Stem cells are a powerful resource for producing a variety of cell types with utility in clinically associated applications, but substantial barriers to clinical translation must be overcome to realize full clinical potential. This concise review describes how microfluidic technologies can contribute to the translation of stem cell research outcomes and provides an update on innovative research efforts in this area.
Keywords: Microfluidics, Stem cells, Investigative techniques, Stem cell research, Translational medical research
Abstract
Stem cells are a powerful resource for producing a variety of cell types with utility in clinically associated applications, including preclinical drug screening and development, disease and developmental modeling, and regenerative medicine. Regardless of the type of stem cell, substantial barriers to clinical translation still exist and must be overcome to realize full clinical potential. These barriers span processes including cell isolation, expansion, and differentiation; purification, quality control, and therapeutic efficacy and safety; and the economic viability of bioprocesses for production of functional cell products. Microfluidic systems have been developed for a myriad of biological applications and have the intrinsic capability of controlling and interrogating the cellular microenvironment with unrivalled precision; therefore, they have particular relevance to overcoming such barriers to translation. Development of microfluidic technologies increasingly utilizes stem cells, addresses stem cell-relevant biological phenomena, and aligns capabilities with translational challenges and goals. In this concise review, we describe how microfluidic technologies can contribute to the translation of stem cell research outcomes, and we provide an update on innovative research efforts in this area. This timely convergence of stem cell translational challenges and microfluidic capabilities means that there is now an opportunity for both disciplines to benefit from increased interaction.
Introduction
Stem Cell Translation Presents a Specific Set of Challenges
Across the globe, many research groups and companies alike are assessing the potential of stem cells as the primary cellular inputs for a range of clinically associated applications, including preclinical drug screening and development, disease and developmental modeling, and regenerative medicine. In terms of translation to the clinic, different stem cell types are of interest depending on the endpoint application and include human pluripotent stem cells (hPSCs), comprising human embryonic stem cells (hESCs) [1] and induced pluripotent stem cells [2]), hematopoietic stem cells (HSCs) [3], mesenchymal stem/stromal cells (MSCs) [4], and tissue-specific stem cells from various tissue sources. hPSCs are of prime interest for deriving specific cell lineages for tissue regeneration and preclinical drug screening. hPSCs derived from patients carrying genetic diseases are also valuable as disease models, and induced pluripotent stem cell technology readily allows these cells to be established readily because access to embryos is not required. MSCs have a more limited differentiation potential but are notable for their anti-immunosurveillance properties and for stimulating tissue regeneration through secretion of therapeutic factors. As for other adult stem cells, HSCs are already in extensive clinical use, neural stem cells (NSCs) are of interest but are somewhat less accessible, and tissue-specific stem cells are likewise of potential interest for certain applications but are still in preliminary investigations.
In the development of a stem cell-based technology, whether that be derivation of mature cells for toxicity screening or progenitors for transplantation to encourage tissue repair, it is recognized that eliciting control over the cellular microenvironment is vital to control of the cellular phenotype through this process. In translating a stem cell-derived technology to a stem cell-derived therapy, in which billions of cells may be required per clinical dose, scaling standard culture protocols to large-scale bioprocessing formats engenders many challenges that, again, relate to being able to elicit microenvironmental control of stem cells or their progeny. Furthermore, the identity, efficacy, safety, and quality of the end product (whether stem cell, progenitor, or terminally differentiated population) must be ensured, overcoming any variation in input donor or cell line properties. Managing all of these issues (as discussed by Serra et al. [5]) to maintain a high-performance bioprocess is critical to support a translational program through preclinical and clinical studies.
Regardless of the type of stem cell involved, with the possible exception of HSCs, substantial barriers to translation still exist and must be overcome to realize the intended outcomes. Some of these barriers represent mainly technical issues, like cell isolation or derivation, expansion, and differentiation, which can be addressed by progress in basic or applied research. Other challenges include ensuring cellular product quality (e.g., efficacy and safety profiles of the process output) or optimizing bioprocess production of cell therapies so as to be economically viable. A generic bioprocess for producing working material to support a translational or clinical program involving a stem cell-derived therapy will include the following steps:
Cell isolation, derivation, and/or enrichment
Ex vivo cell expansion, differentiation, and harvesting
Cell analysis and characterization of viability, potency, and batch quality
Packaging, preservation, and administration
Post-implantation monitoring.
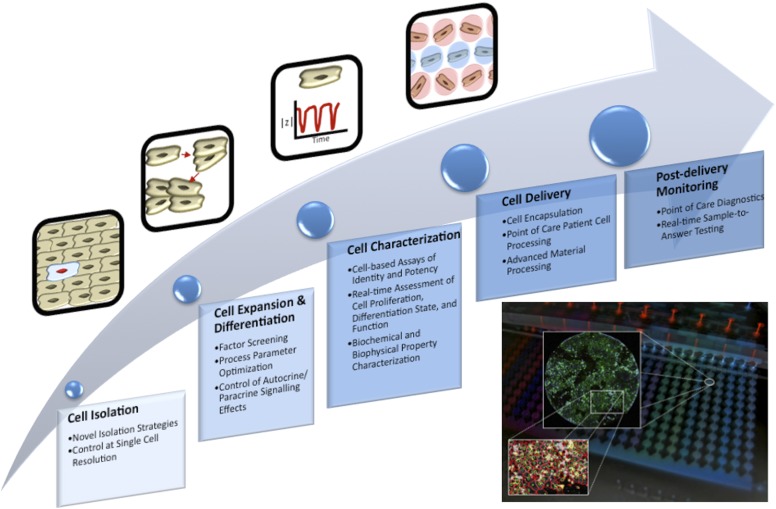
Each of these steps represents a cell-processing operation in which significant uncertainty exists, a strong foundation in both basic research and bioprocess development is required for the steps to operate effectively. Microfluidic systems are emerging as highly useful platforms for addressing gaps in our knowledge in each of these steps and for optimizing applicable bioprocesses within each step. Figure 1 conceptually illustrates this relationship. Bioprocesses to produce large numbers of stem cells, as well as mature cell lineages derived from them, will also be used in preclinical drug screening applications.
Figure 1.
Microfluidic technologies add insight to cell therapy processes. This flow diagram shows a generic cell therapy implementation process. Bullet points in each step highlight areas where microfluidic technologies can add insight. The inset shows how integrated microfluidic platforms provide both control of culture conditions and readout of cellular phenotypes.
Microfluidics Provides Relevant Tools to Address These Problems
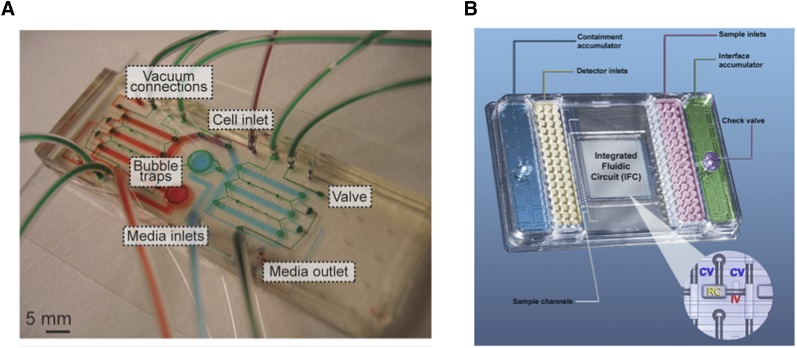
Microfluidic technologies build on a strong history of development of microfabricated bioanalytical systems. “Microfluidics” may be defined as the controlled manipulation of fluids (both gases and liquids) at a length scale of several to hundreds of microns. Such control over fluid flow is enabled through the high-fidelity patterning of three-dimensional microstructured geometries (e.g., channels, chambers, valves, membranes) into devices commonly based on elastic silicone polymer (poly(dimethylsiloxane) [6]), thermoplastic (polystyrene, polycarbonate, polypropylene), and glass or mixtures of these substrates. At these length scales, fluid, mass and energy transport are simplified and are highly predictable, allowing researchers to engineer devices (microfluidic “chips”) that offer unparalleled control over the local environmental conditions, including temperature, pH, dissolved gas concentration, shear stress, and medium exchange rates, at length scales applicable to cellular microenvironments in tissues. The target functions of microfluidic chips are then typically achieved with the design of various microfluidic channel geometries, the integration of various microfluidic components such as valves, and the interface with peripheral equipment such as pumps, incubators, and optical/electrical systems. These devices can be applied at the subcellular, single-cell, or cell-population levels. Figure 2 exemplifies microfluidic chips for various uses.
Figure 2.
Examples of microfluidic chips. (A): A microfluidic chip for perfusion culture of cells. Various connections for cells, medium, and pneumatic valve control are included and are filled with dye for visualization. Modified from [46]. (B): A commercial microfluidic platform for high-throughput gene expression profiling. A central microfluidic chip accepts samples and probes from macroscale wells in a custom multiwell-plate format. The inset shows that the chip contains many microfluidic channels and valves for fluid manipulation. Modified from [70].
Microfluidic technology, a relatively new field of endeavor, has recently begun to be applied to stem cells. The outputs of these studies have already demonstrated the intrinsic capability and flexibility of microfluidic systems to control and interrogate cellular microenvironments and processes with appropriately matched parameters (i.e., accessible timescales, length scales, physical and biochemical conditions). This has particular relevance to addressing translational barriers and achieving the intended outputs of stem cell technologies. Microfluidic systems are poised to accelerate the translation of stem cell outcomes through preliminary testing, regulatory hurdles, and clinical implementation. This timely convergence of stem cell translational challenges and microfluidic capabilities means that there is now an opportunity for both disciplines to benefit from increased interaction.
In this concise review, we will describe how we envision microfluidic technologies contributing to stem cell translation and will provide an update on innovative research making progress in this area, particularly for those who may not be aware of some of these techniques but who stand to benefit from their implementation. For brevity, we limit our scope to systems with “closed” fluidics (i.e., microfluidics) rather than spanning generic microfabricated systems, although many other approaches that leverage microtechnology principles can be useful research tools. Helpful reviews of such technologies are available [7–9]. Although we will focus on the steps associated with a generic stem cell bioprocess, it is important to note that microfluidic systems are also being developed to assist in applications outside of cell therapies, particularly preclinical drug screening. A review explaining the advantages of scaled-down, high-throughput assays in microfluidic systems highlights the advantages provided by microfluidics [10].
Microfluidic Systems Applicable to Stem Cell Translational Barriers
A large suite of microfluidic systems has been developed for a wide variety of biological applications [8]; however, many of these systems have initially been validated with cell lines and have not yet been used to gain biological insight with stem cells. Adaptation of these proof-of-concept systems to accommodate stem cells is largely a technical issue that can be achieved with sufficient demand from the stem cell field; however, some key microfluidic technologies have been developed targeting stem cells from the start and have been validated with stem cells or their progeny [11]. We will now focus our review on these systems because they will be most immediately applicable to stem cell translation. We will categorize each of these systems as addressing barriers within the steps listed previously as associated with a viable cellular therapy bioprocess.
Cell Isolation, Derivation, and Enrichment
Microfluidic technology has been applied to facilitate cell isolation. We highlight several relevant examples and also refer the reader to recent reviews [7, 12] for more detailed information. Microfluidics can act as a label-free tool to isolate specific cell types from heterogeneous cell mixtures (e.g., cord blood and stem cell cultures) based on their differences in size, deformability [13], and adhesion strength [14, 15] as well as cell surface affinity [16]. In addition to single stem cells, embryoid bodies (EBs) formed by the aggregation of spontaneously differentiating mouse embryonic stem cells (mESCs) have been sorted using a microfluidic platform [17]. When injected into a microchannel with micropillars altering the fluid flow pathway, mouse EBs were sorted into size groups of 0–100 μm, 100–200 μm, and 200–300 μm, with purities of 100%, 86%, and 81%, respectively [17]. This microfluidic flow sorting technology is faster, less labor intensive, and more accurate than conventional separation using pipettes and does not cause cell damage compared with separation methods using external force fields (e.g., dielectrophoretic, acoustic, or magnetic fields) [17]. Moreover, when millions or billions of cells are required for cell therapy, the high throughput and parallelization capacity of the microfluidic cell separation device might be a critical advantage compared with the sorting rates achievable by fluorescence-activated cell sorting.
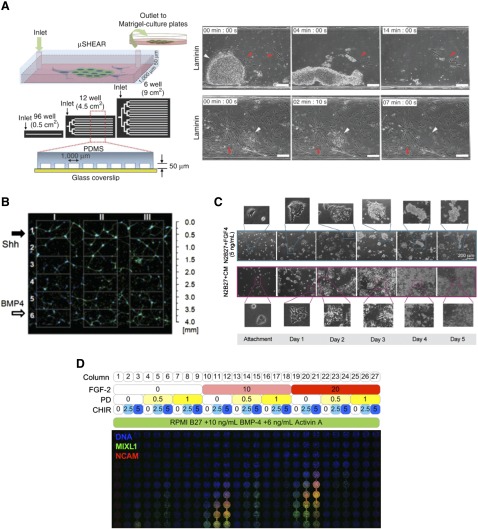
The flow of culture medium over cells results in shear forces being applied to the cells. This phenomenon has recently been generated using a microfluidic device and utilized to isolate intact hPSCs from heterogeneous reprogramming cultures and differentiation processes with an efficiency of 95%–99% (Fig. 3A) [15]. This isolation exploited approximately twofold differences in substrate adhesion strength between hPSCs and non-pluripotent cells in order to selectively detach and collect hPSCs using the shear force of medium flow. Eighty percent of isolated cells were viable and had normal transcriptional profiles, karyotypes, and differentiation potential. This particular microfluidic purification technology has immediate potential for application in hPSC bioprocesses that are based on adherent culture.
Figure 3.
Microfluidic systems for stem cell isolation, expansion, and differentiation. (A): A system for shear force-based purification of human pluripotent stem cells from IMR90 fibroblasts (left panel). Constant medium flow is able to gradually and preferentially detach human pluripotent stem cells for recovery based on differences in adhesion strength relative to fibroblasts. Adapted and reprinted with permission from Macmillan Publishers Ltd. [15]. (B): A microfluidic device was used to apply opposing, overlapping gradients of neural patterning factors Sonic Hedgehog (Shh) and BMP4 to human embryonic stem cell-derived neural progenitors. Patterning of neural network organization was affected by the relative amounts of patterning factors. Adapted from [35]. (C): Continuous microfluidic perfusion of culture medium is used to examine differentiation of mouse embryonic stem cells into Sox1-positive neuroectoderm. FGF4 stimulation with removal of surplus diffusible factors under continuous flow (blue-bordered images) is insufficient for robust cell number expansion through neuroectodermal differentiation, whereas compensation with conditioned medium allows this to proceed (red-bordered images). Adapted from [46]. (D): A continuous-flow microbioreactor array was used to generate various combinations of FGF-2, the MEK inhibitor PD0325901, and the Wnt activator CHIR99021 in each column of the array in a background of mesendoderm-inducing medium containing BMP4 and Activin A. Medium flowed from the top row to the bottom row of each column, gradually accumulating greater levels of soluble factors from top to bottom. Confocal microscopy of the whole array detected MIXL1-GFP expression and immunostaining of NCAM. The various treatments show patterning of hESCs into MIXL1-positive mesendodermal population, also marked by NCAM expression. The effects of the various factors and combinations of factors can be inferred from the responses. Chambers are ∼1.63 mm in diameter. Modified from [39]. Abbreviation: PDMS, poly(dimethylsiloxane).
Ex Vivo Cell Expansion and Differentiation
One intrinsic advantage of microfluidic technology compared with traditional cell culture and analysis platforms is the precision with which fluid flow may be manipulated. This permits unrivalled regulation, spatially and temporally, of both the biophysical parameters (e.g., shear stress due to the convective flow of medium) and biochemical parameters (e.g., nutrient and growth factor level variations due to medium turnover rates) of the cellular microenvironment for implementing cell-based assays and optimizing stem cell culture and differentiation.
Screening Effects of Fluid Shear Stress and Medium Turnover on Stem Cell Fate
Fluid shear stress is recognized as an important biophysical parameter affecting cell cultures. In terms of bioprocessing, fluid shear stress is a significant consideration within large bioreactor formats, particularly for shear-sensitive cells such as hPSCs [18]. It is important to provide scaled-down models of shear stress to investigate and understand the impact of fluid shear stress on stem cell phenotype. Microfluidic devices are ideal systems with which to generate and survey a range of fluid flow rates in parallel that reflect the applied fluid shear stresses and medium turnover rates experienced by cells in a bioprocess. A microfluidic device with shear stress ranging from 0.0016 to 16 dyn/cm2 was applied to show that self-renewing mESCs had the molecular machinery to sense and respond to shear stress [19]. mESCs grown in a microfluidic bioreactor under a logarithmic range of flow rates displayed increasing colony size with increased flow rate [20]. hESCs were grown in a similar system but with a more narrow, linear range of flow rates, and were observed to have a small acceptable range of flow rates for culture [21]. At low flow rates, medium exchange appeared to be limiting the supply of nutrients and removal of wastes, whereas at higher flow rates, cell detachment and altered morphology were observed; both conditions had reduced cell expansion relative to the intermediate flow rates. In other applications, 40% of mESCs exposed to shear stress at 15 dyn/cm2 were reported to differentiate into endothelial cells, compared with the 1% differentiation under static cell culture [22], although the mature functionality of these cells was unclear. A microfluidic device embedded with aligned nanofibers has been applied to study the influence of shear stress and aligned nanofibers on the differentiation of MSCs [23]. Such devices have only just begun to be applied to stem cell culture and differentiation, but these first works exemplify their substantial potential in surveying expansion and differentiation outcomes.
Screening of Molecular Control of Ex Vivo Expansion and Differentiation
Researchers are still optimizing techniques for enhanced expansion of pluripotent cells and pursuing a high-level understanding of the molecular events that support their expansion and maintain their identity [24]. Microfluidic systems can support hPSCs under both static conditions (with periodic feeding) [25] and continuous flow [21, 26]. Microfluidics provides leverage to readily generate a multitude of culture conditions, demonstrated in work from our laboratory in which a hESC pluripotency reporter line was grown in full-factorial array (an array consisting of every combination of three concentration levels each of three factors) of FGF2, transforming growth factor β1, and retinoic acid (RA) [27]. This entailed 27 distinct combinations of soluble factors in total and was used to deconstruct the contributions of FGF2 and transforming growth factor β1, the main maintenance factors in mTeSR-1 hPSC culture medium. Such systems streamline the generation of varied culture conditions and provide a more detailed snapshot of cellular responses to factor treatment regimes than more minimal experiments in static plates. Other microfluidic tools to support these investigations include microfluidic image cytometry [28], in which hPSCs grown under various culture conditions can be analyzed in situ at the single-cell level.
In terms of differentiation, researchers are also continuing to discover routes to the generation of cell lineages of interest by differentiating hPSCs. They may develop a protocol to derive the target phenotype, initially at low efficiency, by selecting several key developmental factors, timing the treatment based on developmental timescales, and crudely recapitulating three-dimensional tissue arrangement using EBs because mimicking development has, so far, provided the best outcomes in vitro [29]. The efficiency can then be improved by optimizing the multistage differentiation trajectory; however, development is inherently programmed to generate an organism of heterogeneous tissues [30], an effect recapitulated by hPSCs in vitro that is particularly demonstrated by self-organizing tissues [31]. This may not necessarily be the trajectory best suited to regenerative and pharmacological purposes, for which it would be preferential to generate pure populations of target lineages. This will require detailed knowledge and artificial exploitation of developmental signals. Robust optimization of instructive differentiation and attenuation of competing differentiating signals to yield the target cell type at a high efficiency and in sufficient numbers for subsequent applications often requires screening and optimization of various formulations of factor combinations as well as concentration ranges, dose timing, cell density, extracellular matrix substrate, and a variety of other microenvironmental factors. This can quickly lead to a large experimental space based on many levels and combinations of multiple parameters. Although the cell source for this screening may not in itself be limiting (hPSCs can be readily generated in high numbers), the sheer number of conditions that must be generated as well as the cost of reagents (often expensive recombinant cytokines) can be obstructive to performing a comprehensive analysis.
Microfluidic systems can assist in this process by miniaturizing the culture system and reducing the amount of cells and reagents required, streamlining the task of generating distinct microenvironmental conditions, and improving the spatial homogeneity and control over the conditions experienced by cells. This was demonstrated by early microbioreactor arrays investigating shear and cell density effects in hESCs undergoing smooth muscle differentiation [32, 33]. More developed microfluidic systems for screening molecular control of expansion and differentiation will help establish best practice cell-processing protocols to efficiently derive target lineages. This has been illustrated by systems demonstrating molecular gradients that drive stem cell differentiation. NSC proliferation and astrocyte differentiation was varied across a gradient of medium containing epidermal growth factor, FGF2, and platelet-derived growth factor, using continuous-flow microfluidics to generate the gradient and remove autocrine factors [34]. The results showed that NSCs proliferated in proportion to the amount of growth factor-containing medium present and, conversely, differentiated toward glial fibrillary acidic protein-positive astrocytes more readily in growth factor-free medium conditions. A gradient-generating microfluidic device has been used to model developmental neural patterning by using nestin-positive hESC-derived neural progenitor cells in overlapping gradients of Sonic Hedgehog, FGF8, and BMP4 [35], showing that a continuum of network organizational features (neuronal cell bodies and neurite bundles) could be patterned in response to the gradient (Fig. 3B). Similarly, overlapping countergradients of the maintenance factor leukemia inhibitory factor and differentiation factor RA were applied to mESC cultures and resulted in gradients of expression of the pluripotency transcription factor Nanog [36], demonstrating spatial control of the factor delivery and cellular phenotype.
Diffusion-based gradients of mesoderm-inducing factors (Wnt3a, Activin A, and BMP4) and countergradients of inhibitory molecules (SB431542 and Dkk1) were applied to a series of immobilized EBs to investigate effects on Wnt activation and mesodermal differentiation tracked by a β-catenin-responsive reporter [37]. This device established distinct molecular conditions, but arguably the phenotypic effects of the molecular gradients were more understated than expected, possibly because of EBs being of variable size and acting as sinks for molecular diffusion. These effects can be overcome by using the convective flow control possible in microfluidics, with more recent work from the same group demonstrating regional control of Nanog expression in mESCs genetically modified to have constitutively silenced but doxycycline-rescuable Nanog expression [38]. Microfluidically generated gradients of doxycycline as well as the maintenance factor leukemia inhibitory factor and differentiation factor RA were then able to pattern a range of expression states of pluripotency and early differentiation genes. Imposition of such artificial conditions on cells is an important tool to deconstruct developmental processes. Work in our own group has also used continuous flow over hESC monolayers to examine paracrine factors in mesendodermal differentiation [39] (detailed below).
Assessment of Dynamic Treatments
Timing of factor stimulation can also be varied readily in closed microfluidic systems (as demonstrated by King et al. [40]), owing to their facile fluidic manipulation. This can also achieve a greater level of automation of cell handling and culture operations, with an array of 96 culture chambers having been used to assay the duration of stimulation with osteogenic medium on MSC proliferation and alkaline phosphatase expression [41]. Similarly, an array of ∼1,600 culture chambers has been used to characterize the response of dynamic Steel factor stimulation on HSCs [42].
Elucidation and Control of Autocrine and Paracrine Signaling
Diffusible autocrine factors (cell to self or same cell type) and paracrine factors (cell to distinct cell type) secreted by various cells along the developmental continuum are critical regulators of differentiation and morphogenesis. These developmental mechanisms also manifest in ex vivo manipulations of hPSC populations and can be manipulated exogenously by selective provision of stimulatory or inhibitory molecules. Yet they remain poorly understood, are difficult to study using conventional techniques, and are often ignored when developing new differentiation protocols toward target cell types.
Traditionally, several approaches have been used to access these autocrine and paracrine effects, including use of conditioned medium, receptor or signaling pathway stimulation or inhibition, factor depletion from medium, manipulation of cell density, or coculturing. Direct analysis of medium supernatants is possible but is limited by access to the molecules, owing to their limited diffusion before binding or their low abundance. Microfluidic systems have now established a new toolbox for assessing paracrine effects (an excellent review is available [43]) by utilizing programmed medium exchange or continuous medium perfusion to regulate the accumulation of these secreted factors. In an early application demonstrating this, programmed medium exchange was used to remove cell-secreted factors from mESC cultures at various frequencies [44]. Better cell viability at lower wash frequencies in minimal medium suggested the presence of cell-secreted, prosurvival factors, whereas addition of serum did not improve viability at higher wash frequencies, so the factors were not likely present in serum. This work was extended by modeling secreted factor transport in various microfluidic flow conditions and was then correlated with experimental outcomes of mESC fate under continuous microfluidic flow [45], providing evidence for the presence of diffusible signals that are critical for stem cells and can be controlled by microfluidics.
A further investigation of the presence and effects of diffusible signals was subsequently performed in a system of mESC differentiation toward neuroectoderm [46]. In this work, autocrine FGF4 was found to be insufficient for robust growth of mESC cultures differentiating into Sox1-positive neuroectoderm because, although the neuroectodermal specification was FGF dependent, other cell-secreted factors were needed to support cell number expansion through this differentiation phase (Fig. 3C). Such results have critical implications for large-scale processing requirements. Related work by the same group used microfluidic perfusion to identify that extracellular matrix remodeling was critical to mESC maintenance and that mESCs were likely kept in a naïve state by secretion of matrix metallo-proteinases that remodeled the ECM to prevent epiblast transition [47].
Recent work from our laboratory has used a microbioreactor array platform to screen exogenous and paracrine signals in early hESC differentiation to a MIXL1-positive, primitive streak-like mesendodermal population [39]. In this work, combinatorially multiplexed medium formulations were continuously flowed over a cell culture array in which serial culture chambers also allowed paracrine signals to accumulate progressively. This spatial dispersion of paracrine factor levels meant that differential effects on cell phenotype could be visualized directly, using fluorescent reporter proteins or in situ immunostaining. This critical function is not possible with traditional approaches. Using this system, temporal variations in paracrine factor accumulation present in a static culture can be stabilized and visualized across a spatial dimension in the dynamic culture (Fig. 3D).
This approach provided evidence of two paracrine phenomena critical for regulating the MIXL1-positive phenotype. The first was a requirement for accumulation of surplus factors over the levels of exogenous BMP4 and Activin A, which could be compensated for by provision of conditioned medium or FGF2 and likewise blocked with the MEK inhibitor PD0325901. The second was a negative feedback loop to the MIXL1-positive population that could be overcome by enforced Wnt activation through the GSK-3β inhibitors CHIR99021 and BIO. This suggested the class of extracellular Wnt antagonists (e.g., Dickkopf proteins and secreted frizzled-related proteins) as candidate effectors of the negative feedback loop. Screening induction of cellular phenotypes with parallel supplementation or inhibition of paracrine factors proved, in this case, to be a powerful strategy for unraveling the hierarchy of factor signals involved in regulating differentiation because the results were also transferable to improving differentiation outcomes in static culture formats.
In the area of MSCs, the role of diffusible signals was investigated in adipogenesis of human adipose-derived stem cells, using a microfluidic perfusion system and conditioned medium [48]. Both conditioned medium (containing secreted factors) and increased cell density were found to enhance adipogenic differentiation, and this finding links endogenous factor secretion to the observed criticality of initial cell density on the success of MSC differentiation. Recent work from our group utilized a microbioreactor array to investigate osteogenic differentiation of mesenchymal precursor cells, a purified subset of bone marrow-derived MSCs that are selected on STRO-1 marker expression, showing that paracrine effects were also likely present in this process and so are key considerations for bioprocessing [49].
Such insights have obvious utility in developing cell-processing regimes for cell-based therapies and confirm that microfluidic systems have been instrumental in identifying and understanding the relative impacts of positive and negative regulation of factors influencing stem cell fate choices, which will be critical for obtaining improved outcomes in macro-scale culture systems. The power of manipulating paracrine regulation of stem cells has been demonstrated in the HSC system, where a macro-scale fed-batch culture system was able to improve HSC expansion through regulation of levels of secreted inhibitory factors by intermittent dilution with fresh medium [50]. Such approaches clearly improve the performance, and thus the economic viability, of stem cell bioprocesses and, in this case, improved the expansion of HSCs, which is still an issue for transplants using these cells, despite their widespread clinical adoption.
Maximizing the stepwise yield of a multistep differentiation process in this manner would be expected to improve the later yield of more mature cell types. In our experience, adoption of the processing conditions that maximize mesendoderm conversion [39] also results in a greater yield of cardiomyocytes in downstream differentiation as well as reduced experimental variability because paracrine factors were regulated exogenously rather than relying on endogenous production levels.
Such approaches might be able to isolate developmental progenitors of interest by identifying conditions that arrest their development and support their expansion because such transient progenitors are likely regulated by microenvironmental signals (particularly paracrine signaling). This aspect is attractive because there has been much discussion within the cellular therapy field regarding the relative value of using progenitor rather than terminally differentiated cell types in certain therapeutic contexts.
Cell Analysis and Characterization of Viability, Potency, and Batch Quality
Long-Term Cell Culture and Analysis
Conventional in vitro cell culture platforms such as tissue culture flasks, which are currently used commercially for stem cell expansion, are intrinsically limited in terms of their scalability, their ability to control features of the cellular microenvironment, and their capacity for in situ analysis of living cells, high-throughput cell analysis, and long-term online monitoring.
Microfluidic technology has recently been applied to achieve controlled, long-term stem cell expansion. A prime example is a self-contained microfluidic system that was developed for automatic cell culture and real-time imaging [51]. A central culture chip in the device is disposable, whereas other components are reusable. The system includes an on-chip micropump for custom medium formulation, which makes the system portable and reduces reliance on external fittings and equipment.
Single-Cell Capture, Culture, and Analysis
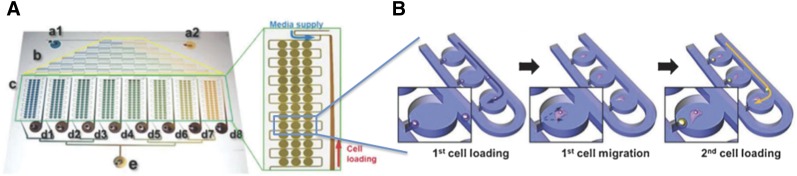
Further understanding of the heterogeneity of stem cell behaviors at the single-cell level will permit further insights into optimization of culture conditions and population quality and phenotype at larger scales. With the increased demand for single-cell-resolution data, microfluidic technologies have turned out to be indispensable because of their unique capabilities in dealing with small volumes of reagents, trapping and capturing single cells with high efficiency and throughput [52, 53], culturing trapped single cells in situ for long periods of time for clonal analysis [54], and performing cell-interaction analyses at single-cell levels (Fig. 4) [55]. As a further development of single-cell-based investigations, a microfluidic device enabling sequential single-cell trapping has been applied to study the interactions between individual mESCs and mouse embryonic fibroblasts [55], that is, coculture of two distinct single cells. Pairs of these cells were then cultured for several generations to study cell-cell interactions (Fig. 4).
Figure 4.
An example of a microfluidic coculture system to study cell-cell interactions at the single-cell level. Adapted from [55] and reproduced with permission from the Royal Society of Chemistry. (A): A coculture system with a chemical gradient generator (from inlets a1 and a2 and region b) and cell culture chambers (region c). After loading of both types of cells, medium is perfused through the system to allow coculture of single cells for extended periods of time. (B): After loaded into wells along the channel, the mouse embryonic fibroblasts adhere to and migrate along the well bottom. Then mouse embryonic stem cells are loaded to form single mouse embryonic fibroblast and mouse embryonic stem cell pairs in each well.
In addition, microfluidic technology has recently permitted medium-throughput single-cell gene expression analysis; with an integrated microfluidic device, one is able to capture and perform high-precision quantitative reverse transcription polymerase chain reaction (PCR) measurements of gene expression on 300 single cells [52]. More recently, Fluidigm Corporation (South San Francisco, CA, http://www.fluidigm.com/) has released a microfluidics-based single-cell isolation and preparation system for downstream gene expression measurements of single cells (C1 Single-Cell Auto Prep System).
Real-Time Measurements of Cell Number, Phenotype, and Function for Preclinical Assessments
Microfluidic systems are generally fabricated with processes that are similar to or compatible with fabrication of microelectromechanical systems components, particularly sensors and actuators, and thus readily allow for their integration. This enables integrated, real-time sensing and control over multiple aspects of the cellular microenvironment (e.g., cell density, temperature, pH, oxygen tension). These systems can often be extended to provide external physical stimulus, if desired, which may prove useful in some stem cell applications.
Integrated microfabricated circuits, for example, allow real-time sensing of cell density and electrophysiological properties. Cell culture microplates have already been commercially developed with integrated, impedance-sensing systems, such as xCELLigence (ACEA Biosciences Inc., San Diego, CA, http://www.aceabio.com; Roche, Switzerland, http://www.roche.com) and Electric Cell-substrate Impedance Sensing or ECIS (Applied BioPhysics Inc., Troy, NY, http://www.biophysics.com), as well as field potential sensing with electrical stimulus such as microelectrode arrays (MEAs; Axion Biosystems, Atlanta, GA, http://www.axionbiosystems.com). Both the xCELLigence system and the MEA sensors have been applied to screening drug-induced proarrhythmia on cardiomyocytes derived from hPSCs [56]. In this trial, 25 cardioactive drugs and 3 negative controls were tested on both systems, and changes to the contractile amplitude and frequency were analyzed, with the impedance signal clearly showing that arrhythmias can be traced in real time. MEAs were also used to correlate drug-induced electrophysiological responses of hPSC-derived cardiomyocytes to existing preclinical drug safety assays, showing that they were effective as a predictive assay platform. In another recent study, cardiomyocytes derived from hPSCs were cultured on MEA devices. They demonstrated that the MEAs showed similar electrical responses to rabbit ventricular wedges when exposed to various cardioactive drugs [57]. This shows the power of using both hPSCs and MEAs for accelerating preclinical work and reducing dependence on in vivo animal models.
Similarly, one study demonstrated that the ECIS platform can distinguish between different lineages of differentiating adipose-derived MSCs based on impedance signal changes [58]. They showed significant differences in the impedance signal of osteo-induced, adipo-induced, and undifferentiated cells in real time. This demonstrates that simple changes in cell dielectric properties can allow for label-free sensing of differentiation status. Furthermore, statistical differences between cell types were detectable on the ECIS system before they were observable under phase contrast imaging by eye. The ECIS also proved useful for measuring cell proliferation in real time.
Microfluidic chips can be packaged with electronic integrated circuits to provide novel instruments. Recently, the use of both impedance analysis and electric field potential was used to screen and sort various electroactive cardiac EBs derived from hPSCs on chip [59]. By doing so, cardiac EBs could be detected (by impedance) and discriminated (by field potential) based on their native electrical properties and electrical response to stimulus in an integrated sorting system based on functional cell properties, a novel capability compared with fluorescence-activated cell sorting. In another flow-through system, the differentiation state of single mouse embryonic carcinoma cells could be discriminated in impedance-based flow cytometry [60]. Using the impedance properties at different frequencies, they were able to determine which cells were differentiated toward neuronal cells and which were undifferentiated, at an efficiency of ∼90%.
Cell Delivery and Postdelivery Monitoring
Microfluidic systems have been developed less intensively toward applications in the cell delivery and postdelivery monitoring stages of the process considered in Figure 1; however, some concepts warrant brief consideration. Microfluidic systems provide cell encapsulation capabilities [61] and capacity for microsized biomaterial synthesis (e.g., of microcarrier beads) [62, 63], which may prove useful in clinical delivery of cells. Likewise, postdelivery monitoring of stem cell persistence or biomarker detection may benefit from development of microfluidic point-of-care diagnostic systems [64], borrowing particularly from cell isolation and detection strategies developed for circulating tumor cells [65], or compact immunoassay systems.
Future Applications, Outlook, and Conclusions
Supportive Data for Clinical Trials and Implementation
With the more widespread emergence of cell therapies, the regulatory environment will develop and engender evolution in the standards of data required to demonstrate safety, efficacy, potency, and mechanism of action for clinical trial and final marketing approvals. Cell therapies from hPSCs or MSCs currently appear to be regulated as biological drugs [66]. The types of assays that will be required as preclinical data, now and in the future, represent a significant departure from those that would be standard for a small molecule drug, with additional focus on processing conditions. This is particularly pertinent to MSC-based therapies, which are approaching clinical trials and applications in multiple, varied therapeutic targets.
In the case of MSCs, cells can form bone, cartilage, and adipose tissue but are also used as infused cells that are thought not to persist by integrating in the body but rather to secrete regenerative or immunosuppressive factors [67]. As highlighted previously, microfluidic systems have the ability to evaluate paracrine signaling and thus can provide new screens to confirm and investigate these processes in more detail. This might also be extended, for example, by coculture with cell types thought to be targeted by MSCs.
Microfluidic culture systems significantly extend the utility of existing assays for coculture or mechanism of action studies, including transwell assays, which require more cells (relevant to the availability of adult stem cells) and are limited to two cell types per well [68]. In the near future, new-generation microfluidic assays that will permit multi-cell type cultures mimicking tissue microenvironments will contribute significant new insights toward elucidating the therapeutic mechanism of action of MSCs.
Quality Testing of Culture Medium Batches and Donor-to-Donor/Line-to-Line Variations
Despite continual refinement of stem cell culture medium compositions, significant variation remains in the inputs into stem cell bioprocesses, including medium additives (e.g., cytokines) and the cells themselves. In the case of adult stem cells, the known donor-to-donor variability, in terms of expansion capacity and propensity for multilineage differentiation, requires batch-to-batch potency testing and represents a significant cost for cell therapy operations. Microfluidic platforms for quality testing, building on some of the platforms described in this review, will have immediate application, offering substantial reductions in required cell numbers and reagents and with real-time readouts of phenotype and time.
Commercialization and Dissemination of Microfluidic Technologies
The microfluidics sphere has historically struggled to streamline the commercialization of its developing technologies; however, many now-ubiquitous instruments are fundamentally enabled by microfluidics (e.g., DNA sequencing, digital PCR, PCR arrays) [69, 70]. Improving the graduation of proof-of-concept technologies to commercial systems will be necessary to fully disseminate the benefits of microfluidics to research and development laboratories. This will be particularly critical in rapidly developing applications such as stem cell therapies, which may simultaneously prove to be a key area in demonstrating the utility of microfluidic technologies.
Acknowledgments
This work was funded in part by the Australian Research Council Special Research Initiatives Scheme (Stem Cells Australia, http://www.stemcellsaustralia.edu.au/) and National Health and Medical Research Council Project Grant APP1041277. The authors would like to apologize to those whose work could not be cited because of space limitations in this concise review. D.M.T. is currently affiliated with the Institute of Medical Biology, Agency for Science, Technology and Research (A*STAR), Singapore.
Author Contributions
D.M.T., H.C., and N.R.G.: conception and design, manuscript writing, final approval of manuscript; J.J.C.-W.: conception and design, financial support, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
D.M.T. and J.J.C-W. are uncompensated inventors on a patent application.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Weissman IL. Stem cells: Units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 4.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 5.Serra M, Brito C, Correia C, et al. Process engineering of human pluripotent stem cells for clinical application. Trends Biotechnol. 2012;30:350–359. doi: 10.1016/j.tibtech.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Duffy DC, McDonald JC, Schueller OJ, et al. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 7.Titmarsh DM, Chen H, Wolvetang EJ, et al. Arrayed cellular environments for stem cells and regenerative medicine. Biotechnol J. 2013;8:167–179. doi: 10.1002/biot.201200149. [DOI] [PubMed] [Google Scholar]
- 8.Salieb-Beugelaar GB, Simone G, Arora A, et al. Latest developments in microfluidic cell biology and analysis systems. Anal Chem. 2010;82:4848–4864. doi: 10.1021/ac1009707. [DOI] [PubMed] [Google Scholar]
- 9.Park JY, Takayama S, Lee S-H. Regulating microenvironmental stimuli for stem cells and cancer cells using microsystems. Integr Biol (Camb) 2010;2:229–240. doi: 10.1039/c000442a. [DOI] [PubMed] [Google Scholar]
- 10.Neuži P, Giselbrecht S, Länge K, et al. Revisiting lab-on-a-chip technology for drug discovery. Nat Rev Drug Discov. 2012;11:620–632. doi: 10.1038/nrd3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesher-Perez SC, Frampton JP, Takayama S. Microfluidic systems: A new toolbox for pluripotent stem cells. Biotechnol J. 2013;8:180–191. doi: 10.1002/biot.201200206. [DOI] [PubMed] [Google Scholar]
- 12.Gossett DR, Weaver WM, Mach AJ, et al. Label-free cell separation and sorting in microfluidic systems. Anal Bioanal Chem. 2010;397:3249–3267. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McFaul SM, Lin BK, Ma H. Cell separation based on size and deformability using microfluidic funnel ratchets. Lab Chip. 2012;12:2369–2376. doi: 10.1039/c2lc21045b. [DOI] [PubMed] [Google Scholar]
- 14.Didar TF, Tabrizian M. Adhesion based detection, sorting and enrichment of cells in microfluidic Lab-on-Chip devices. Lab Chip. 2010;10:3043–3053. doi: 10.1039/c0lc00130a. [DOI] [PubMed] [Google Scholar]
- 15.Singh A, Suri S, Lee T, et al. Adhesion strength-based, label-free isolation of human pluripotent stem cells. Nat Methods. 2013;10:438–444. doi: 10.1038/nmeth.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi S, Karp JM, Karnik R. Cell sorting by deterministic cell rolling. Lab Chip. 2012;12:1427–1430. doi: 10.1039/c2lc21225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lillehoj PB, Tsutsui H, Valamehr B, et al. Continuous sorting of heterogeneous-sized embryoid bodies. Lab Chip. 2010;10:1678–1682. doi: 10.1039/c000163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung HW, Chen A, Choo AB, et al. Agitation can induce differentiation of human pluripotent stem cells in microcarrier cultures. Tissue Eng Part C Methods. 2011;17:165–172. doi: 10.1089/ten.TEC.2010.0320. [DOI] [PubMed] [Google Scholar]
- 19.Toh Y-C, Voldman J. Fluid shear stress primes mouse embryonic stem cells for differentiation in a self-renewing environment via heparan sulfate proteoglycans transduction. FASEB J. 2011;25:1208–1217. doi: 10.1096/fj.10-168971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim L, Vahey MD, Lee HY, et al. Microfluidic arrays for logarithmically perfused embryonic stem cell culture. Lab Chip. 2006;6:394–406. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 21.Titmarsh D, Hidalgo A, Turner J, et al. Optimization of flowrate for expansion of human embryonic stem cells in perfusion microbioreactors. Biotechnol Bioeng. 2011;108:2894–2904. doi: 10.1002/bit.23260. [DOI] [PubMed] [Google Scholar]
- 22.Ahsan T, Nerem RM. Fluid shear stress promotes an endothelial-like phenotype during the early differentiation of embryonic stem cells. Tissue Eng Part A. 2010;16:3547–3553. doi: 10.1089/ten.tea.2010.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong W, Zhang W, Wang S, et al. Regulation of fibrochondrogenesis of mesenchymal stem cells in an integrated microfluidic platform embedded with biomimetic nanofibrous scaffolds. PLoS One. 2013;8:e61283. doi: 10.1371/journal.pone.0061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pera MF, Tam PPL. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–720. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 25.Kamei K-i, Guo S, Yu ZT, et al. An integrated microfluidic culture device for quantitative analysis of human embryonic stem cells. Lab Chip. 2009;9:555–563. doi: 10.1039/b809105f. [DOI] [PubMed] [Google Scholar]
- 26.Villa-Diaz LG, Torisawa YS, Uchida T, et al. Microfluidic culture of single human embryonic stem cell colonies. Lab Chip. 2009;9:1749–1755. doi: 10.1039/b820380f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titmarsh DM, Ovchinnikov DA, Wolvetang EJ, et al. Full factorial screening of human embryonic stem cell maintenance with multiplexed microbioreactor arrays. Biotechnol J. 2013;8:822–834. doi: 10.1002/biot.201200375. [DOI] [PubMed] [Google Scholar]
- 28.Kamei K-i, Ohashi M, Gschweng E, et al. Microfluidic image cytometry for quantitative single-cell profiling of human pluripotent stem cells in chemically defined conditions. Lab Chip. 2010;10:1113–1119. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Tam PPL, Loebel DAF. Gene function in mouse embryogenesis: Get set for gastrulation. Nat Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- 31.Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Cimetta E, Figallo E, Cannizzaro C, et al. Micro-bioreactor arrays for controlling cellular environments: Design principles for human embryonic stem cell applications. Methods. 2009;47:81–89. doi: 10.1016/j.ymeth.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Figallo E, Cannizzaro C, Gerecht S, et al. Micro-bioreactor array for controlling cellular microenvironments. Lab Chip. 2007;7:710–719. doi: 10.1039/b700063d. [DOI] [PubMed] [Google Scholar]
- 34.Chung BG, Flanagan LA, Rhee SW, et al. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 35.Park JY, Kim SK, Woo DH, et al. Differentiation of neural progenitor cells in a microfluidic chip-generated cytokine gradient. Stem Cells. 2009;27:2646–2654. doi: 10.1002/stem.202. [DOI] [PubMed] [Google Scholar]
- 36.Kawada J, Kimura H, Akutsu H, et al. Spatiotemporally controlled delivery of soluble factors for stem cell differentiation. Lab Chip. 2012;12:4508–4515. doi: 10.1039/c2lc40268h. [DOI] [PubMed] [Google Scholar]
- 37.Cimetta E, Sirabella D, Yeager K, et al. Microfluidic bioreactor for dynamic regulation of early mesodermal commitment in human pluripotent stem cells. Lab Chip. 2013;13:355–364. doi: 10.1039/c2lc40836h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YS, Sevilla A, Wan LQ, et al. Patterning pluripotency in embryonic stem cells. Stem Cells. 2013;31:1806–1815. doi: 10.1002/stem.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Titmarsh DM, Hudson JE, Hidalgo A, et al. Microbioreactor arrays for full factorial screening of exogenous and paracrine factors in human embryonic stem cell differentiation. PLoS One. 2012;7:e52405. doi: 10.1371/journal.pone.0052405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King KR, Wang S, Jayaraman A, et al. Microfluidic flow-encoded switching for parallel control of dynamic cellular microenvironments. Lab Chip. 2008;8:107–116. doi: 10.1039/b716962k. [DOI] [PubMed] [Google Scholar]
- 41.Gómez-Sjöberg R, Leyrat AA, Pirone DM, et al. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 42.Lecault V, Vaninsberghe M, Sekulovic S, et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat Methods. 2011;8:581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- 43.Przybyla L, Voldman, J Probing embryonic stem cell autocrine and paracrine signaling using microfluidics. Annu Rev Anal Chem (Palo Alto Calif) 2012;5:293–315. doi: 10.1146/annurev-anchem-062011-143122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellison D, Munden A, Levchenko A. Computational model and microfluidic platform for the investigation of paracrine and autocrine signaling in mouse embryonic stem cells. Mol Biosyst. 2009;5:1004–1012. doi: 10.1039/b905602e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moledina F, Clarke G, Oskooei A, et al. Predictive microfluidic control of regulatory ligand trajectories in individual pluripotent cells. Proc Natl Acad Sci USA. 2012;109:3264–3269. doi: 10.1073/pnas.1111478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blagovic K, Kim LY, Voldman J. Microfluidic perfusion for regulating diffusible signaling in stem cells. PLoS One. 2011;6:e22892. doi: 10.1371/journal.pone.0022892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Przybyla LM, Voldman J. Attenuation of extrinsic signaling reveals the importance of matrix remodeling on maintenance of embryonic stem cell self-renewal. Proc Natl Acad Sci USA. 2012;109:835–840. doi: 10.1073/pnas.1103100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemmingsen M, Vedel S, Skafte-Pedersen P, et al. The role of paracrine and autocrine signaling in the early phase of adipogenic differentiation of adipose-derived stem cells. PLoS ONE. 2013;8:e63638. doi: 10.1371/journal.pone.0063638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frith J, Titmarsh D, Padmanabhan H, et al. Microbioreactor array screening to elucidate the role of Wnt signaling and microenvironmental factors in osteogenic differentiation of mesenchymal progenitor cells. PLoS. doi: 10.1371/journal.pone.0082931. One 2013 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Csaszar E, Kirouac DC, Yu M, et al. Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell. 2012;10:218–229. doi: 10.1016/j.stem.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Skafte-Pedersen P, Hemmingsen M, Sabourin D, et al. A self-contained, programmable microfluidic cell culture system with real-time microscopy access. Biomed Microdevices. 2012;14:385–399. doi: 10.1007/s10544-011-9615-6. [DOI] [PubMed] [Google Scholar]
- 52.White AK, VanInsberghe M, Petriv OI, et al. High-throughput microfluidic single-cell RT-qPCR. Proc Natl Acad Sci USA. 2011;108:13999–14004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawrenz A, Nason, F, Cooper-White, JJ Geometrical effects in microfluidic-based microarrays for rapid, efficient single-cell capture of mammalian stem cells and plant cells. Biomicrofluidics. 2012;6:24112–2411217. doi: 10.1063/1.4704521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H, Li J, Zhang H, et al. Microwell perfusion array for high-throughput, long-term imaging of clonal growth. Biomicrofluidics. 2011;5:44117–4411713. doi: 10.1063/1.3669371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong S, Pan Q, Lee LP. Single-cell level co-culture platform for intercellular communication. Integr Biol (Camb) 2012;4:374–380. doi: 10.1039/c2ib00166g. [DOI] [PubMed] [Google Scholar]
- 56.Guo L, Abrams RM, Babiarz JE, et al. Estimating the risk of drug-induced proarrhythmia using human induced pluripotent stem cell-derived cardiomyocytes. Toxicol Sci. 2011;123:281–289. doi: 10.1093/toxsci/kfr158. [DOI] [PubMed] [Google Scholar]
- 57.Harris K, Aylott M, Cui Y, et al. Comparison of electrophysiological data from human-induced pluripotent stem cell-derived cardiomyocytes to functional preclinical safety assays. Toxicol Sci. 2013;134:412–426. doi: 10.1093/toxsci/kft113. [DOI] [PubMed] [Google Scholar]
- 58.Bagnaninchi PO, Drummond N. Real-time label-free monitoring of adipose-derived stem cell differentiation with electric cell-substrate impedance sensing. Proc Natl Acad Sci USA. 2011;108:6462–6467. doi: 10.1073/pnas.1018260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myers FB, Zarins CK, Abilez OJ, et al. Label-free electrophysiological cytometry for stem cell-derived cardiomyocyte clusters. Lab Chip. 2013;13:220–228. doi: 10.1039/c2lc40905d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song H, Wang Y, Rosano JM, et al. A microfluidic impedance flow cytometer for identification of differentiation state of stem cells. Lab Chip. 2013;13:2300–2310. doi: 10.1039/c3lc41321g. [DOI] [PubMed] [Google Scholar]
- 61.Velasco D, Tumarkin E, Kumacheva E. Microfluidic encapsulation of cells in polymer microgels. Small. 2012;8:1633–1642. doi: 10.1002/smll.201102464. [DOI] [PubMed] [Google Scholar]
- 62.Allazetta S, Hausherr TC, Lutolf MP. Microfluidic synthesis of cell-type-specific artificial extracellular matrix hydrogels. Biomacromolecules. 2013;14:1122–1131. doi: 10.1021/bm4000162. [DOI] [PubMed] [Google Scholar]
- 63.Chung BG, Lee KH, Khademhosseini A, et al. Microfluidic fabrication of microengineered hydrogels and their application in tissue engineering. Lab Chip. 2012;12:45–59. doi: 10.1039/c1lc20859d. [DOI] [PubMed] [Google Scholar]
- 64.Chin CD, Linder V, Sia SK. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip. 2012;12:2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 65.Dong Y, Skelley AM, Merdek KD, et al. Microfluidics and circulating tumor cells. J Mol Diagn. 2013;15:149–157. doi: 10.1016/j.jmoldx.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Sipp D, Turner L. Stem cells. U.S. regulation of stem cells as medical products. Science. 2012;338:1296–1297. doi: 10.1126/science.1229918. [DOI] [PubMed] [Google Scholar]
- 67.Bianco P, Cao X, Frenette PS, et al. The meaning, the sense and the significance: Translating the science of mesenchymal stem cells into medicine. Nat Med. 2013;19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Domenech M, Yu H, Warrick J, et al. Cellular observations enabled by microculture: Paracrine signaling and population demographics. Integr Biol (Camb) 2009;1:267–274. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanchez-Freire V, Ebert AD, Kalisky T, et al. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat Protoc. 2012;7:829–838. doi: 10.1038/nprot.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]