crystal violet stained colonies in MDA-MB-231 cells transfected with miR-107 or NC. B: Cloning efficiency, P<0.05.
miR-107 inhibited MDA-MB-231 cell migration
Absorbance at 573 nm showed that tumor cells migrating out of chamber in 50 nM miR-107 mimics-treated group (0.216 ± 0.0046) was statistically reduced than negative control group (0.436 ± 0.0041), (P<0.01), indicating that miR-107 could inhibit tumor cell invasion (Figure 4). Cell scratch assay showed that cell transfected with miR-107 mimics migrated slowly. Scratch in control group was almost healed 48 h after scratch had been made but not in miR-107 mimics group. These data showed that miR-107 inhibited MDA-MB-231 cell migration (Figure 5).
Figure 4.
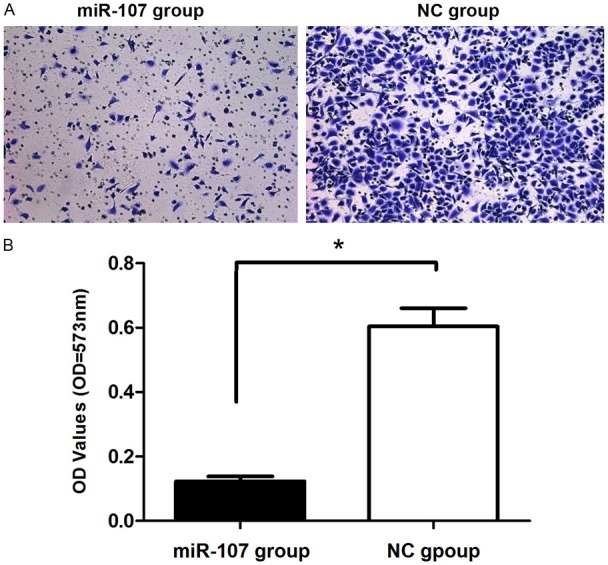
miR-107 inhibited migration of MDA-MB-231 breast cancer cells. Cell migration ability was analyzed by transwell chamber assay 18 h after miR-107 or NC transfection. A: Representative images of crystal violet stained MDA-MB-231 migratory cells transfected with miR-107 or NC. B: Quantification of the migratory cells by solubilization of crystal violet and spectrophotometric reading at OD 573 nm. Data represented mean + SD, p<0.01.
Figure 5.
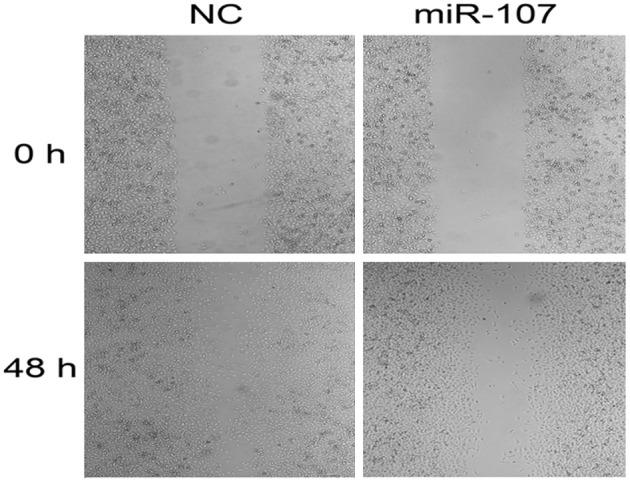
miR-107 inhibited scratch healing of MDA-MB-231 cells. Cell scratch assay was used to detect the migration of MDA-MB-231 cells. Scratch healed in NC group after 48 h, while scratch in miR-107 transfected group did not.
Cell cycle analysis
Flow cytometry analysis of MDA-MB-231 cells transfected with 150 nM miR-107 mimics for 48 h showed that cells in G0/G1 phase increased significantly while cells in G2/M phase or S phase cells had no significant change, suggesting that miR-107 arrested cell cycle mainly in G0/G1 phase (P<0.05) (Figure 6).
Figure 6.
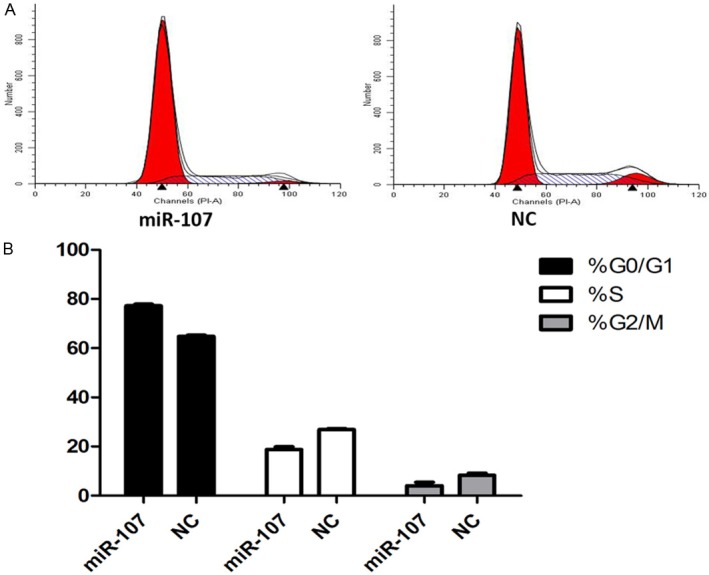
MiR-107 affected cell cycle distribution. A: Cell cycle distribution was analyzed by flow cytometry 48 h after transfection of MDA-MB-231 breast cancer cells with 100 nM miR-107 mimics or NC. B: The respective proportion of G0/G1 phase, S-phase and G2/M phase of miR-107 and NC groups, P<0.05.
Luciferase reporter gene assay
Comparison of luciferase activity in experimental group with negative control group showed that luciferase activity in MDA-MB-231 cells co-transfected with pMIR-REPORT and miR-107 mimics (3.183 ± 0.0336) was 34.7% of that in pMIR-REPORT and NC co-transfected group (9.166 ± 0.0904). This difference was statistically significant (P<0.01). This data showed that there was specific binding between miR-107 and 3’-UTR in CDK8 gene (Figure 7).
Figure 7.
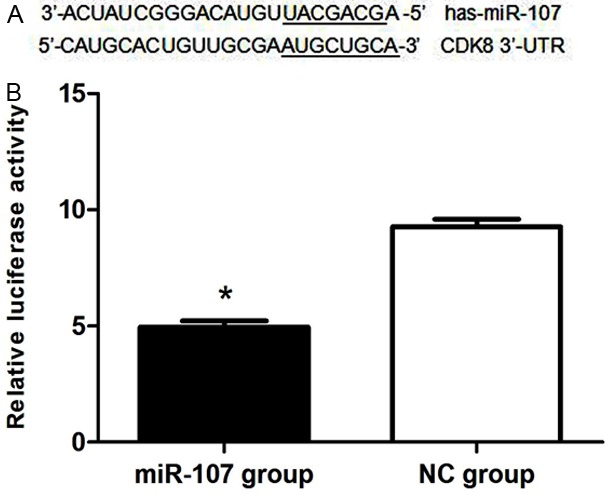
CDK8 was a direct target of miR-107. A: The binding site for miR-107 in the 3’-UTR of CDK8 mRNA. B: The relative luciferase activity (firefly/renilla) was measured in HEK293 cells after co-transfection of the CDK8 luciferase construct with either miR-107 or NC, p<0.01.
miR-107 negatively regulated CDK8 protein expression
CDK8 protein expression measured by Western blotting in miR-107 transfected MDA-MB-231 cells was significantly decreased compared with control group. Grayscale analysis showed that CDK8 protein expression was inhibited after transfection with the inhibition rate of 46.3% (P<0.05) (Figure 8).
Figure 8.
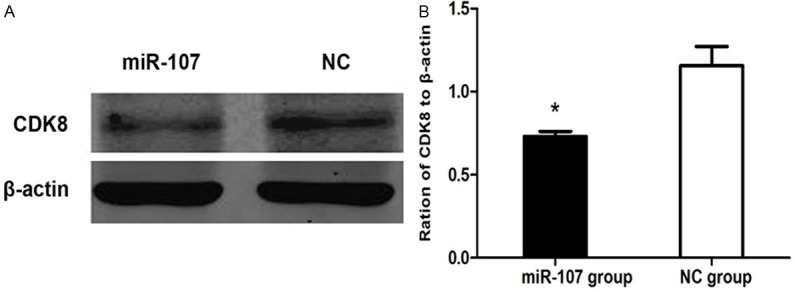
miR-107 inhibited CDK8 expression. A: Western blotting analysis of CDK8 protein level. β-actin was used as a loading control. B: The expression level of CDK8 in MDA-MB-231 cells, normalized by β-actin expression, P<0.05.
Discussion
Breast cancer is one of the most common malignant tumors in female. Currently, surgery and chemotherapy are the main treatments, but some patients accepted chemotherapy have early recurrence and metastasis resulting poor prognosis, especially the patients with negatively expressed estrogen, progesterone, and human epidermal growth factor (triple negative breast cancer, TNBC) [10]. Therefore it is especially important to explore TNBC-targeting treatment. It has been a global research hotspot to looking for new therapeutic targets for breast cancer treatment [11]. The discovery of the first miRNA, lin-4 in Caenorhabditis elegans initiated a new era of miRNA biology. Since then, thousands of miRNAs have been identified and annotated. Furthermore, an increasing body of evidence indicates that miRNAs are differentially expressed between normal and tumor tissues, suggesting that dysregulation of miRNA expression is a key factor underlying tumorigenesis [12-16]. A large number of studies suggest that understanding of miRNA function will provide us broad prospects to understand and overcome tumor. The key to study miRNA function is to determine its target. CDK8 is a member of CDK family (CDKs), which is a group of serine-threonine protein kinase and consists of 10 members with different homology. In the past decade, It has been showed that CDKs are excessively activated in different tumors [17]. Preclinical studies have proved that CDKs can promote gene transcription, cell differentiation and angiogenesis [18]. Recent study has shown that miR-107 targeting to CDK protein can inhibit cancer cell invasion [5]. But the role of miR-107 in breast cancer has not been reported.
miRNA functions through interacting with target gene thus the key to explore the mechanism of miRNA is to study the interaction between miRNA and its target gene. In this study, CDK8 was predicted to be the target gene of miR-107 by online biological software, then luciferase reporter vectors containing CDK8 gene 3’-UTR region with miR-107 binding site was constructed and specific binding between miR-107 and CDK8 was verified. However, it is still not clear about the exact function of miR-107, the impact of miR-107 on the biological behavior of breast cancer cells, and the underlying molecular mechanism. We over-expressed miR-107 in triple negative breast cancer cells MDA-MB-231, and Western blotting showed that CDK8 was significantly decreased in miR-107 over-expressed cells, indicating that miR-107 may affect CDK8 protein levels. This data confirmed that CDK8 is miR-107 target gene, and proved prediction data from bioinformatics software. Over-expression of miR-107 was found to be able to significantly inhibit breast cancer cell proliferation in the MTT assay, inhibit breast cancer cell migration in transwell and cell scratch experiments, and arrest cell cycle to G0/G1 phase. miRNA has now become a global cancer research hotspot [19,20], and this study strongly suggested that miR-107 could affect the function of TNBC cells line MDA-MB-231, although it needs to be studied further that if miR-107 functions by targeting to CDK8. In general, the research focusing on miR-107 and its target gene is expected to become a new direction of TNBC treatment exploration.
Conclusion
In summary, our study demonstrated that miR-107 is downregulated in breast cancer specimens compared with normal tissue. Up-regulation of miR-107 expression causes cellular growth inhibition, migration and G1 phase arrest by targeting CDK8. These data indicate that miR-107 may serve as a tumor suppressor gene involved in breast cancer pathogenesis.
Acknowledgements
This work was partially supported by grants from the National Natural Science Foundation of China (No. 81272240).
Disclosure of conflict of interest
None.
References
- 1.Nieto Y, Nawaz F, Jones RB, Shpall EJ, Nawaz S. Advanced breast cancer and breast cancer mortality in randomized controlled trials on mammography screening. J. Clin. Oncol. 2009;27:5919–5923. doi: 10.1200/JCO.2009.22.7041. [DOI] [PubMed] [Google Scholar]
- 2.Wu M, Jolicoeur N, Li Z, Zhang L, Fortin Y, L’Abbe D, Yu Z, Shen SH. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis. 2008;29:1710–1716. doi: 10.1093/carcin/bgn073. [DOI] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Chen PS, Su JL, Cha ST, Tarn WY, Wang MY, Hsu HC, Lin MT, Chu CY, Hua KT, Chen CN, Kuo TC, Chang KJ, Hsiao M, Chang YW, Chen JS, Yang PC, Kuo ML. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J Clin Invest. 2011;121:3442–3455. doi: 10.1172/JCI45390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol. 2012;29:856–863. doi: 10.1007/s12032-011-9823-1. [DOI] [PubMed] [Google Scholar]
- 6.Moncini S, Salvi A, Zuccotti P, Viero G, Quattrone A, Barlati S, De Petro G, Venturin M, Riva P. The role of miR-103 and miR-107 in regulation of CDK5R1 expression and in cellular migration. PLoS One. 2011;6:e20038. doi: 10.1371/journal.pone.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler AS, McCleland ML, Truong T, Lau S, Modrusan Z, Soukup TM, Roose-Girma M, Blackwood EM, Firestein R. CDK8 maintains tumor dedifferentiation and embryonic stem cell pluripotency. Cancer Res. 2012;72:2129–2139. doi: 10.1158/0008-5472.CAN-11-3886. [DOI] [PubMed] [Google Scholar]
- 8.Adler AS, McCleland ML, Truong T, Lau S, Modrusan Z, Soukup TM, Roose-Girma M, Blackwood EM, Firestein R. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, Chheda MG, Tamayo P, Finn S, Shrestha Y, Boehm JS, Jain S, Bojarski E, Mermel C, Barretina J, Chan JA, Baselga J, Tabernero J, Root DE, Fuchs CS, Loda M, Shivdasani RA, Meyerson M, Hahn WC. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan BB, Schnitt SJ, Collins LC. Ductal carcinoma in situ with basal-like phenotype: a possible precursor to invasive basal-like breast cancer. Mod Pathol. 2006;19:617–621. doi: 10.1038/modpathol.3800570. [DOI] [PubMed] [Google Scholar]
- 11.Ljungberg BJ, Jacobsen J, Rudolfsson SH, Lindh G, Grankvist K, Rasmuson T. Different vascular endothelial growth factor (VEGF), VEGF-receptor 1 and -2 mRNA expression profiles between clear cell and papillary renal cell carcinoma. BJU international. 2006;98:661–667. doi: 10.1111/j.1464-410X.2006.06387.x. [DOI] [PubMed] [Google Scholar]
- 12.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 13.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumor spheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposiscoli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Gong YH, Chao TF, Peng XZ, Yuan JG, Ma ZY, Jia G, Zhao JZ. Identification of differentially expressed microRNAs by microarray: a possible role for microRNAs gene in medulloblastomas. Chin Med J (Engl) 2009;122:2405–2411. [PubMed] [Google Scholar]
- 17.Sharma PS, Sharma R, Tyagi R. Inhibitiors of cyclin depentent kinases: useful targets for cancer treatment. Curr Cancer Drug Targets. 2008;8:53–75. doi: 10.2174/156800908783497131. [DOI] [PubMed] [Google Scholar]
- 18.Senderowicz AM, Sausville EA. Preclinical and clinical development of cyclin-dependent kinase modulators. J Natl Cancer Inst. 2000;92:376–387. doi: 10.1093/jnci/92.5.376. [DOI] [PubMed] [Google Scholar]
- 19.Tryfonopoulos D, Walsh S, Collins DM, Flanagan L, Quinn C, Corkery B, McDermott EW, Evoy D, Pierce A, O’Donovan N, Crown J, Duffy MJ. Src: a potential target for the treatment of triple-negative breast cancer. Ann Oncol. 2011;22:2234–2240. doi: 10.1093/annonc/mdq757. [DOI] [PubMed] [Google Scholar]
- 20.Oliveras-Ferraros C, Vazquez-Martin A, Lopez-Bonet E, Martin-Castillo B, Del Barco S, Brunet J, Menendez JA. Growth and molecular interactions of the anti-EGFR antibody cetuximab and the DNA cross-linking agent cisplatin in gefitinib-resistant MDA-MB-468 cells: new prospects in the treatment of triple-negative/basal-like breast cancer. Int J Oncol. 2008;33:1165–1176. [PubMed] [Google Scholar]


