Abstract
Brugada syndrome (BrS) is an inheritable sudden cardiac death disease mainly caused by SCN5A mutations. Traditional approaches can be costly and time-consuming if all candidate variants need to be validated through in vitro studies. Therefore, we developed a new approach by combining multiple in silico analyses to predict functional and structural changes of candidate SCN5A variants in BrS before conducting in vitro studies. Five SCN5A non-synonymous variants (1651G>A, 1776C>G, 1673A>G, 3269C>T and 3578G>A) were identified in 14 BrS patients using direct DNA sequencing. Several bioinformatics algorithms were applied and predicted that 1651G>A (A551T) and 1776C>G (N592K) were high-risk SCN5A variants (odds ratio 59.59 and 23.93). The results were validated by Mass spectrometry and in vitro electrophysiological assays. We concluded that integrating sequence-based information and secondary protein structures elements may help select highly potential variants in BrS before conducting time-consuming electrophysiological studies and two novel SCN5A mutations were validated.
Brugada syndrome (BrS) is a rare inheritable cardiac disorder associated with a high risk of sudden cardiac death (SCD), mostly affecting young men1,2. The prevalence of BrS is estimated to be 5 per 10,000 in western countries and 12 per 10,000 in Southeast Asia3,4. In our previous study, the prevalence of Brugada-type electrocardiogram (ECG) pattern in Taiwan was reported to be 0.13%5. In 1998, SCN5A was found as the first putative causal gene that encodes the cardiac sodium channel hNav1.56,7.
In the past two decades, SCN5A mutations were identified through traditional sequencing technology8. However, a bunch of candidate genetic variants are being identified using high-throughput genomic technologies such as microarrays or next-generation sequencing. Validation of all of them through electrophysiological functional studies can be costly and time-consuming. One possible strategy to overcome this challenge is to evaluate each candidate mutated locus using a bioinformatics approach9,10. Different prediction algorithms and statistical models have been developed to explore the association between genetic variants and diseases by considering protein stability, sequence conservation and physical and chemical property11. Among them, a growing number of reports have demonstrated that such functional prediction algorithms have been utilized and applied in several diseases, including X-linked mental retardation syndrome12, amyotrophic lateral sclerosis13, and prostate cancer14. In addition, a genetic variant may affect the normal function of one protein by changing the hydrogen bond network, pH dependence, and conformational dynamics15. Several studies have taken protein structural variations and free energy changes into consideration when assessing the deregulatory effects16,17,18, which have shown the effectiveness of predicting the possible influences of amino acid changes. To sum up, it is well known that sequence mutation is a primary contributor to protein structural variations, which may interfere with correct folding and/or docking. Therefore, combining the bioinformatics results of genetic variants with both protein structure and function analyses may improve the predictions and facilitate the characterization of the pathogenesis of BrS.
In this study, we hypothesized that utilizing online sequence-based predictors and protein structure predictors concurrently could help predict the protein functional and structural changes of SCN5A variants in BrS. To validate the predicted results, two assays including both mass spectrometry and in vitro electrophysiological experiments were performed.
Results
Clinical characteristics of BrS patients and associations between clinical characteristics and SCN5A variants in patients with BrS
Fourteen BrS patients (13 men) had nucleotide alterations of SCN5A compared to the reference sequences from NCBI website (CCDS46799.1), and clinical data of these patients were summarized in Table S1. The average age at the time of syncope, seizure or cardiac arrest was 42 ± 12 years. Four patients experienced sudden cardiac death (SCD), and 6 presented with seizure or syncope. All of them had normal physical examinations, and none had significant biochemical abnormalities correlated with the electrical events. Cardiac catheterization and echocardiography of these patients did not reveal any coronary artery disease or obvious structural heart disease.
Five variants of SCN5A were identified, and resulted in distinct amino acid changes. The association between clinical presentations and SCN5A variants of each patient with BrS is shown in Table 1. We found that none of the BrS patients carrying the H558R variant presented with SCD whereas more than half of those carrying other 4 variants presented with SCD or seizure. This suggested that the 4 variants were associated with more severe clinical presentations of BrS than the H558R variant. Furthermore, the BrS patients carrying homozygous A551T or N592K variants experienced SCD clinically and had longer PR interval or QRS duration of ECG, respectively, when being compared with the other BrS patients carrying other SCN5A variants. The two homozygous variants caused delay depolarization of action potential resulting in prolonged electrical conduction velocity.
Table 1. Clinical presentation, electrocardiographic parameters and SCN5A variants of 14 patients with Brugada syndrome.
| Number of Patient | Gender | Age (yrs) | Presentation | PR interval | QRS duration | Type of Brugada ECG | Type of SCN5A variants |
|---|---|---|---|---|---|---|---|
| 1 | M | 52 | SCD | 285 | 114 | Spontaneous Type I | A551T |
| 2 | M | 35 | SCD | 156 | 131 | Spontaneous Type I | N592K |
| 3 | M | 34 | Syncope | 182 | 106 | Drug induced Type I | R1193Q |
| 4 | M | 48 | Severe chest pain and dizziness | 130 | 118 | Drug induced Type I | R1193Q |
| 5 | M | 50 | Seizure | 198 | 95 | Spontaneous Type I | P1090L |
| 6 | F | 24 | Seizure | 207 | 127 | Spontaneous Type I | P1090L |
| 7 | M | 70 | Syncope | 183 | 129 | Spontaneous Type I | P1090L |
| 8 | M | 45 | Syncope | 190 | 98 | Spontaneous Type I | P1090L |
| 9 | M | 24 | SCD | 207 | 127 | Spontaneous Type I | P1090L |
| 10 | M | 40 | SCD | 171 | 85 | Spontaneous Type I | P1090L |
| 11 | M | 38 | Chest tightness and palpitation | 168 | 116 | Spontaneous Type I | H558R |
| 12 | M | 44 | Dizziness | 145 | 121 | Drug induced Type I | H558R |
| 13 | M | 36 | Syncope | 165 | 103 | Spontaneous Type I | H558R |
| 14 | M | 51 | Chest tightness with hypotension | 147 | 115 | Spontaneous Type I | H558R |
SCD: sudden cardiac death; ECG: electrocardiogram.
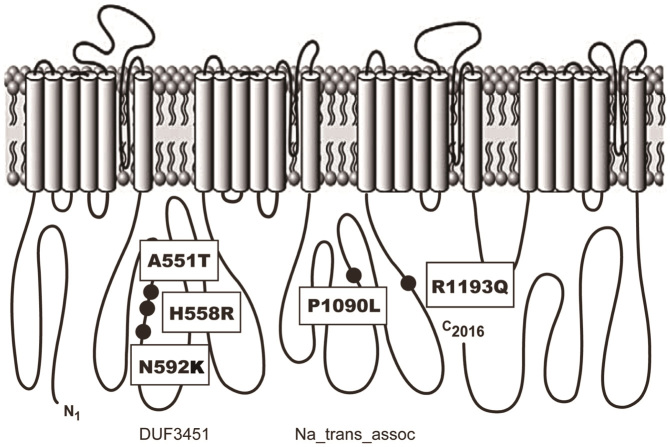
Three variants (1651G>A (A551T), 1776C>G (N592K) and 1673A>G (H558R)) were located in DUF3451 domain and exon 12 of Nav1.5 whereas the other two variants (3269C>T (P1090L) and 3578G>A (R1193Q)) were located in Na_trans_assoc domain and exons 18 and 20, respectively (Figure 1). To assess the substitution deficits of the 5 variants, we retrieved the genomic evolutionary rate profiling (GERP) scores from the UCSC genome browser28,29. As shown in Table 2, the GERP scores of the two variants (A551T and N592K) were high (3.35 and 4.12, respectively) which indicated that they have a low chance of substituting nucleotides (P < 0.001). On the other hand, the other 3 variants showed relatively lower GERP scores, especially the score of P1090L was negative. The negative GERP score of the P1090L variant indicated that the corresponding mutation in this locus may not have significant impact on protein function. Therefore, the results of GERP scores suggested that A551T and N592K may have higher probabilities of causing malfunction of Nav1.5 than the other 3 variants.
Figure 1. Location of five amino acid differences in the linear topology of the human cardiac sodium channel α- subunit.
Table 2. Summary of the results of prediction and EP studies of the 5 SCN5A variants in BrS patients.
| Functional Change | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pt ID (Count) | Exon | cDNAa and aa Changes | pfam Domain | SNP number | Allele Freq (CHB) | Allele Freq (CEU) | GERP Score | SIFT | Polyphen-2 | Align-GVGD | Structural Change | EP studies |
| 1 (1) | E12 | c:1651G>A p:A551T | DUF3451 | NA | NA | NA | 3.35 | T | D | Del1 | + | Pathogenic (our study) |
| 2–5 (4) | E12 | c.1673A>G p.H558R | DUF3451 | rs1805124 | 0.866–0.919 | 0.82 | 2.44 | T | B | N1 | Excluded | Neutral30,33,34,36,37 |
| 6 (1) | E12 | c.1776C>G p.N592K | DUF3451 | NA | NA | NA | 4.12 | Int | D | Del2 | − | Pathogenic (our study) |
| 7–12 (6) | E18 | c.3269C>T p.P1090L | Na_trans _assoc | rs1805125 | NA | 0.989–1 | −0.553 | Int | D | N2 | − | Neutral (our study and37) |
| 13–14 (2) | E20 | c.3578G>A p.R1193Q | Na_trans _assoc | rs41261344 | NA | 0.875–1 | 1.58 | Int | B | N1 | + | Pathogenic30,36,37 |
aa: amino acid; B: Benign; BrS: Brugada syndrome; CHB: Han Chinese in Beijing, China; CEU: Utah residents with Northern and Western European ancestry from the CEPH collection; D: damaging; Del1: Deleterious 1; Del2: Deleterious 2; EP: electrophysiological; Freq: Frequency; GERP: Genomic Evolutionary Rate Profiling; GVGD, Grantham Variation Grantham Deviation; Int: intolerant; N1: Neutral 1; N2: Neutral 2; PolyPhen-2, Polymorphism phenotyping-2; Pt: Patient; SIFT: Sorting Intolerant From Tolerant; T: tolerant.
aRelative to CCDS46799.1.
In silico functional analyses of SCN5A
To explore the functional impact of the five variants in Nav1.5, we utilized three bioinformatics algorithms and the results are shown in Table 2. No significant changes were predicted in the H558R variant, which concurred with previous reports indicating it as a neutral variant without harmful effects30,31,32. In contrast, all three algorithms indicated N592K as a potentially pathogenic variant with high probability scores. One algorithm indicated A551T and P1090L as tolerant substitutions but two algorithms predicted them as possibly damaging variants. However, the probability scores of predictions were much lower than those for the H558R variant (A551T (P = 0.44) vs. H558R (P = 1) and P1090L (GD = 63.55) vs. H558R (GD = 0)). This suggested the bioinformatics algorithms had lower confidence in reporting these two variants as neutral changes. For R1193Q variant, only PolyPhen-2 predicted it as a benign substitution. Since all three algorithms indicated the probability of H558R being a causal variant for BrS was very low, we excluded H558R variant for further studies.
In silico secondary structure analysis of SCN5A variants
To explore potential regions of structural disruption in Nav1.5, we performed the RONN algorithm in assessing the wild type of Nav1.5 (2016 amino acids). As shown in Figure 2A, the two regions with the highest probabilities of disorder were the DUF3451 and Na_trans_assoc domains, which contained all of the five variants. This result suggested that one nucleotide alternation in these two domains of Nav1.5 would have higher chance of causing structural changes of Nav1.5 than mutations in other regions do. To further evaluate the secondary structure of each variant, except the H558R variant, we compared the wild-type sequences with each altered amino acid sequence using the Phyre server25. The Phyre server indicated that the two variants, A551T and R1193Q, were more likely to cause structural changes of Nav1.5 but the N592K and P1090L variants may not result in secondary structural changes (Figure 2B–E). The most altered structure occurred in the R1193Q variant, for which the association with BrS has been reported previously, especially in the Han Chinese population (Figure 2E)33,34. Next, consecutive secondary structural disruptions were shown in the A551T variant from amino acid position 560 to 563 (Figure 2B), indicating a beta sheet conformation was adopted in the neighborhood of the mutated locus. Since such structural variations of A551T and R1193Q variants may affect the transportation efficiency of sodium ions, we decided to further evaluate the biological impacts of them. In addition, N592K and P1090L variants were also included due to their high probabilities in functional disruptions of Nav1.5.
Figure 2. Prediction of secondary structural variations at sequence alteration sites in Nav1.5.
(A) Probability of disorder in Nav1.5 was estimated by the RONN algorithm. The X-axis represents amino acid residue position. The yellow bars represent the two identified domains, DUF3451 and Na_trans_assoc. (B–F) The Phyre server was utilized to evaluate possible secondary structural changes around the five sequence alteration sites. The upper panel shows predictions for the wild-type sequence, and the lower panel shows predictions for each mutant. The predicted results are shown in three-state forms: alpha helix (red), beta strand (blue), and coil (gray), and they are summarized in the consensus row. The yellow bars indicate regions which were changed after sequence substitution.
Risk prediction of the four SCN5A variants
To evaluate whether these four SCN5A variants were associated with BrS, a large-scale screening was performed in 551 healthy individuals using mass spectrum technology. The results are summarized in Table 3, and revealed that the allele frequencies of one non-synonymous variant, 3578G>A (R1193Q), did not reach a significant level (Trend test: P-values ≈ 0.8). In contrast, the trend test indicated the other three variants, 1651G>A (A551T), 1776C>G (N592K) and 3269C>T (P1090L), were significantly enriched in BrS patients (P-value < 0.001). To evaluate the 4 variants, we calculated estimated prediction values (EPVs) and obtained their allele frequencies by using the Exome Sequencing Project (ESP) from National Heart, Lung, and Blood Institute35. As shown in Table 3, 1651G>A (A551T) and 1776C>G (N592K) showed high EPVs and no corresponding variants were identified in the ESP, suggesting they had higher prevalence in BrS patients than the normal controls. On the other hand, 3269C>T (P1090L) and 3578G>A (R1193Q) showed relatively lower EPVs and were reported in the ESP, suggesting these two variants were not specific to BrS patients. In addition to EPVs, ORs were utilized to measure the quantitative effect of each variant. Similarly, the results demonstrated that 1651G>A (A551T) and 1776C>G (N592K) were extremely high-risk in BrS patients (OR: 59.59 and 23.93, respectively) whereas 3269C>T (P1090L) was relatively low-risk (OR: 3.35). To sum up, the results of EPVs and ORs indicated that BrS patients are more likely to have the A551T and the N592K variants. Furthermore, 3269C>T (P1090L) and 3578G>A (R1193Q) were annotated as SNPs in the dbSNP database among Caucasians, suggesting they might have a minor impact on the functions of SCN5A.
Table 3. Risk predictions of the four SCN5A variants for Brugada syndrome.
| Healthy Controls | NHLBI ESP Allele Freq | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | No. of Patient | Het | Homo | WT | Het | Homo | MAF | EPV (95%CI) | Odds Ratioa | p-valueb | EuA | AfA |
| 1651G>A (A551T) | 1 | 0 | 1 | 551 | 0 | 0 | 0 | 100 (64.28–100) | 59.59 | 1.41E-04 | NA | NA |
| 1776C>G (N592K) | 1 | 0 | 1 | 550 | 1 | 0 | 0.001 | 97.46 (61.84–99.83) | 23.93 | 4.33E-04 | NA | NA |
| 3269C>T (P1090L) | 6 | 6 | 0 | 529 | 22 | 0 | 0.020 | 90.68 (80.55–95.54) | 3.35 | 3.08E-08 | 0 | 0.00025 |
| 3578G>A (R1193Q) | 2 | 0 | 2 | 485 | 63 | 3 | 0.063 | 16.15 (0–77.19) | 0.66 | 8.09E-01 | 0.0013 | 0 |
Het: Heterozygote; Homo: Homozygote; MAF: Minor Allele Frequency; WT: Wild type; EPV: Estimated Prediction Value; NHLBI ESP: National Heart, Lung, and Blood Institute Exome Sequencing Project; EuA: European American; AfA: African American; NA: Not available;
aAllelic odds ratio was calculated.
bCochran-Armitage trend test was used.
To sum up the above results, we proposed that 1651G>A (A551T) and 1776C>G (N592K) were possible pathogenic variants with high confidence; 3269C>T (P1090L) and 3578G>A (R1193Q) might cause minor functional changes due to their equivocal data.
Electrophysiological characterization of SCN5A variants using patch clamp
Previous functional studies have demonstrated that R1193Q accelerated fast inactivation of the sodium channel and resulted in reduced sodium current (“loss of function”) in Xenopus oocytes and is an intolerant genetic variant in BrS whereas H558R is not30,33,34,36,37. Thus, electrophysiological experiments were performed to evaluate the functional impact caused by the remaining three variants (A551T (1651G>A), N592K (1776C>G) and P1090L (3269C>T)).
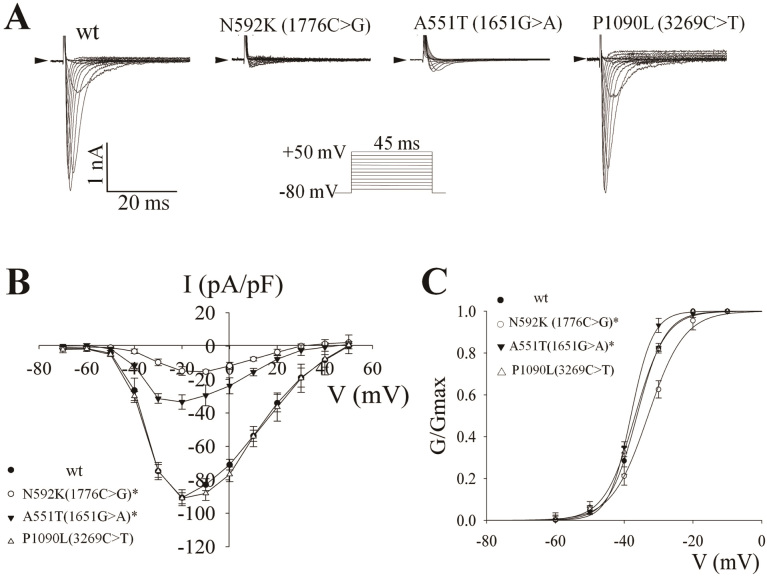
The activation of cloned sodium channels was characterized by a whole-cell voltage clamp technique, and a range of sodium currents was elicited by the voltage protocol (Figure 3A). The sodium current amplitudes of both A551T and N592K variants were significantly lower than that of wild-type, whereas no obvious difference was observed in the P1090L variant.
Figure 3. Effect of the mutations on the electrophysiological function of the human cardiac sodium channel.
(A) Representative current traces. Sodium currents were elicited by a family of step pulses for 45 ms from holding potential of −80 mV to +50 mV with an increase of +10 mV at every step. (B) The voltage-current curves of human cardiac sodium currents of wild-type and mutant proteins. Data are represented as mean ± SEM (n = 3 in N592K (1776C>G) and A551T (1651G>A); n = 4 in wild-type (wt) and P1090L (3269C>T)). Asterisks indicate the statistical significance of the difference between wild-type and the indicated mutants (P < 0.05, by On-way ANOVA with Bonferroni post-hoc test at the same testing membrane potentials). (C) The voltage-dependent steady-state activation curves of sodium currents. Currents were converted to conductance and fit to the Boltzmann equation.
The current densities of the SCN5A constructs were calculated by dividing the current amplitude by the cell capacitance. The relationship of current density to the corresponding activation potential was plotted in Figure 3B. Compared with wild-type protein, the current densities of A551T and N592K were significantly decreased, whereas no significant alteration was found in P1090L. The half-activation potential (V0.5,act) was calculated via the Boltzmann equation, and the results are listed in Table 4 and illustrated in Figure 3C. The steady-state activation of N592K was significantly shifted to the positive potential rightward by +3 mV as compared with wild-type (−33.1 ± 0.3 versus −36.3 ± 0.2, P < 0.01), and the steady-state activation of A551T was significantly shifted to the negative potential leftward by −2 mV (−38.1 ± 0.3 versus −36.3 ± 0.2, P < 0.01). However, no significant shifts were observed in P1090L (−36.8 ± 0.3 versus −36.3 ± 0.2, P = 0.238). In addition, the slope of activation of A551T was significantly smaller than that of wild-type (3.2 ± 0.2 versus 4.1 ± 0.2, P < 0.05).
Table 4. The parameters of steady-state activation and inactivation of wild-type and SCN5A variants.
| WT | N592K (1776C>G) | A551T (1651G>A) | P1090L (3269C>T) | |
|---|---|---|---|---|
| V0.5, act | −36.3 ± 0.2 | −33.1 ± 0.3** | −38.1 ± 0.2** | −36.8 ± 0.3 |
| s, act | 4.1 ± 0.2 | 5.3 ± 0.6 | 3.2 ± 0.2* | 4.5 ± 0.2 |
| V0.5, inact | −86.4 ± 0.7 | −85.2 ± 0.9 | −87.8 ± 0.9 | −87.9 ± 0.7 |
| s, inact | −7.5 ± 0.6 | −7 ± 0.8 | −7.1 ± 0.8 | −8.6 ± 0.6 |
WT: Wild type; V0.5, act is the half-activation potential and s, act is the slope of the activation curve. V0.5, inact is the half-inactivation potential and s, inact is the slope of the inactivation curve. Data are mean ± SEM and the asterisks indicate the statistical significances by One-way ANOVA with Bonferroni post-hoc test (**P < 0.01; *P < 0.05; n = 4 in wt and 3269C>T and n = 3 in 1776C>G and 1651G>A).
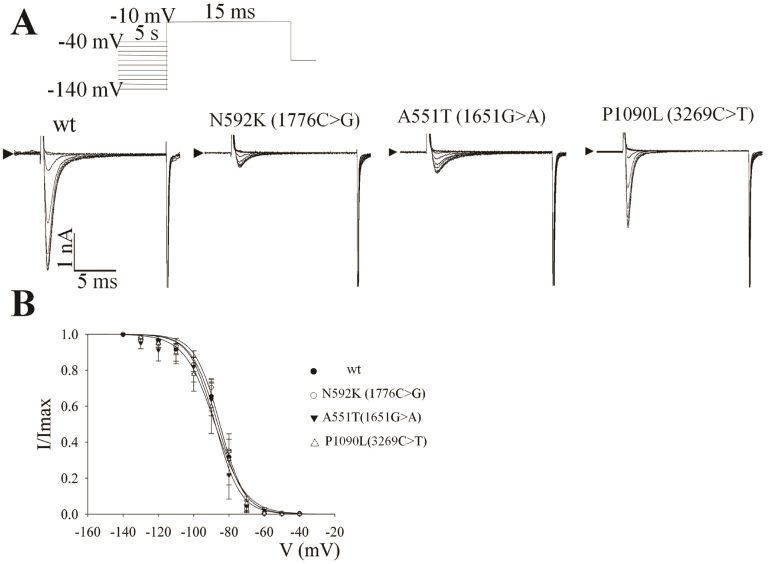
The voltage-dependent steady-state inactivations of all three SCN5A variants were analyzed by the voltage protocol (Figure 4A), and the maximum sodium currents of N592K and A551T elicited at −140 mV were significantly lower than that of either wild-type or P1090L. However, no significant differences in the half-inactivation potentials and the slopes of the inactivation curves were observed among these SCN5A variants.
Figure 4. Effect of the mutations on the steady-state inactivation of the human cardiac sodium channel.
(A) Representative current traces. The protocol used is shown in the inset. (B) The steady-state inactivation curves of the wild-type and mutant Nav1.5. The typical current traces of wild-type protein were elicited by a brief 25 msec step pulse to −10 mV after a 3 sec prepulse to clamp the sodium channel to a steady-state inactivation status. The current amplitude was normalized to the maximum value elicited at −130 or −140 mV and the normalized curves were fit to the Boltzmann equation.
Discussion
In this study, we demonstrated that utilizing online sequence-based predictors and protein structure predictors concurrently may facilitate identifying SCN5A variants with possible functional impact in BrS before performing in vitro studies. In silico analyses of their structural and functional effects predicted which variants would differ from the wild-type protein in functional assays. The electrophysiological study validated the predicted functional defects of the A551T and N592K variants, which caused a decrease in sodium current density without altering the voltage-dependent steady-state inactivation. This could lead to defects in cardiac cellular membrane excitability, resulting in the occurrence of life-threatening arrhythmias. In addition, these two variants were identified in BrS patients resuscitated successfully from ventricular fibrillation with sudden cardiac arrest.
The functional prediction results suggested that H558R caused no harmful effects on the function of Nav1.5. Poelzing et al. showed that co-expressing the R282H-SCN5A mutant with the H558R variant produced significantly greater current than coexpressing the mutant with the wild-type gene, demonstrating the H558R variant restores trafficking of a BrS mutation30. Therefore, our prediction results were compatible with those previous functional studies.
Our in silico results predicted that the P1090L variant may have a functional impact on Nav1.5, but may not result in secondary structural changes. Furthermore, our electrophysiological data showed that the P1090L variant did not affect the function of Nav1.5. However, Tan et al. reported that the P1090L variant showed a significant negative shift of the activation midpoint in patients with the Q1077del mutant (−5 mV), but interestingly, no such shifting was observed in patients without the Q1077 deletion. This suggested that the functional effect of the P1090L variant depends on an interaction with Q1077 splice variants37.
In a recent publication, Takahata et al.38 identified the R1193Q variant in one of six BrS patients. In electrophysiological experiments, the R1193Q variant has been shown to inactivate more rapidly, and the onset of inactivation was three times faster. These functional changes are able to decrease the extent of sodium conductance, which may also be associated with BrS39. In our data, although only one bioinformatics algorithm predicted the severe functional impact of the R1193Q variant, the structural prediction demonstrated it had the most altered secondary structure (Figure 2E). These data implied that performing bioinformatics analyses on both protein function and structure is able to compensate for the insufficiency of examining either function or structure alone. Furthermore, two BrS patients with this variant suffered from sudden cardiac death in our study. Thus, these results suggest that the R1193Q variant may affect the secondary structure of Nav1.5 and increase susceptibility to life-threatening arrhythmia in BrS.
To further evaluate the performances of combining multiple prediction algorithms in predicting SCN5A variants in BrS, we performed a literature search based on the variants reported by Kapplinger et al40. Among those mutations, 12 of them were validated by in vitro studies (Table S2). For each variant, we used the three prediction algorithms (SIFT, PolyPhen-2, and Align-GVGD) to predict the potential functional effect. As shown in Table S2, the prediction results generally matched the results of electrophysiological studies. Ten of the 12 mutations were predicted correctly. The accuracy of combining the three prediction algorithms was 83.33%, which suggested that combining multiple prediction algorithms is an effective approach in screening and prioritizing SCN5A variants in BrS before conducting time-consuming in vitro studies.
Regarding the structure of Nav1.5, the two domains (DUF3451 and Na_trans_assoc) were shown to have the highest probability of disorder (Figure 2A). DUF3451 (pfam11933) is a presumed protein domain with uncharacterized function in eukaryotes. This domain has been found to contain the two members, PF06512 and PF0052041. PF06512 is a member of the eukaryotic sodium transporter associated family, whereas PF00520 is a member of the transmembrane ion channel family. In the 3D structure of sodium channel proteins, such as Nav1.5, the loop flanked by two C-terminal transmembrane helices may determine the ion selectivity of the channel pores42, which suggests that DUF3451 may be associated with the selectivity of the sodium channel. Moreover, the second domain (Na_trans_assoc) we identified is the sodium ion transport-associated domain, PF06512, itself, which contains a region found exclusively in eukaryotic voltage-gated sodium channels. Eight BrS patients had one of two different amino acid changes in this domain, and mutations in this domain may interfere with the transportation by affecting the stability or structure of the sodium channel.
Although genetic testing is not involved in the diagnosis of BrS43, the identification of a causative mutation may help confirm a clinically uncertain diagnosis44. For genetic counseling, knowledge of a patient's BrS-causal gene defect can help physicians identify activities and behaviors that should be avoided and may be useful in determining your risk of experiencing a life-threatening cardiac event. For family screening (all first-degree relatives (children, siblings, parents)), genetic testing in families with an underlying causal gene defect may play a decisive role in who should take precautions in appropriate conditions (ex. avoid fever and drugs that decrease sodium channel availability/functionality) and who should be followed. In most of clinical molecular genetic testing laboratories, however, it is not feasible to perform in vitro functional studies for all identified genetic variants. To compensate this limitation, the prediction results of genetic variants using these algorithms may provide a quick and low-cost reference to physicians.
Even though integrated bioinformatics approaches did have merits, there are some limitations of them. First, most of them require established or well-defined data to develop prediction models, which may overlook the importance of some proteins that have not been investigated in previous studies. For instance, DUF3451, a domain of unknown function, lacks a 3D crystal structure in SWISS-MODEL and further investigations are needed to characterize its biological roles45,46. Second, contradictory results are sometimes obtained across different algorithms, which may be attributed to their use of statistical models that consider different aspects of biological and chemical properties. To reduce the complexity, one of the possible solutions is to perform in silico analyses with multiple prediction algorithms and select the most consistent results among them. For example, the N592K variant was regarded as a harmful allele based on three prediction algorithms; its severe functional changes were then validated by the electrophysiological assay. Lastly, regarding the secondary structure predictions, only sequentially proximal regions were evaluated; that is, the potential long-range effects of these variants on protein structure were not considered. However, these amino acid changes may be physically close to sequentially distant protein domains after folding. Therefore, incorporating 3D structural data into the analysis is essential for improving the prediction results in the future.
In conclusion, with the advancement in high-throughput experiments, processing large numbers of genetic variants is very challenging, even in a rare disease. We demonstrated that concurrently utilizing online sequence-based predictors and protein structure predictors concurrently helps select candidate variants. Two functional pathogenic loci of SCN5A in BrS were identified through this strategy, and thus performing computational approaches facilitates selecting highly potential disease-causative genetic variants for future studies before performing costly and time-consuming electrophysiological studies.
Methods
Study subjects
We identified 14 BrS patients with SCN5A variants in Han Chinese population in Taiwan from 2000 to 2010 (COBRA_ChiP registry, Cohort Of Brugada syndrome in an Asian Chinese Population). This cohort is the largest cohort with symptomatic BrS in Han Chinese population. Symptomatic BrS was diagnosed according to the current consensus shown in supplementary data2. Five hundred fifty-one healthy individuals with no cardiac arrhythmia and normal ECG were genotyped as a control group. This study was approved by the Institutional Review Board of National Taiwan University Hospital and written informed consent was obtained from all subjects. All investigations and experiments followed the Declaration of Helsinki.
Genetic screening for SCN5A variants
DNAs were extracted from peripheral blood by standard procedure (Qiagen, Hilden, Germany). PCR and direct sequencing were performed according to a previous study19. We screened variants on 27 translated exons and exon-intron boundaries of SCN5A. Amplicons were purified and sequenced on a PE Biosystems 373A/3100 sequencer (PE Biosystems, Foster City, CA).
In silico functional analyses of SCN5A variants
To evaluate the potential functional impact of the identified amino acid alterations, we utilized three widely used algorithms to perform in silico analyses: Sorting Intolerant From Tolerant (SIFT)20, Polymorphism phenotyping-2 (PolyPhen-2)21, and Align Grantham Variation Grantham Deviation (Align-GVGD)22,23. Generally, these three algorithms all have high prediction accuracy rates, with reports showing their values of 0.69, 0.76, and 0.88, respectively21,23. The detailed analysis procedures were described in supplementary data. Briefly, SIFT predicts amino acid substitution effects using sequence homology; that is, whether a non-synonymous substitution occurring in a conserved locus among the homologous sequences might be deleterious20. Normalized SIFT scores of the non-synonymous alterations were reported, and a score of less than 0.05 was regarded as a deleterious change (i.e., intolerant). PolyPhen-2 integrates sequence-based and structure-based features to predict amino acid substitution effects using a naïve Bayes classifier21. An amino acid change was classified as “probably damaging” if its probability score was greater than 0.85 or as “possibly damaging” if the score was between 0.85 and 0.15. Lastly, we used Align-GVGD to evaluate the potential physical and chemical changes caused by these variants. Align-GVGD performs multiple sequence alignment to model 3D coordinates of an amino acid by considering its composition, polarity, and volume22,23, and one variant is classified as neutral 1, neutral 2, deleterious 1, deleterious 2, and unclassified based on the computed GV and GD scores. A variant classified into the “neutral 2” category suggests that the Align-GVGD algorithm has lower confidence in predicting it as a tolerant change than that classified into the “neutral 1” category. Similar definition can be applied to the “deleterious 1” and “deleterious 2” categories.
In silico secondary structure analysis of SCN5A variants
To explore the effects of potentially disordered regions on protein structural characteristics, Regional Order Neural Network (RONN)24 algorithm was utilized. RONN is a structural bioinformatics program based on a pattern recognition algorithm, which collects entries from the Molecular Structure Database to create prediction models. We also performed protein structure predictions of Nav1.5 in the Phyre server25, which predicted their secondary structures using three independent algorithms.
Validations by using mass spectrometric technology in a large healthy population
To examine whether the five variants of SCN5A were specific to BrS patients, we performed a genetic screening in 551 healthy individuals (mean age = 41 ± 12, range 9–80 years, Male/Female = 470/30) enrolled from Han Chinese population using mass spectrometric methodology (Sequenom, San Diego, CA). All of their ancestors moved to Taiwan from southeastern China about 400 years ago, and no major population immigration or admixture has subsequently occurred. They were not Taiwanese aborigines. Since BrS affects more frequently in males, the male-female ratio for the control group was consistent with the ratio in our 14 BrS patients (M/F = 13:1). All experimental procedures were described in supplementary data, which followed the standard protocols provided by the manufacturer and were similar to a previous study26. Cochran-Armitage trend test was used to assess the statistical significances, and odds ratios (ORs) were estimated using the additive model. We also calculated estimated prediction values (EPVs) for each variant. EPV was applied in another rare inheritable arrhythmic disease, Long-QT syndrome27. The formula for calculating EPV was (case frequency – control frequency)/(case frequency) and for the confidence interval (CI) of EPV was CI = 1 − 1/(ê{ln(RR) ± z*[SE(log RR)]}). RR is the relative ratio (case frequency/control frequency) and SE[log(RR)] was the standard error around the log of RR.
Validations by performing in-vitro electrophysiological experiments
The details about cloning of SCN5A and cell culture were shown in supplementary data. HEK293T cells were transfected with wide-type or mutant SCN5A constructs, respectively. The eletrophysiological study was performed 48 h after transfection. A droplet of the HEK293T cell suspension was placed in a chamber (1 mL) mounted on the stage of an inverted microscope (Nikon, Ti1, Japan). Cells were incubated in 1.8 mmol·L−1 Ca2+-containing HEPES buffer (NaCl 137.0, KCl 5.4, MgCl2 1.22, CaCl2 1.8, HEPES 10, NaH2PO4 0.33, glucose 5.5, pH 7.4) in the voltage clamp experiments. Membrane currents were recorded at room temperature by using a patch clamp amplifier (WPC-100, E.S.F. Electronic, Göttingen, Germany) via a digital-to-analog converter (Digidata 1322, Axon Instruments) controlled by pClamp software. Patch electrodes were made from borosilicate glass (WPI, Sarasota, Fla., USA). Cell capacitance was assessed by pClamp software immediately after forming whole cell configuration via charging the membrane with a 10 mV square wave for 3 ms to calculate the divided value of current decaying time constant versus the access resistance. The data were filtered by a low pass bessel filter at 10 kHz and were digitized at sampling rate of 100 k Hz. For recording of hNav1.5, the pipette was filled with the internal solution containing (mmol·L−1): CsCl 130, NaCl 10, Mg-ATP 5, MgCl2 2, EGTA 11, CaCl2 1, TEA.Cl 15, and HEPES 10; pH was adjusted to 7.2 using CsOH. For constructing the current and voltage relation curves, sodium currents were elicited by a family of depolarizing pulses from holding potential (−80 mV) stepped by 10 mV to the indicated depolarization potentials every step once per 5 s. The general access resistance (Ra) was about 2–5 MΩ in most of our experimental conditions. The series resistance was electronically compensated by 60–80% to reduce the voltage-clamp error. The pipette tip potential was adjusted to zero for each experiment8. The details of measuring the steady-state activation and inactivation were in supplementary data.
Author Contributions
J.M.J.J., L.P.L., T.P.L., W.P.C. and E.Y.C. conceived and designed the experiments; and J.M.J.J., W.P.C., S.F.S.Y. and W.Y.C. performed the experiments. J.M.J.J., W.P.C. and T.P.L. analyzed the data. J.M.J.J., L.P.L., J.L.L., Y.B.L., C.T.T., C.H.H., J.J.H., F.T.C., C.D.T., C.Y.C., L.W.L. and E.Y.C. contributed reagents, materials, and/or analysis tools. J.M.J.J., T.P.L., W.P.C. and E.Y.C. wrote the paper.
Supplementary Material
Supplementary Data
Acknowledgments
We were sincerely grateful to many cardiologists for referring patients to our hospital, and to the staff of the Sixth Core Lab, Department of Medical Research, National Taiwan University Hospital for technical support and Melissa Stauffer, PhD, of Scientific Editing Solutions, for editing the manuscript. This work was supported partially through grants NTUH 98-N1266, NTUH100-N1775, VN100-08, NTUH101-N2010, NTUH101-S1780, VN101-04, NTUH 101-S1784, NTUH 102-M2224, NTUH.102-S2099, NTUH.102-S2035, UN102-019 and NSC 101-2314-B-002-168-MY2, NSC 101-2314-B-002-173-MY2.
References
- Brugada P. & Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol 20, 1391–6 (1992). [DOI] [PubMed] [Google Scholar]
- Berne P. & Brugada J. Brugada syndrome 2012. Circ J 76, 1563–71 (2012). [DOI] [PubMed] [Google Scholar]
- Brugada P., Brugada R. & Brugada J. The Brugada syndrome. Curr Cardiol Rep 2, 507–14 (2000). [DOI] [PubMed] [Google Scholar]
- Nademanee K. et al. Arrhythmogenic marker for the sudden unexplained death syndrome in Thai men. Circulation 96, 2595–600 (1997). [DOI] [PubMed] [Google Scholar]
- Juang J. M. et al. Brugada-type electrocardiogram in the Taiwanese population--is it a risk factor for sudden death? J Formos Med Assoc 110, 230–8 (2011). [DOI] [PubMed] [Google Scholar]
- Chen Q. et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature 392, 293–6 (1998). [DOI] [PubMed] [Google Scholar]
- Alings M. & Wilde A. “Brugada” syndrome: clinical data and suggested pathophysiological mechanism. Circulation 99, 666–73 (1999). [DOI] [PubMed] [Google Scholar]
- Hsueh C. H. et al. Distinct functional defect of three novel Brugada syndrome related cardiac sodium channel mutations. J Biomed Sci 16, 23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Wang L., Wang Q., Kruger W. D. & Dunbrack R. L. Jr Testing computational prediction of missense mutation phenotypes: functional characterization of 204 mutations of human cystathionine beta synthase. Proteins 78, 2058–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Michonova-Alexova E. & Alexov E. Approaches and resources for prediction of the effects of non-synonymous single nucleotide polymorphism on protein function and interactions. Curr Pharm Biotechnol 9, 123–33 (2008). [DOI] [PubMed] [Google Scholar]
- Schwartz C. E. & Chen C. F. Progress in detecting genetic alterations and their association with human disease. J Mol Biol 425, 3914–8 (2013). [DOI] [PubMed] [Google Scholar]
- Budny B. et al. Novel missense mutations in the ubiquitination-related gene UBE2A cause a recognizable X-linked mental retardation syndrome. Clin Genet 77, 541–51 (2010). [DOI] [PubMed] [Google Scholar]
- Daoud H. et al. Contribution of TARDBP mutations to sporadic amyotrophic lateral sclerosis. J Med Genet 46, 112–4 (2009). [DOI] [PubMed] [Google Scholar]
- Tischkowitz M. D. et al. Identification and characterization of novel SNPs in CHEK2 in Ashkenazi Jewish men with prostate cancer. Cancer Lett 270, 173–80 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl S., Nishi H., Petukh M., Panchenko A. R. & Alexov E. Molecular mechanisms of disease-causing missense mutations. J Mol Biol 425, 3919–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Teng S., Wang L., Schwartz C. E. & Alexov E. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum Mutat 31, 1043–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. et al. A Y328C missense mutation in spermine synthase causes a mild form of Snyder-Robinson syndrome. Hum Mol Genet 22, 3789–97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccuto L. et al. A mutation in a ganglioside biosynthetic enzyme, ST3GAL5, results in salt & pepper syndrome, a neurocutaneous disorder with altered glycolipid and glycoprotein glycosylation. Hum Mol Genet (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li Z., Shen J. & Keating M. T. Genomic organization of the human SCN5A gene encoding the cardiac sodium channel. Genomics 34, 9–16 (1996). [DOI] [PubMed] [Google Scholar]
- Kumar P., Henikoff S. & Ng P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4, 1073–81 (2009). [DOI] [PubMed] [Google Scholar]
- Adzhubei I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian S. V. et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 43, 295–305 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe E. et al. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res 34, 1317–25 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. R., Thomson R., McNeil P. & Esnouf R. M. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21, 3369–76 (2005). [DOI] [PubMed] [Google Scholar]
- Kelley L. A. & Sternberg M. J. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4, 363–71 (2009). [DOI] [PubMed] [Google Scholar]
- Chen P. C. et al. Use of germline polymorphisms in predicting concurrent chemoradiotherapy response in esophageal cancer. Int J Radiat Oncol Biol Phys 82, 1996–2003 (2012). [DOI] [PubMed] [Google Scholar]
- Kapa S. et al. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation 120, 1752–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A. S. & Kent W. J. The UCSC Genome Browser. Curr Protoc Bioinformatics Chapter 1, Unit 1 4 (2007). [DOI] [PubMed] [Google Scholar]
- Cooper G. M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 15, 901–13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelzing S. et al. SCN5A polymorphism restores trafficking of a Brugada syndrome mutation on a separate gene. Circulation 114, 368–76 (2006). [DOI] [PubMed] [Google Scholar]
- Ackerman M. J. et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm 1, 600–7 (2004). [DOI] [PubMed] [Google Scholar]
- Tan B. H. et al. Common human SCN5A polymorphisms have altered electrophysiology when expressed in Q1077 splice variants. Heart Rhythm 2, 741–7 (2005). [DOI] [PubMed] [Google Scholar]
- Hwang H. W. et al. R1193Q of SCN5A, a Brugada and long QT mutation, is a common polymorphism in Han Chinese. J Med Genet 42, e7; author reply e8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Zhao J., Barrane F. Z., Champagne J. & Chahine M. Nav1.5/R1193Q polymorphism is associated with both long QT and Brugada syndromes. Can J Cardiol 22, 309–13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W. et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman M. J. et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart rhythm: the official journal of the Heart Rhythm Society 1, 600–7 (2004). [DOI] [PubMed] [Google Scholar]
- Tan B. H. et al. Common human SCN5A polymorphisms have altered electrophysiology when expressed in Q1077 splice variants. Heart rhythm: the official journal of the Heart Rhythm Society 2, 741–7 (2005). [DOI] [PubMed] [Google Scholar]
- Takahata T. et al. Nucleotide changes in the translated region of SCN5A from Japanese patients with Brugada syndrome and control subjects. Life sciences 72, 2391–9 (2003). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. The common SCN5A mutation R1193Q causes LQTS-type electrophysiological alterations of the cardiac sodium channel. Journal of medical genetics 41, e66 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapplinger J. D. et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm 7, 33–46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic acids research 39, D225–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S. Potassium channel structures. Nature reviews. Neuroscience 3, 115–21 (2002). [DOI] [PubMed] [Google Scholar]
- Antzelevitch C. et al. Brugada syndrome: report of the second consensus conference: endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation 111, 659–70 (2005). [DOI] [PubMed] [Google Scholar]
- Ackerman M. J. et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace 13, 1077–109 (2011). [DOI] [PubMed] [Google Scholar]
- Bateman A., Coggill P. & Finn R. D. DUFs: families in search of function. Acta Crystallogr Sect F Struct Biol Cryst Commun 66, 1148–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J. & Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data