Abstract
The aim of this study was to determine the correlation between expression of HPV16 E6, p53 and p21 proteins and the physical state of HPV16 in cervical cytologies without squamous intraepithelial lesions (Non-SIL) and with low grade squamous intraepithelial lesions (LSIL), both with HPV16 infection. 101 liquid-based cytological samples were analyzed. 50 samples were without squamous intraepithelial lesions (Non-IL) and 51 samples of low grade squamous intraepithelial lesions (LSIL), both with HPV16 infection. HPV16 infection was determined by PCR-RFLP, and the physical state of HPV16 by in situ hybridization with tyramide-amplification. The expression of E6, p53 and p21 proteins was evaluated by immunocytochemistry. The expression of HPV16 E6 protein was significantly higher in LSIL that in Non-SIL samples (p=0.006). We found a significant correlation between E6 expression and the physical state of HPV16 in Non-SIL (p=0.049). Our results suggest that high expression of E6 in LSIL is an early event of cervical carcinogenesis and perhaps can be used as an early marker.
Keywords: E6, p53, p21, HPV16, cervical cancer, LSIL, physical state, liquid-based cytological, immunocytochemistry, in situ hybridization
Introduction
Cervical cancer is characterized by the progression through well characterized squamous intraepithelial lesions (SIL) of the cervix. Based on cytopathologic characteristics, these lesions are divided into low grade (LSIL) and high grade (HSIL), and invasive cervical cancer [1]. Cervical cancer is a multistep process that slowly develops upon persistent infection with oncogenic types of human papillomavirus (HPV) [2,3]. In Mexico, HPV16 is the most frequent genotype found in cervical cancer [4]. Moreover, HPV16 is one of the most frequent genotypes found in LSIL, HSIL and in women without SIL [5].
HPV16 genome encodes two oncoproteins, E6 and E7. Both proteins are able to cause transformation of the host cell [6]. The E6 protein binds to p53 tumor suppressor and cause its degradation by 26S proteasome, resulting in its inactivation and the impairment of p53-induced cellular apoptosis [7]. Expression of E6 has been proposed as a useful diagnostic and/or prognostic marker in cervical carcinogenesis [8], although there are contradictory results [9]. p53 protein is known as the guardian of the genome, and plays an important role in cellular response to genotoxic stress [10]. This protein acts as a tumor suppression by a variety of mechanisms, including cell cycle arrest, induction of apoptosis, and cellular senescence [11]. Previous studies have evaluated p53 protein expression in cervical intraepithelial lesions and in invasive carcinomas, however the results are contradictory [12-15], making it difficult to establish whether p53 expression is a good biomarker in cervical carcinogenesis. The p21 protein is member of the Cip/Kip family, and is responsible for cell cycle control, blocking the transition from G1-phase to S-phase. The p21 gene is regulated through two different pathways, a p53-dependent pathway and a p53-independent way, through platelet-derived, fibroblast and epidermal growth factors [16,17]. Reduced expression of p21 protein by immunohistochemistry has been reported in invasive squamous cell carcinoma [18], and it has been suggested as a biomarker.
The HPV16 genome can be found in the host cell in episomal, integrated or mixed forms. HPV16 integration into the host genome results in increased levels of E6 and E7 proteins, and this event is considered a critical late-event in cervical carcinogenesis. The prevalence of episomal and integrated forms of HPV16 genome in cervical SIL, varies with severity of disease [19,20]. In general, the integration of HPV16 genome is considered a late event in cervical carcinogenesis [21].
Several studies have suggested that the immunocytochemical or immunohistochemical detection of p16 [9], p53, p21 [22], cyclin A, cyclin E [23], Ki-67 [24], telomerase [25], E6 [9] and the detection of physical state of HPV16 by ISH [23] in smears or cervical samples may provide useful diagnostic and prognostic information. The aim of this study was to determine the correlation between expression of E6 HPV16, p53 and p21, and the physical state of HPV16 in cervical cytologies without squamous intraepithelial lesions (Non-SIL) and with low grade squamous intraepithelial lesions (LSIL), both with infection by HPV16, to identify possible biomarkers of early cervical lesion.
Materials and methods
Subjects and specimen collection
101 liquid-based cervical cytology samples were collected from women residents in the State of Guerrero, in Southern Mexico. The study population consisted of 50 women diagnosed with Non-SIL and 51 diagnosed with LSIL, all positive to HPV16 by PCR. Exo-endocervical exfoliated cell samples were collected by sampling the ectocervix with an Ayre spatula and endocervix with a cytobrush. Immediately after sample collection smears were prepared for cytomorphological examination through conventional Papanicolaou staining. The remaining cellular content was preserved in liquid base liquid-PREPTM (LPT) and used for immunocytochemistry and ISH. A second sample was collected for DNA extraction. All samples Pap smears were evaluated by an experienced cytopathologist and were classified according to the Bethesda System [26]. All patients signed an informed consent and filled a questionnaire to obtain demographic data and information about gynecological risk factors. This project was approved by the Bioethics Committee of the Autonomous University of Guerrero, Mexico, and all procedures where in accordance with the ethical guidelines of the 2008 Helsinki Declaration.
HPV detection and genotyping
Genomic DNA was extracted from cervical cells by the phenol chloroform method [27]. Purified DNA was used to PCR-amplify a 450 pb conserved region of the HPV L1 gene using consensus primers MY09 and MY11 [28,29]. The reaction mixtures (50 μl) contained 0.8 μM of each primer, 2 mM MgCl2, 1X PCR buffer, 150 μM of each dNTP, 1.25 unit of AmpliTaqGoldTM (Applied Biosystems, Foster City, CA) and 500 ng of target DNA. DNA was amplified in GeneAmp PCR System 2400 (Applied Biosystems, Foster City, CA), under the following conditions: 95°C, 10 min; 40 amplification cycles (95°C, 1 min; 58°C, 1 min and 72°C, 1 min) and 72°C, 10 min. Integrity of DNA specimens was verified by amplification of a 268 bp region of the human β-globin gene using PC04 and GH20 primers [30]. HPV16 plasmid, and genomic DNA from CaSki and HeLa cells were used as positive controls. Genomic DNA without HPV DNA and water were used as negative controls. For HPV genotyping, amplified PCR products were digested with restriction enzymes BamHI, DdeI, HaeIII, HinfI, PstI, RsaI and Sau3AI (Invitrogen, Carlsbad, CA) and RFLP analysis was performed to identify more than 40 genital types of HPV [31].
Immunocytochemistry
The presence of E6, p53 and p21 proteins was determined by the streptavidin-biotin-peroxidase immunocytochemical method, utilizing the Cytoscan HRP/DAB Cell detection system (Cell Marque Corporation, Hot Springs, AR, USA). The monoclonal antibodies used were anti-E6 (clone C1P5; 1:50; Santa Cruz Biotechnology and Chemicon International, Inc.) anti-p53 (clone DO-7; 1:50; Dako, Carpinteria, CA, USA) and anti-p21 (clone SX118; 1:50; Dako, Carpinteria, CA, USA). The cytology slides in liquid base were subjected to antigen retrieval (Immuno DNA Retrieval with citrate, Bio SB Inc., Santa Barbara, CA, USA) for 3 minutes at 120°C. The primary antibody was added for 1 hour, and then the secondary antibody coupled with biotin was added followed by incubation with streptavidin peroxidase. The reaction was developed using the chromogen DAB and samples were counterstained with Mayer’s hematoxylin. Cervical cancer samples with HPV16 and normal cervical tissue were used as positive and negative controls for E6, p53 and p21 expression, respectively. Negative controls were processed without the primary antibody. The brown staining in the nucleus indicated positivity for E6 HPV16. The E6 expression was classified into four groups: negative, 0-9%; mild, 10-25%; moderate, 26-50% and intense, >51% positive cells [32]. The brown staining of the nucleus indicated positivity for p53 and p21. The p53 [33] and p21 [18] immunostaining were considerate as positive when >10% of cells were staining.
In situ hybridization
Detection of the viral genome was done with a system of tyramide signal amplification (GenPoint Dako Cytomation, Carpinteria, CA, USA). The monolayer smears were digested for 1 minute with proteinase K (1:1000). A drop of test reagent (biotinylated viral DNA) with probes for 13 HR-HPV genotypes (16, 18, 31, 33, 39, 45, 51, 52, 56, 58, 59 and 68) was added to each slide. The slides were denatured for 10 minutes and subjected to hybridization for 20 hours (Hybridizer Dako, Carpinteria, CA, USA). Samples were placed in an astringent solution, incubated with primary streptavidin peroxidase, followed by biotinyl– tyramide and then secondary streptavidin. The reaction was developed with DAB was added and samples were counterstained with Mayer’s hematoxylin (Merck). Positive reaction was visualized as a brown color inside the nucleus as diffuse (episomal state), punctate (integrated state) or mixed (diffuse and punctate) pattern. SiHa cell lines (HPV-16) which carry integrated HPV16 genome, were used as positive controls; the same cell line without the probe was used as negative control.
Statistical analysis
Statistical analysis was performed using the STATA 10.0 software package (Stat corporation, College Station, TX, USA). Comparison of proteins expression among Non-SIL and LSIL was done by Χ2 or Fisher exact test. All the correlations were analyzed by the Fisher exact test. A p value of <0.05 was considerate statistically significant.
Results
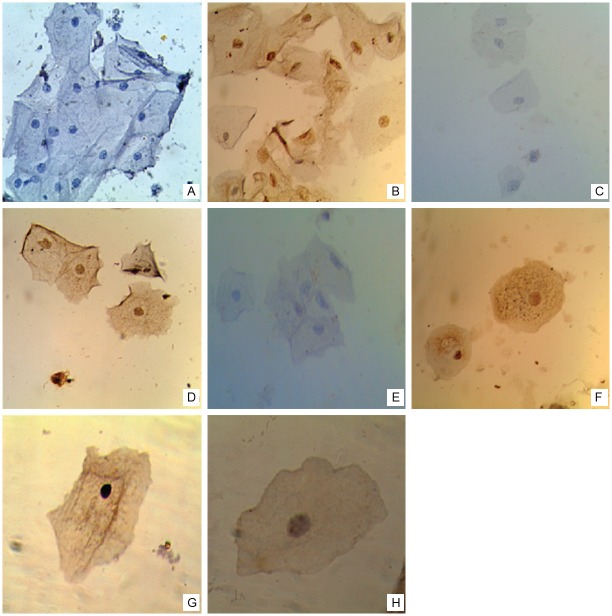
We analyzed the physical state of HPV16 and the expression of E6, p53 and p21 proteins in cervical samples from 101 women diagnosed with Non-SIL (50) or LSIL (51). The mean age of the study population was 41.2±9.6 years (range, 25-66 years) for Non-SIL and 36.7±12.5 years (range, 20-66 years) for LSIL. Table 1 shows the expression of E6, p53 and p21 proteins and the HPV16 physical state in Non-SIL and LSIL samples. The expression of HPV16 E6 protein was significantly higher in LSIL that Non-SIL (p=0.006). We found no significant differences in p53 and p21 expression or HPV16 physical state between Non-SIL and LSIL. Representative images of immunostaining for E6, p53, p21 and in situ hybridization for HPV16 physical state in Non-SIL and LSIL samples are shown in Figure 1.
Table 1.
Expression of E6, p53 and p21 proteins and HPV16 physical state in Non-SIL and LSIL
| Non-IL | LSIL | ||||
|---|---|---|---|---|---|
| n=50 | % | n=51 | % | P | |
| Expression of E6 | |||||
| Negative | 9 | 18.0 | 0 | 0.0 | 0.006a |
| Mild | 27 | 54.0 | 31 | 60.8 | |
| Moderate | 13 | 26.0 | 17 | 33.3 | |
| Intense | 1 | 2.0 | 3 | 5.9 | |
| Expression of p53 | |||||
| Negative | 29 | 58.0 | 34 | 66.7 | 0.369b |
| Positive | 21 | 42.0 | 17 | 33.3 | |
| Expression of p21 | |||||
| Negative | 41 | 82.0 | 45 | 88.2 | 0.378b |
| Positive | 9 | 18.0 | 6 | 11.8 | |
| HPV16 Physical state | |||||
| Episomal | 13 | 26.0 | 8 | 15.7 | 0.357b |
| Integrated | 11 | 22.0 | 10 | 19.6 | |
| Mixed | 26 | 52.0 | 33 | 64.7 |
HPV human papillomavirus, SIL squamous intraepithelial lesion, LSIL low-grade squamous intraepithelial lesion.
Fisher exact test.
Χ 2.
Significant value is indicated in bold.
Figure 1.
Expression of E6, p53 and 21, and physical state of HPV16 genome in cervical smears. Representative images of immunocytochemical staining for E6 (A, B), p53 (C, D) and p21 (E, F) proteins (40X). (A) Negative in Non-IL and (B) positive immunostaining for E6 in LSIL. (C) Negative and (D) positive immunostaining for p53 in Non-IL. (E) Negative and (F) positive immunostaining for p21 in LSIL. (G, H) Representative images of in situ hybridization for HPV16 genome (100X). (G) Diffuse signal pattern in Non-SIL, and (H) punctate signal pattern in LSIL.
The correlation between expression of E6 and the HPV16 physical state in Non-SIL and LSIL is shown in Table 2. We found a significant correlation between E6 expression and the HPV16 physical state in Non-SIL (p=0.049) but not in LSIL (p=0.335). The correlation between expression of E6 and p21 and expression of p53 in Non-SIL and LSIL is shown in Table 3. We did not find any correlation between expression of E6 and p21 and expression of p53 in Non-SIL or LSIL.
Table 2.
Correlation between expression of E6 and the HPV16 physical state in Non-SIL and LSIL
| Non-SIL n=50 | LSIL n=51 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Physical state | Physical state | |||||||||||||
|
|
|
|||||||||||||
| Episomal n | % | Integrated n | % | Mixed n | % | P | Episomal n | % | Integrated n | % | Mixed n | % | P | |
| Expression of E6 | ||||||||||||||
| Negative | 4 | 30.8 | 1 | 9.1 | 4 | 15.4 | 0.049 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0.335 |
| Mild | 9 | 69.2 | 6 | 54.5 | 12 | 46.1 | 7 | 87.5 | 4 | 40.0 | 20 | 60.6 | ||
| Moderate | 0 | 0.0 | 3 | 27.3 | 10 | 38.5 | 1 | 12.5 | 5 | 50.0 | 11 | 33.3 | ||
| Intense | 0 | 0.0 | 1 | 9.1 | 0 | 0.0 | 0 | 0.0 | 10.0 | 2 | 6.1 | |||
SIL squamous intraepithelial lesion, LSIL low-grade squamous intraepithelial lesion. P was calculate using Fisher exact test. Significant value is indicated in bold.
Table 3.
Correlation between expression of E6 and p21 and expression of p53 in Non-SIL and LSIL
| Non-SIL n=50 | LSIL n=51 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p53 | p53 | |||||||||
|
|
|
|||||||||
| Negative n | % | Positive n | % | P | Negative n | % | Positive n | % | P | |
| Expression of E6 | ||||||||||
| Negative | 3 | 10.3 | 6 | 28.6 | 0.128 | 0 | 0.0 | 0 | 0.0 | 0.563 |
| Mild | 19 | 65.5 | 8 | 38.1 | 19 | 55.9 | 12 | 70.6 | ||
| Moderate | 7 | 24.2 | 6 | 28.6 | 12 | 35.3 | 5 | 29.4 | ||
| Intense | 0 | 0.0 | 1 | 4.8 | 3 | 8.8 | 0 | 0.0 | ||
| p21 | ||||||||||
| Negative | 24 | 82.8 | 17 | 80.9 | 1.000 | 30 | 88.2 | 15 | 88.2 | 1.000 |
| Positive | 5 | 17.2 | 4 | 19.1 | 4 | 11.8 | 2 | 11.8 | ||
SIL squamous intraepithelial lesion, LSIL low-grade squamous intraepithelial lesion. P was calculate using Fisher exact test.
Discussion
In this study, we analyzed the expression of E6, p53 and p21 proteins and the physical state of HPV16 in Non-SIL and LSIL, both with HPV16 infection.
Several reports have shown the important role of E6 protein in the genesis and development of cervical cancer. The E6 protein from HPV16 is sufficient for the induction and maintenance of cellular transformation [34]. This is mainly due to E6-induced degradation of p53 and p73 by 26S proteasome [7]. The inactivation of p53 compromises the integrity of the cellular genome, causes DNA damage and chromosomal instability, these abnormalities result in increased cell proliferation and tumor development [35-37]. In this work, we found that E6 expression was significantly higher in LSIL that in Non-SIL (p=0.006). These results suggest that elevated E6 expression may be an early marker of cervical cancer progression. To our knowledge few studies have evaluated the E6 expression in precancerous lesion and cervical cancer by immunocytochemistry. However, other cellular proteins with high expression in LSIL have been shown to be useful as early markers in cervical cancer [9,16,22,38].
In this work, we found no significant differences in p53 expression between Non-SIL and LSIL. We detected the p53 expression in 42% Non-SIL and 33.3% LSIL samples. In a previous study, Graspa et al analyzed the expression of p53 in normal cervical tissue and LSIL [39], however, in contrast with our data, they did not detect p53 expression in either Non-SIL or LSIL samples. Ours results can be explained by the fact that all Non-SIL samples are positive to HPV16 infection, whereas in their work, samples where negative to HPV16 because this infection could represent a stress response to viral infection [40]. Furthermore p53 expression could occur as a result of p53 stabilization by E6 protein binding [41,42].
As a consequence of the E6-mediated p53 inactivation, p21 gene transcription is inhibited [43], also E7 oncoprotein from high risk HPV, can target p21 for degradation during carcinogenesis [44]. In this work, we found no significant differences in p21 expression between Non-SIL and LSIL. p21 protein was detected 18% Non-SIL samples and 11.8% LSIL samples. In contrast to our results, previous reports found p21 expression in 15% of samples from normal cervical tissue without HPV and in 100% cases of NICI (equivalent to LSIL) [18]. These differences can be explained by the number of samples and by HPV16 infection. Also, the inactivation of p21 via reduced expression has been reported in various human tumors [45].
Our results show HPV16 integration in 22% of Non-SIL and 19.6% of LSIL. These results suggest that viral integration is an early event in the progression of cervical cancer. Furthermore, it has been demonstrated that cell populations with integrated HPV16 posses a selective growth advantage compared to cells that maintain HPV16 viral genomes as episomes [46]. We found a significant correlation between E6 expression and the HPV16 physical state in Non-SIL (p=0.049). Integration of HPV16 into the host genome can results in elevated expression levels of E6 and E7 viral oncoproteins, with subsequent interaction between these proteins and the cell cycle machinery [19,20]. In agreement with these observations, we found that the E6 expression level was higher in HPV16 integrated genome samples.
In conclusion, our results suggest that high E6 expression is an early event of cervical carcinogenesis and perhaps can be used as an early marker. However, compared with previous reports our study includes small number of samples; therefore, it is necessary to validate our results in a larger population. Moreover, including high grade squamous intraepithelial lesions (HSIL) in the study will provide important information to validate the usefulness of E6 expression as an early marker in cervical cancer progression.
Acknowledgements
Diana Karen Jiménez Tagle was recipient of a master fellowship from Consejo Nacional de Ciencia y tecnología (CONACyT), belongs to Programa de Maestría en Ciencias Biomédicas, UAG. We would like to thank to Programa de Fortalecimiento Académico del Posgrado de Alta Calidad of CONACyT.
References
- 1.Benedet JL, Bender H, Jones H 3rd, Ngan HY, Pecorelli S. FIGO staging classifications and clinical practice guidelines in the management of gynecologic cancers. FIGO Committee on Gynecologic Oncology. Int J Gynaecol Obstet. 2000;70:209–262. [PubMed] [Google Scholar]
- 2.Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208:152–164. doi: 10.1002/path.1866. [DOI] [PubMed] [Google Scholar]
- 3.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 4.Illades-Aguiar B, Cortes-Malagon EM, Antonio-Vejar V, Zamudio-Lopez N, Alarcon-Romero Ldel C, Fernandez-Tilapa G, Hernandez-Sotelo D, Teran-Porcayo MA, Flores-Alfaro E, Leyva-Vazquez MA. Cervical carcinoma in Southern Mexico: Human papillomavirus and cofactors. Cancer Detect Prev. 2009;32:300–307. doi: 10.1016/j.cdp.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Illades-Aguiar B, Alarcon-Romero Ldel C, Antonio-Vejar V, Zamudio-Lopez N, Sales-Linares N, Flores-Alfaro E, Fernandez-Tilapa G, Vences-Velazquez A, Munoz-Valle JF, Leyva-Vazquez MA. Prevalence and distribution of human papillomavirus types in cervical cancer, squamous intraepithelial lesions, and with no intraepithelial lesions in women from Southern Mexico. Gynecol Oncol. 2010;117:291–296. doi: 10.1016/j.ygyno.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 6.Tungteakkhun SS, Duerksen-Hughes PJ. Cellular binding partners of the human papillomavirus E6 protein. Arch Virol. 2008;153:397–408. doi: 10.1007/s00705-007-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganguly N, Parihar SP. Human papillomavirus E6 and E7 oncoproteins as risk factors for tumorigenesis. J Biosci. 2009;34:113–123. doi: 10.1007/s12038-009-0013-7. [DOI] [PubMed] [Google Scholar]
- 8.Lin HP, Wang YP, Chiang CP. Expression of p53, MDM2, p21, heat shock protein 70, and HPV16/18 E6 proteins in oral verrucous carcinoma and oral verrucous hyperplasia. Head Neck. 2011;33:334–340. doi: 10.1002/hed.21452. [DOI] [PubMed] [Google Scholar]
- 9.Roncaglia MT, Fregnani JH, Tacla M, DE Campos SG, Caiaffa HH, Ab’saber A, DA Motta EV, Alves VA, Baracat EC, Longatto Filho A. Characterization of p16 and E6 HPV-related proteins in uterine cervix high-grade lesions of patients treated by conization with large loop excision. Oncol Lett. 2013;6:63–68. doi: 10.3892/ol.2013.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 11.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 12.Huang LW, Chou YY, Chao SL, Chen TJ, Lee TT. p53 and p21 expression in precancerous lesions and carcinomas of the uterine cervix: overexpression of p53 predicts poor disease outcome. Gynecol Oncol. 2001;83:348–354. doi: 10.1006/gyno.2001.6397. [DOI] [PubMed] [Google Scholar]
- 13.Hunt CR, Hale RJ, Buckley CH, Hunt J. p53 expression in carcinoma of the cervix. J Clin Pathol. 1996;49:971–974. doi: 10.1136/jcp.49.12.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosun G, Sendag F, Zeybek B, Cosan Terek M, Guven C, Zekiogly O, Bilgin O. Immunohistochemical expressions of p16 and p53 proteins in cervical intraepithelial neoplasia and in benign cervical tissue. Eur J Gynaecol Oncol. 2010;31:627–631. [PubMed] [Google Scholar]
- 15.Vassallo J, Derchain SF, Pinto GA, Martinez EZ, Syrjanen KJ, Andrade LA. High risk HPV and p53 protein expression in cervical intraepithelial neoplasia. Int J Gynaecol Obstet. 2000;71:45–48. doi: 10.1016/s0020-7292(00)00248-4. [DOI] [PubMed] [Google Scholar]
- 16.Masumoto N, Fujii T, Ishikawa M, Saito M, Iwata T, Fukuchi T, Susumu N, Mukai M, Kubushiro K, Tsukazaki K, Nozawa S. P16 overexpression and human papillomavirus infection in small cell carcinoma of the uterine cervix. Hum Pathol. 2003;34:778–783. doi: 10.1016/s0046-8177(03)00284-3. [DOI] [PubMed] [Google Scholar]
- 17.Motoyama S, Ladines-Llave CA, Luis Villanueva S, Maruo T. The role of human papilloma virus in the molecular biology of cervical carcinogenesis. Kobe J Med Sci. 2004;50:9–19. [PubMed] [Google Scholar]
- 18.Bahnassy AA, Zekri AR, Alam El-Din HM, Aboubakr AA, Kamel K, El-Sabah MT, Mokhtar NM. The role of cyclins and cyclins inhibitors in the multistep process of HPV-associated cervical carcinoma. J Egypt Natl Canc Inst. 2006;18:292–302. [PubMed] [Google Scholar]
- 19.Jeon S, Allen-Hoffmann BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon S, Lambert PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersson S, Safari H, Mints M, Lewensohn-Fuchs I, Gyllensten U, Johansson B. Type distribution, viral load and integration status of high-risk human papillomaviruses in pre-stages of cervical cancer (CIN) Br J Cancer. 2005;92:2195–2200. doi: 10.1038/sj.bjc.6602648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giannoudis A, Herrington CS. Differential expression of p53 and p21 in low grade cervical squamous intraepithelial lesions infected with low, intermediate, and high risk human papillomaviruses. Cancer. 2000;89:1300–1307. doi: 10.1002/1097-0142(20000915)89:6<1300::aid-cncr15>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 23.Zubillaga-Guerrero MI, Illades-Aguiar B, Leyva-Vazquez MA, Flores-Alfaro E, Castaneda-Saucedo E, Munoz-Valle JF, Alarcon-Romero LC. The integration of HR-HPV increases the expression of cyclins A and E in cytologies with and without low-grade lesions. J Cytol. 2013;30:1–7. doi: 10.4103/0970-9371.107504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva-Filho AL, Traiman P, Triginelli SA, Reis FM, Pedrosa MS, Miranda D, Abreu ES, Macarenco RS, Cunha-Melo JR. Expression of p53, Ki-67, and CD31 in the vaginal margins of radical hysterectomy in patients with stage IB carcinoma of the cervix. Gynecol Oncol. 2004;95:646–654. doi: 10.1016/j.ygyno.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 25.Bravaccini S, Sanchini MA, Amadori A, Medri L, Saragoni L, Calistri D, Monti F, Volpi A, Amadori D. Potential of telomerase expression and activity in cervical specimens as a diagnostic tool. J Clin Pathol. 2005;58:911–914. doi: 10.1136/jcp.2004.024158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solomon D, Davey D, Kurman R, Moriarty A, O’Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T Jr, Young N. The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA. 2002;287:2114–2119. doi: 10.1001/jama.287.16.2114. [DOI] [PubMed] [Google Scholar]
- 27.Davis LG, Kuehl WM, Battey JF. Basic methods in molecular biology. Norwalk, Conn: Appleton & Lange; 1994. [Google Scholar]
- 28.Herrington CS, McGee JOD. Diagnostic molecular pathology: a practical approach. Oxford; New York: IRL Press at Oxford University Press; 1992. [Google Scholar]
- 29.Persing DH. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. [Google Scholar]
- 30.Vossler JL, Forbes BA, Adelson MD. Evaluation of the polymerase chain reaction for the detection of human papillomavirus from urine. J Med Virol. 1995;45:354–360. doi: 10.1002/jmv.1890450321. [DOI] [PubMed] [Google Scholar]
- 31.Bernard HU, Chan SY, Manos MM, Ong CK, Villa LL, Delius H, Peyton CL, Bauer HM, Wheeler CM. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis. 1994;170:1077–1085. doi: 10.1093/infdis/170.5.1077. [DOI] [PubMed] [Google Scholar]
- 32.Halloush RA, Akpolat I, Jim Zhai Q, Schwartz MR, Mody DR. Comparison of ProEx C with p16INK4a and Ki-67 immunohistochemical staining of cell blocks prepared from residual liquid-based cervicovaginal material: a pilot study. Cancer. 2008;114:474–480. doi: 10.1002/cncr.23951. [DOI] [PubMed] [Google Scholar]
- 33.Norimatsu Y, Miyamoto M, Kobayashi TK, Moriya T, Shimizu K, Yanoh K, Tsukayama C, Miyake Y, Ohno E. Diagnostic utility of phosphatase and tensin homolog, beta-catenin, and p53 for endometrial carcinoma by thin-layer endometrial preparations. Cancer. 2008;114:155–164. doi: 10.1002/cncr.23495. [DOI] [PubMed] [Google Scholar]
- 34.Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng YW, Wu MF, Wang J, Yeh KT, Goan YG, Chiou HL, Chen CY, Lee H. Human papillomavirus 16/18 E6 oncoprotein is expressed in lung cancer and related with p53 inactivation. Cancer Res. 2007;67:10686–10693. doi: 10.1158/0008-5472.CAN-07-1461. [DOI] [PubMed] [Google Scholar]
- 36.Cooper B, Brimer N, Vande Pol SB. Human papillomavirus E6 regulates the cytoskeleton dynamics of keratinocytes through targeted degradation of p53. J Virol. 2007;81:12675–9. doi: 10.1128/JVI.01083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaeffer AJ, Nguyen M, Liem A, Lee D, Montagna C, Lambert PF, Ried T, Difilippantonio MJ. E6 and E7 oncoproteins induce distinct patterns of chromosomal aneuploidy in skin tumors from transgenic mice. Cancer Res. 2004;64:538–546. doi: 10.1158/0008-5472.can-03-0124. [DOI] [PubMed] [Google Scholar]
- 38.Skomedal H, Kristensen GB, Lie AK, Holm R. Aberrant expression of the cell cycle associated proteins TP53, MDM2, p21, p27, cdk4, cyclin D1, RB, and EGFR in cervical carcinomas. Gynecol Oncol. 1999;73:223–228. doi: 10.1006/gyno.1999.5346. [DOI] [PubMed] [Google Scholar]
- 39.Grapsa D, Frangou-Plemenou M, Kondi-Pafiti A, Stergiou E, Nicolopoulou-Stamati P, Patsouris E, Chelidonis G, Athanassiadou P. “Immunocytochemical expression of P53, PTEN, FAS (CD95), P16INK4A and HPV L1 major capsid proteins in ThinPrep cervical samples with squamous intraepithelial lesions”. Diagn Cytopathol. 2013 doi: 10.1002/dc.23003. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 40.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Cox LS. Multiple pathways control cell growth and transformation: overlapping and independent activities of p53 and p21Cip1/WAF1/Sdi1. J Pathol. 1997;183:134–140. doi: 10.1002/(SICI)1096-9896(199710)183:2<134::AID-PATH960>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 42.Seavey SE, Holubar M, Saucedo LJ, Perry ME. The E7 oncoprotein of human papillomavirus type 16 stabilizes p53 through a mechanism independent of p19(ARF) J Virol. 1999;73:7590–7598. doi: 10.1128/jvi.73.9.7590-7598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho NH, Kim YT, Kim JW. Alteration of cell cycle in cervical tumor associated with human papillomavirus: cyclin-dependent kinase inhibitors. Yonsei Med J. 2002;43:722–728. doi: 10.3349/ymj.2002.43.6.722. [DOI] [PubMed] [Google Scholar]
- 44.Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volgareva G, Zavalishina L, Andreeva Y, Frank G, Krutikova E, Golovina D, Bliev A, Spitkovsky D, Ermilova V, Kisseljov F. Protein p16 as a marker of dysplastic and neoplastic alterations in cervical epithelial cells. BMC Cancer. 2004;4:58. doi: 10.1186/1471-2407-4-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peitsaro P, Johansson B, Syrjanen S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol. 2002;40:886–891. doi: 10.1128/JCM.40.3.886-891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]