Abstract
Background:
In traditional medicine, Tridax procumbens Linn. is used in the treatment of injuries and wounds. The bacterial endophytes (BEs) of medicinal plants could produce medicinally important metabolites found in their hosts; and hence, the involvement of BEs in conferring wound healing properties to T. Procumbens cannot be ruled out. But, we do not know which types of BEs are associated with T. Procumbens.
Objective:
The objective of this study was to investigate the fast growing and cultivable BEs associated with T. procumbens.
Materials and Methods:
Leaves and stems of healthy T. Procumbens plants were collected and cultivable BEs were isolated from surface-sterilized leaf and stem tissue samples using Luria-Bertani (LB) agar (medium) at standard conditions. A polymerase chain reaction was employed to amplify 16S rRNA coding gene fragments from the isolates. Cultivable endophytic bacterial isolates (EBIs) were identified using 16S rRNA gene nucleotide sequence similarity based method of bacterial identification.
Results:
Altogether, 50 culturable EBIs were isolated. 16S rRNA gene nucleotide sequences analysis using the Basic Local Alignment Search Tool (BLAST) revealed identities of the EBIs. Analysis reveals that cultivable Bacillus spp., Cronobacter sakazakii, Enterobacter spp., Lysinibacillus sphaericus, Pantoea spp., Pseudomonas spp. and Terribacillus saccharophilus are associated with T. Procumbens.
Conclusion:
Based on the results, we conclude that 24 different types of culturable BEs are associated with traditionally used medicinal plant, T. Procumbens, and require further study.
KEY WORDS: 16S rRNA, herbs, injury, traditional medicine, Tridax, wound healing
INTRODUCTION
In many tropical countries, a few indigenous plants are utilized traditionally as remedies by various rural and tribal communities in the treatment of disease or injury. Tridax procumbens is one of the most common plants used by rural and tribal communities to cure various health ailments.[1,2,3]
Tridax procumbens is a well-known traditional medicinal plant.[4,5] Traditionally, the juice from leaves of T. Procumbens has been used for healing dermal wounds or injury due to its prohealing properties.[3] The results published by Yaduvanshi et al.,[6] show that T. Procumbens leaf juice possesses dose-dependent prohealing property.
The research findings on experimentally induced dermatophytic lesion in mice reported by Sharma et al.,[7] indicate that T. Procumbens plant extract is an excellent topical antifungal agent for the cure of dermatophytosis. This plant also known to have antibacterial properties.[5,8]
Bacteria that live inside plants are called as endophytes (bacterial). Some reports suggest that endophytic bacteria can produce antifungal[9,10] and antibacterial[11] compounds. Due to their ability to produce antibiotics, bacterial endophytes (BEs) are also considered as one of the potential sources of novel antibiotics. The plants that have antimicrobial properties are the potential sources of novel antibiotics producing BEs. Tridax procumbens is known for its antimicrobial properties; hence, it can be a source of novel antibiotics producing BEs. The aim of this brief study was to investigate types of fast growing and cultivable BEs associated with T. Procumbens. The isolated and identified fast growing and cultivable BEs are reported in this paper.
MATERIALS AND METHODS
No specific permissions were required to collect the plant material samples. The T. Procumbens plant species used in this study for the isolation of fast growing BEs is not an endangered or a protected species in Malaysia.
Surface-sterilization of plant tissues
The stem and leaf samples were collected from 35 individual healthy-looking T. Procumbens plants from their natural habitat at Taman Tasek Indah (Semeling, Kedah, Malaysia) and AIMST University campus, Kedah, Malaysia. Collected stem and leaf samples were washed under plenty of running tap water. To avoid contamination, surface-sterilization of leaves and stem pieces samples was carried out carefully as described elsewhere.[12,13] In brief, plant material samples were cleaned with detergent (5% Teepol) and washed with distilled water. Samples were taken into laminar hood and immersed in 70% ethanol for 30 s. The samples were treated with sodium hypochlorite (5%) solution for 3 min; finally, samples were dipped in absolute alcohol for about 10 s before washing with (at least 3 times) autoclaved distilled water.
Inoculation and incubation of tissues
About sixty (60) leaf-discs (~1 cm2) and pieces of stem tissue from 35 plant material samples were inoculated aseptically in the petri plates containing Luria-Bertani (LB) agar. Water from the last round of samples washing was tested for bacterial presence to verify effectiveness of surface-sterilization process. The petri plates (with inoculated tissue samples) were incubated for 18-20 h at 37°C (±3°C) in the dark.
Isolation and identification of BEs
The isolation and cultivation of endophytic bacterial isolates (EBIs), amplification of 16S rRNA coding gene fragments, and identification of isolates was carried out as described elsewhere.[12] In brief, well-grown colonies of cultivable BEs that are visible on (leaf and stem tissue margins) the LB medium at standard conditions were chosen randomly for further analysis. Pure culture of each isolate was cultivated separately in universal bottles containing 10 mL LB medium. Each isolate-cultivation was carried out under standard conditions: 37°C, 160 rpm for 18 h. Glycerol stocks were prepared and kept at -80°C to preserve the endophytic isolates for future research.
The lysate of bacterial cells was used in polymerase chain reaction (PCR) to amplify 16S rDNA. The 16S rDNA PCR was carried out in a total volume of 50 μL containing PCR buffer, 2.5 mM MgCl2, 0.16 mM dNTPs, 0.75 U Taq DNA polymerase, and 0.2 pmoL forward [Bak11W-F; 5’-AGT TTG ATC MTG GCT CAG-3’] and reverse [Bak-R; 5’-GGA CTA CHA GGG TAT CTA AT-3’] primer. The PCR conditions were as follows: Initial hot start at 95°C for 3 min, 30 amplification cycles of 95°C for 30 sec (denaturation), 52°C for 30 sec (annealing), and 72°C for 30 sec (extension), and finally one cycle of 72°C for 5 min.[12]
The amplified PCR products were purified and both strands of amplified 16S rDNA were sequenced. Finally, the isolates were identified based on hits analysis from megablast (highly similar sequences) output.
RESULTS
The incubation of inoculated leaf and stem tissue samples on the LB agar enabled culturable BEs to grow. By the end of the incubation, the colonies of BEs were visible on the margins of the inoculated leaf discs and stem tissues. Altogether, 50 (34 from leaves tissue and 16 from stem tissue) EBIs were analyzed.
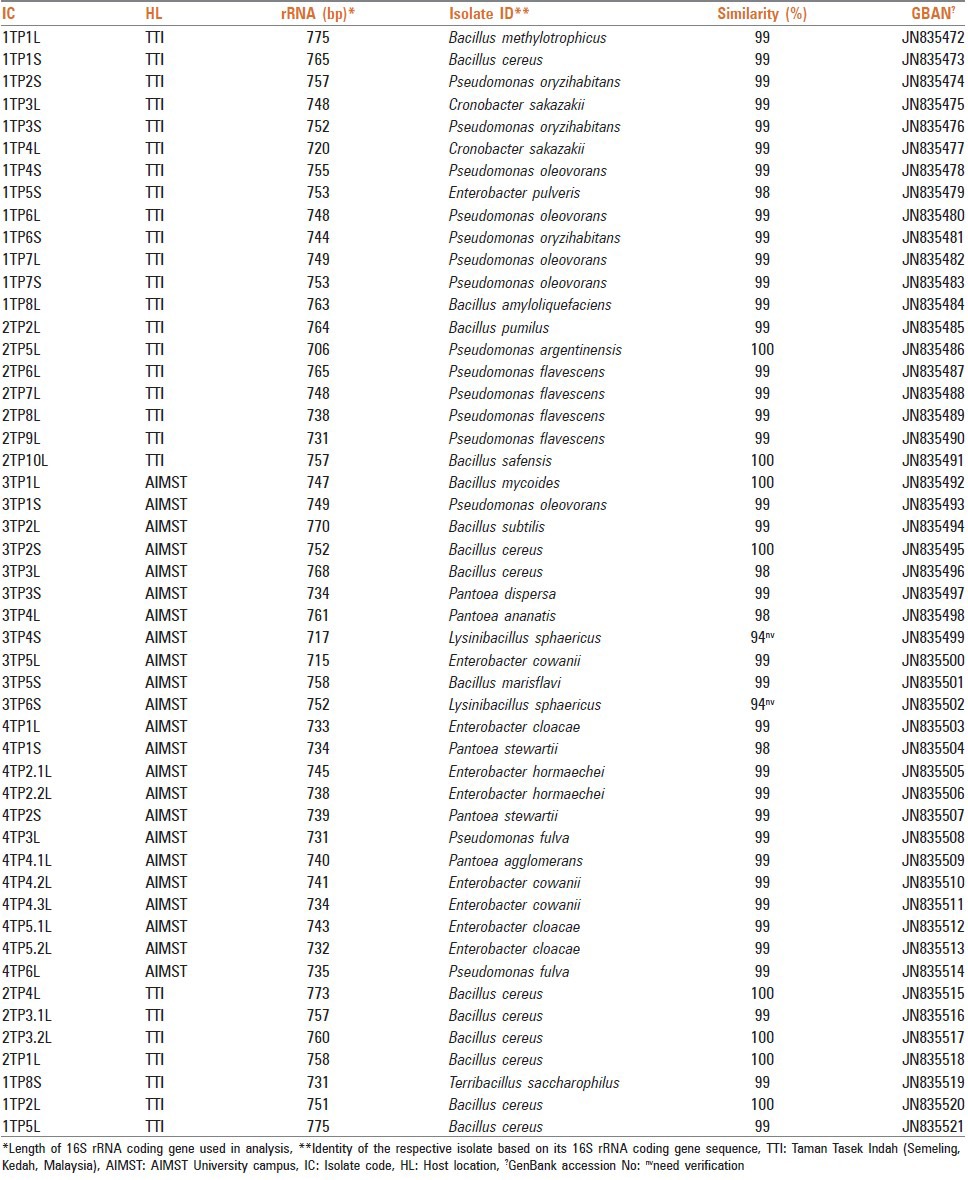
Each EBI was identified based on its 16S rRNA coding gene nucleotide sequence BLAST (megablast) hits analysis. Annotated, 16S rRNA coding gene fragments nucleotide sequences of all 50 EBIs have been deposited in the GenBank/DDBJ/EMBL DNA database under accession numbers: JN835472 - JN835521.
The scrutiny of the identified 50 isolates revealed that there were 24 different types of bacterial species namely, Bacillus amyloliquefaciens, B. cereus, B. marisflavi, B. methylotrophicus, B. mycoides, B. pumilus, B. safensis, B. subtilis, Cronobacter sakazakii, Enterobacter cloacae, E. cowanii, E. hormaechei, E. pulveris, Lysinibacillus sphaericus, Pantoea agglomerans, P. ananatis, P. dispersa, P. stewartii, Pseudomonas argentinensis, P. flavescens, P. fulva, P. oleovorans, P. oryzihabitans, and Terribacillus saccharophilus. Thirty-two (32)% of the isolates represented Bacillus genus. The accession numbers of deposited 16S rRNA coding gene fragments and codes of 50 EBIs are depicted in Table 1.
Table 1.
Isolated and identified fifty (50) EBIs associated with T. procumbens Linn.
DISCUSSION
Tridax procumbens is a well-known traditional medicinal plant used in the treatment of wounds and injuries.[3] In order to explore the potential applications of BEs from this pant, it was necessary to understand its associated BEs. We identified 50 fast growing cultivable EBIs that were associated with leaf and stem tissues of T. Procumbens. However, the EBIs have been reported from various other traditional medicinal plants; for instance, Gynura procumbens,[12] Piper nigrum L.,[14] Trifolium repens,[15] and Artemisia annua.[16] But, to our knowledge, this study is the first to show diverse types of BEs in T. Procumbens.
The 16S rRNA gene nucleotide sequences do provide bacterium species-specific signature and hence, 16S rRNA gene sequence-based bacterial identification is considered as a precise method of bacterial identification.[17] Thus, we used this method for the rapid and accurate identification of EBIs.
It is strongly believed that nearly all terrestrial and aquatic plants do harbor BEs.[18] These BEs can produce valuable metabolites which can be utilized in modern pharmaceutical industry. Mehanni and Safwat[19] suggested that BEs isolated from medicinal plants can produce the same metabolites as their hosts. Therefore, BEs do have a great potential as a source of therapeutic agents including antifungal and antibacterial compounds. T. Procumbens is known to have antimicrobial properties; but recently, Appiah-Opong et al.[20] have reported that this plant do have activity against drug-resistant pathogens. This further elevates the importance of T. Procumbens as a pharmacologically important candidate. The ability of isolated fast growing cultivable BEs of T. Procumbens to produce novel antibiotics needs to be studied. Novel antibiotics can be useful in dealing with drug resistant pathogens, for instance chloroquine-resistant P. falciparum.[13]
Bacillus spp., Pseudomonas spp., and Azospirillum spp. are abundantly found in soil and commonly associated with plants as endophytes. However, in this study, we did not find any Azospirillum spp. in isolated 50 cultivable EBIs. It can be argued that the culture medium used for isolating cultivable BEs might be influencing the type of BEs that can be isolated from the plant tissues. In addition, the leaf and stem tissues were incubated only for 18-20 h, this limited incubation period might have allowed only fast growing cultivable BEs to grow. Some reports have suggested that a seasonal fluctuation of the endophytes does occur in plants;[21,22] therefore, we strongly believe that some other types of cultivable BEs are likely to inhabit T. Procumbens.
Hardoim et al.,[23] have reported that soil type is a major factor that determines the diversity of BEs in plants. In addition, most of the plants are also known to harbor endophytic fungi; therefore, T. Procumbens plants are likely to have some unique endophytic fungi.[12,24]
The novel antibiotics producing ability of the cultivable BEs isolated from other plants has been reported.[9,10,11,25,26,27,28] The extracts from T. Procumbens are known to have antimicrobial activity; but, its activity against chloroquine-resistant P. falciparum (Dd2) is also reported.[13]
Based on the results, we conclude that T. Procumbens does contain diverse (24) types of culturable BEs and requires further study. Our research findings could serve as a foundation for further research on T. Procumbens in determining potential roles of its BEs in producing novel therapeutic compounds.
ACKNOWLEDGEMENT
Authors are grateful to the Ministry of Agriculture and Agro-Based Industry (MoA), Malaysia for financial support (Research grant code number: 05-02-16-SF1001).
Footnotes
Source of Support: Authors are grateful to the Ministry of Agriculture and Agro-Based Industry (MoA), Malaysia for financial support (Research Grant Code Number: 05-02-16-SF1001).
Conflict of Interest: None declared.
REFERENCES
- 1.Parthipan M, Aravindhan V, Rajendran A. Medico-botanical study of Yercaud hills in the eastern Ghats of Tamil Nadu, India. Anc Sci Life. 2011;30:104–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Udupa SL, Udupa AL, Kulkarni DR. Influence of Tridax procumbens on lysyl oxidase activity and wound healing. Planta Med. 1991;57:325–7. doi: 10.1055/s-2006-960108. [DOI] [PubMed] [Google Scholar]
- 3.Diwan PV, Tilloo LD, Kulkarni DR. Steroid depressed wound healing and Tridax procumbens. Indian J Physiol Pharmacol. 1983;27:32–6. [PubMed] [Google Scholar]
- 4.Dhandapani R, Sabna B. Phytochemical constituents of some Indian medicinal plants. Anc Sci Life. 2008;27:1–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Perumal Samy R, Ignacimuthu S, Raja DP. Preliminary screening of ethnomedicinal plants from India. J Ethnopharmacol. 1999;66:235–40. doi: 10.1016/s0378-8741(99)00038-0. [DOI] [PubMed] [Google Scholar]
- 6.Yaduvanshi B, Mathur R, Mathur SR, Velpandian T. Evaluation of wound healing potential of topical formulation of leaf juice of Tridax procumbens L. In mice. Indian J Pharm Sci. 2011;73:303–6. doi: 10.4103/0250-474X.93523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma B, Kumar P, Joshi SC. Topical treatment of dermatophytic lesion on mice (Mus musculus) model. Indian J Microbiol. 2011;51:217–22. doi: 10.1007/s12088-011-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naqash SY, Nazeer RA. Anticoagulant, antiherpetic and antibacterial activities of sulphated polysaccharide from Indian medicinal plant Tridax procumbens L. (Asteraceae) Appl Biochem Biotechnol. 2011;165:902–12. doi: 10.1007/s12010-011-9307-y. [DOI] [PubMed] [Google Scholar]
- 9.Harrison L, Teplow DB, Rinaldi M, Strobel G. Pseudomycins, a family of novel peptides from Pseudomonas syringae possessing broad-spectrum antifungal activity. J Gen Microbiol. 1991;137:2857–65. doi: 10.1099/00221287-137-12-2857. [DOI] [PubMed] [Google Scholar]
- 10.Miller CM, Miller RV, Garton-Kenny D, Redgrave B, Sears J, Condron MM, et al. Ecomycins, unique antimycotics from Pseudomonas viridiflava. J Appl Microbiol. 1998;84:937–44. doi: 10.1046/j.1365-2672.1998.00415.x. [DOI] [PubMed] [Google Scholar]
- 11.Ding L, Maier A, Fiebig HH, Lin WH, Hertweck C. A family of multicyclic indolosesquiterpenes from a bacterial endophyte. Org Biomol Chem. 2011;9:4029–31. doi: 10.1039/c1ob05283g. [DOI] [PubMed] [Google Scholar]
- 12.Bhore SJ, Ravichantar N, Loh CY. Screening of endophytic bacteria isolated from leaves of Sambung Nyawa [Gynura procumbens (Lour.) Merr.] for cytokinin-like compounds. Bioinformation. 2010;5:191–7. doi: 10.6026/97320630005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher PJ, Petrini O, Lappin Scott HM. The distribution of some fungal and bacterial endophytes in maize (Zea mays L.) New Phytol. 1992;122:299–305. doi: 10.1111/j.1469-8137.1992.tb04234.x. [DOI] [PubMed] [Google Scholar]
- 14.Aravind R, Kumar A, Eapen SJ, Ramana KV. Endophytic bacterial flora in root and stem tissues of black pepper (Piper nigrum L.) genotype: Isolation, identification and evaluation against Phytophthora capsici. Lett Appl Microbiol. 2009;48:58–64. doi: 10.1111/j.1472-765X.2008.02486.x. [DOI] [PubMed] [Google Scholar]
- 15.Burch G, Sarathchandra U. Activities and survival of endophytic bacteria in white clover (Trifolium repens L) Can J Microbiol. 2006;52:848–56. doi: 10.1139/w06-039. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zhao GZ, Huang HY, Qin S, Zhu WY, Zhao LX, et al. Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Antonie Van Leeuwenhoek. 2012;101:515–27. doi: 10.1007/s10482-011-9661-3. [DOI] [PubMed] [Google Scholar]
- 17.Clarridge JE., 3rd Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–62. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehanni MM, Safwat MS. Endophytes of medicinal plants. Acta Hort (ISHS) 2010;854:31–9. [Google Scholar]
- 20.Appiah-Opong R, Nyarko AK, Dodoo D, Gyang FN, Koram KA, Ayisi NK. Antiplasmodial activity of extracts of Tridax procumbens and Phyllanthus amarus in in vitro Plasmodium falciparum culture systems. Ghana Med J. 2011;45:143–50. [PMC free article] [PubMed] [Google Scholar]
- 21.Gao XX, Zhou H, Xu DY, Yu CH, Chen YQ, Qu LH. High diversity of endophytic fungi from the pharmaceutical plant, Heterosmilax japonica Kunth revealed by cultivation-independent approach. FEMS Microbiol Lett. 2005;249:255–66. doi: 10.1016/j.femsle.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Forchetti G, Masciarelli O, Izaguirre MJ, Alemano S, Alvarez D, Abdala G. Endophytic bacteria improve seedling growth of sunflower under water stress, produce salicylic acid, and inhibit growth of pathogenic fungi. Curr Microbiol. 2010;61:485–93. doi: 10.1007/s00284-010-9642-1. [DOI] [PubMed] [Google Scholar]
- 23.Hardoim PR, Hardoim CC, van Overbeek LS, van Elsas JD. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One. 2012;7:e30438. doi: 10.1371/journal.pone.0030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vieira ML, Hughes AF, Gil VB, Vaz AB, Alves TM, Zani CL, et al. Diversity and antimicrobial activities of the fungal endophyte community associated with the traditional Brazilian medicinal plant Solanum cernuum Vell. (Solanaceae) Can J Microbiol. 2012;58:54–66. doi: 10.1139/w11-105. [DOI] [PubMed] [Google Scholar]
- 25.Castillo UF, Strobel GA, Ford EJ, Hess WM, Porter H, Jensen JB, et al. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology. 2002;148:2675–85. doi: 10.1099/00221287-148-9-2675. [DOI] [PubMed] [Google Scholar]
- 26.Castillo UF, Strobel GA, Mullenberg K, Condron MM, Teplow DB, Folgiano V, et al. Munumbicins E-4 and E-5: Novel broad-spectrum antibiotics from Streptomyces NRRL 3052. FEMS Microbiol Lett. 2006;255:296–300. doi: 10.1111/j.1574-6968.2005.00080.x. [DOI] [PubMed] [Google Scholar]
- 27.Castillo U, Harper JK, Strobel GA, Sears J, Alesi K, Ford E, et al. Kakadumycins, novel antibiotics from Streptomyces sp NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Microbiol Lett. 2003;224:183–90. doi: 10.1016/S0378-1097(03)00426-9. [DOI] [PubMed] [Google Scholar]
- 28.Waring MJ, Wakelin LP. Echinomycin: A bifunctional intercalating antibiotic. Nature. 1974;252:653–7. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]


