Summary
Nkx2.2 is a homeodomain-containing transcriptional regulator necessary for the appropriate differentiation of ventral neuronal populations in the spinal cord and hindbrain, and endocrine cell populations in the pancreas and intestine. In each tissue, Nkx2.2 inactivation leads to reciprocal cell fate alterations. To confirm the cell fate changes are due to respecification of Nkx2.2-expressing progenitors and to provide a novel tool for lineage tracing in the pancreas and CNS, we generated an Nkx2.2:Cre mouse line by knocking in a Cre-EGFP cassette into the Nkx2.2 genomic locus and inactivating endogenous Nkx2.2. The R26R-CAG-LSL-tdTomato reporter was used to monitor the specificity and efficiency of Nkx2.2:Cre activity; the tomato reporter faithfully recapitulated endogenous Nkx2.2 expression and could be detected as early as embryonic day (e) 9.25 in the developing CNS and was initiated shortly thereafter at e9.5 in the pancreas. Lineage analyses in the CNS confirmed the cell populations thought to be derived from Nkx2.2-expressing progenitor domains. Furthermore, lineage studies verified Nkx2.2 expression in the earliest pancreatic progenitors that give rise to all cell types of the pancreas; however they also revealed more robust Cre activity in the dorsal versus ventral pancreas. Thus, the Nkx2.2:Cre line provides a novel tool for gene manipulations in the CNS and pancreas. genesis 00:00-00.
Keywords: Nkx2.2, Cre-lox, CNS, pancreas, lineage
INTRODUCTION
Nkx2.2 is a tissue specific homeodomain transcription factor expressed in the central nervous system (CNS), pancreas, and intestine. In each of these tissues, Nkx2.2 is dynamically expressed and plays essential roles in cell fate decisions. In the developing neural tube, Nkx2.2 is expressed in a ventral stripe along the anterior-posterior axis at the ~13 somite stage where it specifies the fates of neural progenitors in response to sonic hedgehog (Shh) signaling (Briscoe et al., 1999; Ericson et al., 1997a; Pattyn et al., 2003a; Shimamura et al., 1995). Nkx2.2 function has been most extensively characterized in the hindbrain and spinal cord regions, where disruption of Nkx2.2 causes progenitor populations to undergo fate switches to produce ectopic motor neurons (Briscoe et al., 1999; Pattyn et al., 2003a,b).
In the developing pancreas, Nkx2.2 expression can first be detected in the dorsal pancreatic rudiment around embryonic day (e) 8.75 and subsequently in the ventral pancreatic rudiment at e9.5 (Jorgensen et al., 2007). At the earliest stages, Nkx2.2 is expressed in the Pancreatic duodenal homeobox 1 (Pdx1)—expressing pancreatic progenitor cells, but is subsequently restricted to the Neurogenin3 (Neurog3) —expressing endocrine progenitor cells. Ultimately, Nkx2.2 is localized to specific endocrine populations within the islet, including insulin-producing beta cells, glucagon-producing alpha cells, PP-producing PP cells, and ghrelin-producing epsilon cells (Arnes et al., 2012; Jorgensen et al., 2007; Sussel et al., 1998). In mice lacking Nkx2.2, islet cell specification is disrupted; beta cells are not specified, alpha and PP cells form in reduced numbers, and the majority of endocrine cells express ghrelin (Prado et al., 2004). Nkx2.2 is also expressed in several enteroendocrine cell populations within the small intestine. Disruption of Nkx2.2 function also leads to the loss of many hormone expressing cell types in the duodenum at the expense of increased ghrelin cell numbers (Desai et al. 2008).
The well-defined expression of Nkx2.2 in several essential progenitor domains makes it a good candidate to study progenitor cell fates in normal and perturbed developmental conditions. To provide a tool for manipulating gene expression in the Nkx2.2+ progenitor cells of the CNS and pancreas, we expressed Cre recombinase from the endogenous Nkx2.2 regulatory elements. Although caveats exist, the Cre/LoxP site-specific recombinase system remains a powerful tool to facilitate cell-specific gene inactivation and lineage analyses in the mouse (Harno et al., 2013; Magnuson and Osipovich, 2013; Smedley et al., 2011). Previously, a Nkx2.2-Cre transgenic mouse line was generated to express Cre recombinase under the transcriptional control of a Nkx2.2 CNS regulatory element; however, this element does not drive expression in the pancreas (Lei et al., 2006; Wang et al., 2011). In this study, we knocked Cre into the first coding exon of Nkx2.2 to recapitulate Cre expression in the entire Nkx2.2 expression domain and allow for the study of lineage decisions in the presence and absence of Nkx2.2 function. This novel mouse line provides a valuable tool for studying coordinated gene functions and lineage decisions in the CNS, pancreas and intestine.
RESULTS AND DISCUSSION
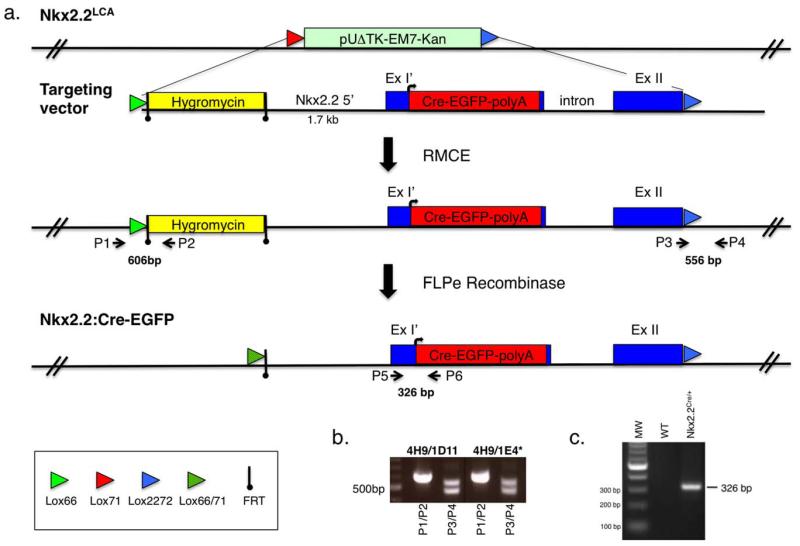
The Nkx2.2:Cre-EGFP knockin line was generated using recombination mediated cassette exchange (RMCE) technology in Nkx2.2 acceptor allele (Nkx2.2LCA) ES cells (Fig. 1a) (Arnes et al., 2012; Chen et al., 2011). To generate the Nkx2.2:Cre-EGFP allele, we engineered a Cre-EGFP fusion protein into the first coding exon of Nkx2.2 such that Cre would be expressed from the endogenous Nkx2.2 start codon and the majority of coding exon 1 would be deleted (Fig. 1a). This strategy allowed for inactivation of Nkx2.2, while retaining all potential non-coding regulatory elements. We confirmed the Cre-mediated exchange event in Nkx2.2LCA ES cells by PCR analysis (Fig. 1c) and ES cell clone 4H9/IE4 was used to generate mice carrying the Nkx2.2CreEGFP allele. Following germline transmission of the Nkx2.2CreEGFP allele, the hygromycin selection cassette was excised from the genome by mating to FLPe mice (Dymecki, 1996), as described previously (Arnes et al., 2012). Subsequent PCR genotyping was performed using primers P5 and P6 (326 bp PCR product) to determine the presence of the Nkx2.2CreEGFP allele (Fig. 1c).
FIG. 1.
Generation of the Nkx2.2:CreEGFP knockin allele. (a) Schematic of the strategy used to create the Nkx2.2:CreEGFP knockin allele. Nkx2.2LCA refers to the RMCE Nkx2.2 acceptor allele integrated into the genomic locus of the 4H9 ES cell line (Arnes et al., 2012). The RMCE lox sites are indicated by colored arrows. The targeting vector includes a FRT flanked hygromycin cassette for positive selection of the targeting event. The CreEGFP gene was recombinered in frame at the Nkx2.2 ATG codon and deleted most of coding exon 1. ExI’ signifies the truncated region of Nkx2.2 coding exon 1 and ExII refers to the Nkx2.2 coding exon 2. The CreEGFP cassette replaces the Nkx2.2LCA allele using RMCE. P1 and P2 indicate the 5′ PCR primers that verify integration of the hygromycin cassette into the correct genomic locus. P3 and P4 indicate the 3′ primers that distinguish the wild-type Nkx2.2 allele and the newly integrated Nkx2.2:CreEGFP allele containing the 2722 lox site. The Flpe-recombinase is used to remove the FRT-flanked hygromycin cassette. P5 and P6 refer to the genotyping primer set used to determine for the presence of the Nkx2.2:CreEGFP allele. (b) Representative PCR amplification results of Nkx2.2LCA ES cells containing RMCE-mediated recombination of the Nkx2.2:CreEGFP allele. For each RMCE ES cell clone, two sets of primers (P1/P2 and P3/P4) were used. Primers 1 and 2 detect a 606 bp product. Primers 3 and 4 detect a 556 bp for the Nkx2.2:CreEGFP allele and 479 bp for the Nkx2.2 wild-type allele. (c) Representative PCR amplification results of Nkx2.2:CreEGFP mice and wild-type littermates. Primer P5 and P6 amplify a 326 bp band from only the mice carrying the Nkx2.2:CreEGFP allele.
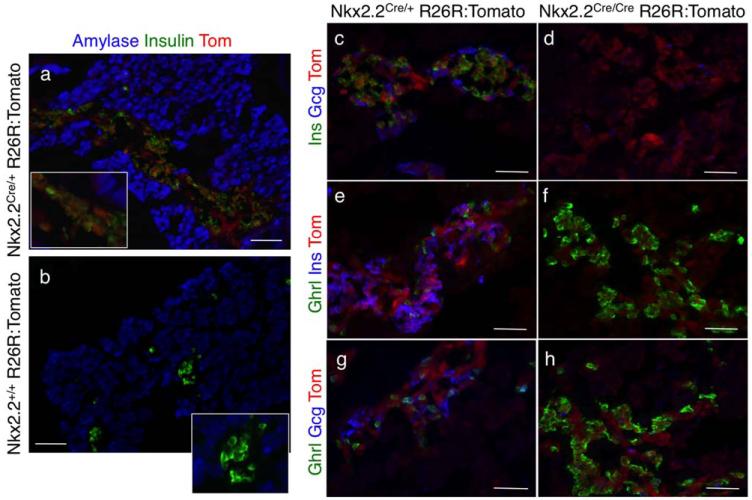
Similar to the conventional Nkx2.2+/− mice (Sussel et al., 1998), heterozygous Nkx2.2CreEGFP/+ mice were viable and did not display overt developmental or postnatal defects (data not shown). To determine the activity and integrity of the novel Nkx2.2CreEGFP/+ allele, we crossed the Nkx2.2CreEGFP/+ mice to R26R-tdTomato Cre-inducible reporter mice (Madisen et al., 2010). Nkx2.2Cre/+; R26R:Tomato embryos and mice showed robust Cre activity; we could detect reporter expression in all previously characterized Nkx2.2-expressing tissues, whereas reporter expression was undetectable in littermates lacking the Nkx2.2-Cre allele (Fig. 2a,b, data not shown). Although Cre appeared to be highly active, we could only detect extremely low levels of EGFP by direct fluorescence and FAC sorting (Supporting Information Fig. 1a-c). Furthermore, any potential EGFP nuclear signal was quenched during tissue processing and therefore did not interfere with subsequent immunofluorescence analyses (Supporting Information Fig. 1d-i). Analogous to mice deleted for both copies of Nkx2.2 (Nkx2.2−/−; (Prado et al., 2004; Sussel et al., 1998), homozygous Nkx2.2CreEGFP/CreEGFP mice died shortly after birth and displayed pancreatic phenotypes indistinguishable from the Nkx2.2−/− null mice, including the absence of insulin-producing beta cells, decreased glucagon-producing alpha cells and a concomitant increase in ghrelin-producing epsilon cells (Fig. 2c-h).
FIG. 2.
Characterization of the Nkx2.2CreEGFP; R26R:Tomato mice. (a,b) Pancreata derived from Nkx2.2CreEGFP/+; R26R:Tomato e18.5 embryos and Nkx2.2+/+; R26R:Tomato littermate controls were sectioned and immunolabeled with antibodies against amylase (blue) and insulin (green). Tomato reporter expression (red; direct fluorescence) could only be visualized in the embryos containing the Nkx2.2:CreEGFP allele and not the Nkx2.2+/+ littermate controls, verifying the Tomato reporter is only activated in the presence of Cre recombinase. Magnification: 20×; Scale bar = 100 μm. Insets are 40×. (c-h) Pancreata derived from Nkx2.2CreEGFP/+; R26R:Tomato e18.5 embryos and Nkx2.2CreEGFP/EGFP; R26R:Tomato littermate controls were sectioned and immunolabeled with antibodies against endocrine hormones as indicated. Similar to the phenotype of the Nkx2.2−/− mice, insulin+ cells are absent, glucagon+ cells are reduced and ghrelin+ cells are elevated. Magnification: 40×; Scale bar = 500 μm.
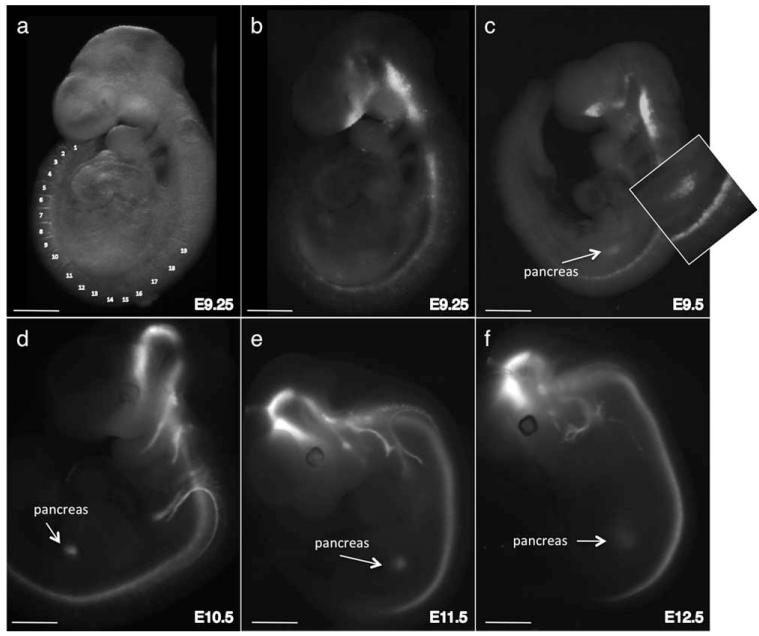
To determine the onset of Nkx2.2-Cre activity in Nkx2.2Cre/+; R26R:Tomato embryos, we visualized the conditional tomato reporter using direct fluorescence of whole mount embryos. We could first detect Cre activity along the entire A/P axis of the CNS by e9.25 (19 somites; Fig. 3a, b). By e9.5, shortly after detection of reporter expression in the CNS, we could visualize tomato expression in the pancreatic rudiments (Fig. 3c). Robust reporter expression in Nkx2.2-expressing domains and in Nkx2.2-derived lineages was maintained at subsequent embryonic stages, confirming the contribution of Nkx2.2-expressing progenitor cells in the ventral hindbrain to vagal motor neuron populations (Ericson et al., 1997b; Fig. 3d-e).
FIG. 3.
Characterization of the onset of Nkx2.2-Cre activity. (a) Whole mount bright field image of an ~9.25 (19 somites) Nkx2.2CreEGFP/+; R26R:Tomato embryo with the somite numbers indicated. (b) Detection of the tomato reporter in the same embryo shown in (a) using direct fluorescence imaging. The reporter is visible along the A/P axis of the CNS. (c) Fluorescence imaging of an e9.5 Nkx2.2CreEGFP/+; R26R:Tomato embryo. Tomato reporter expression is now visible in the pancreas. The inset shows higher magnification of the pancreas and spinal cord. (d,e) Fluorescence imaging of tomato expression in whole mount Nkx2.2CreEGFP/+; R26R:Tomato embryos at successively later embryonic stages. The tomato reporter is also visible in the cells and axons derived from Nkx2.2-expressing progenitor domains in the CNS. Scale bars = 250 μm (E9.25); 450 μm (E9.5); 600 μm (E10.5); 1 mm (E11.5, E12.5).
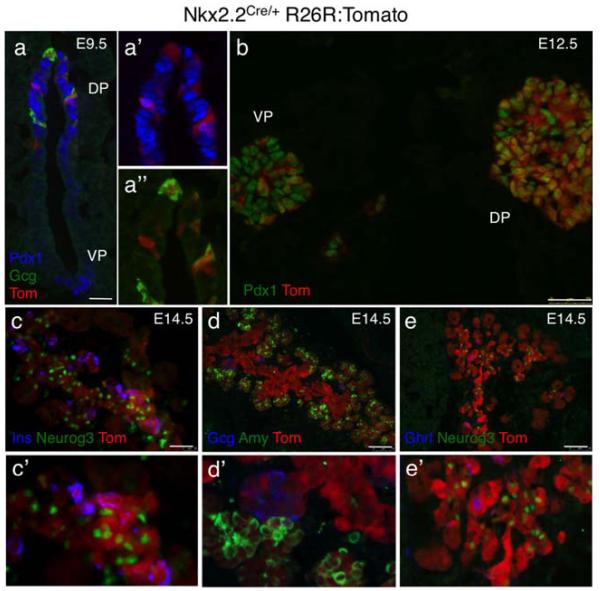
To monitor the efficiency and specificity of the Nkx2.2-Cre allele in each of the organs expressing Nkx2.2, we assessed tomato expression in tissue sections. At e9.5, tomato expression was visible in the dorsal but not ventral pancreatic rudiment, consistent with the previously reported delay of Nkx2.2 expression in the ventral bud (Jorgensen et al., 2007; Fig. 4a). In the dorsal bud at this stage, tomato was co-expressed with all glucagon-producing cells and a subset of Pdx1-expressing cells (Fig. 4a’, a”’). Interestingly, the two populations do not appear to overlap, suggesting that the “early” alpha cells are derived from Nkx2.2-expressing populations that are negative for Pdx1. By e12.5, Nkx2.2-Cre was active in the majority, but not all Pdx1-expressing cells of the dorsal pancreatic bud. Surprisingly, however, even at this later stage of pancreas development, the tomato reporter was only expressed in a small subset of cells in the ventral pancreatic bud, perhaps indicating Cre is not expressed or there is lower Nkx2.2-Cre activity in these cells (Fig. 4b). By 14.5, we can detect tomato activity throughout the dorsal and ventral pancreas in the central and tip domains. Tomato expression appeared most robust in the central epithelium where it was coexpressed with Neurog3 in the endocrine progenitors and insulin, glucagon, and ghrelin in the differentiated endocrine populations (Fig. 4c-e). Tomato expression could also be detected in the amylase-positive acinar cells (Fig. 4d). Endogenous Nkx2.2 expression is normally extinguished from the differentiating acinar cells, however tomato staining of the exocrine lineage is consistent with expression of Nkx2.2 in the pancreatic precursors that give rise to all pancreatic lineages, including the acini (Gu et al., 2003).
FIG. 4.
Assessment of Nkx2.2-Cre activity in the developing pancreas. Combined direct fluorescence (tomato reporter; red) and immunofluorescence of different pancreatic markers in Nkx2.2CreEGFP/+; R26R:Tomato embryos to illustrate cell type specific Cre activity. (a) In a transverse section through an e9.5 embryo, tomato expression can be visualized in the dorsal pancreatic epithelium (DP) and not in the ventral pancreas epithelium (VP). In the dorsal pancreas, tomato is co-expressed with Pdx1 (blue) and glucagon (green). Scale bar = 25 μm. (a’) Higher magnification two channel image showing that tomato is co-expressed with a subset of Pdx1+ cells. (a”) Higher magnification two channel image showing tomato is co-expressed with glucagon+, Pdx1− cells. (b) Tomato is extensively co-expressed with Pdx1 (green) in the dorsal (DP) pancreatic bud at e12.5; only a small subset of Pdx1 cells does not express the tomato reporter. Scale bar = 75 μm. (c-e) In sections through E14.5 pancreas, tomato is co-expressed with Neurog3, insulin, glucagon, ghrelin, and amylase. Magnification 20×; Scale bars = 75 μm. (c’-e’) Higher power images showing co-expressing cells within the pancreas (40×).
In the adult pancreas, tomato-expressing cells are present throughout the exocrine and endocrine compartments. In the islet, tomato is coexpressed with glucagon, insulin, and PP in the alpha, beta, and PP cell populations, as previously reported for endogenous Nkx2.2 (Sussel et al., 1998; Fig. 5a and data not shown). Nkx2.2 is also expressed in the enteroendocrine cells of the small intestine (Desai et al. 2008). Consistently, tomato is expressed in scattered enteroendocrine cell populations within the villi of the duodenum at birth (Fig. 5b).
FIG. 5.
Nkx2.2-Cre activity in the postnatal pancreas and intestine. Combined direct fluorescence (tomato reporter; red) and immunofluorescence of in Nkx2.2CreEGFP/+; R26R:Tomato mice. (a) The tomato reporter is expressed in the exocrine and endocrine compartments of the adult pancreas. Within the islet, tomato is co-expressed with insulin and glucagon. White squares indicate magnified fields in a’ (islet cells) and a” (acinar cells). (b) Tomato expression can be detected in enteroendocrine cells within the villi of the small intestine. Dapi staining (blue) labels all cell nuclei to help visualize the intestinal morphology. Magnification 20×; Scale bars = 100 μm.
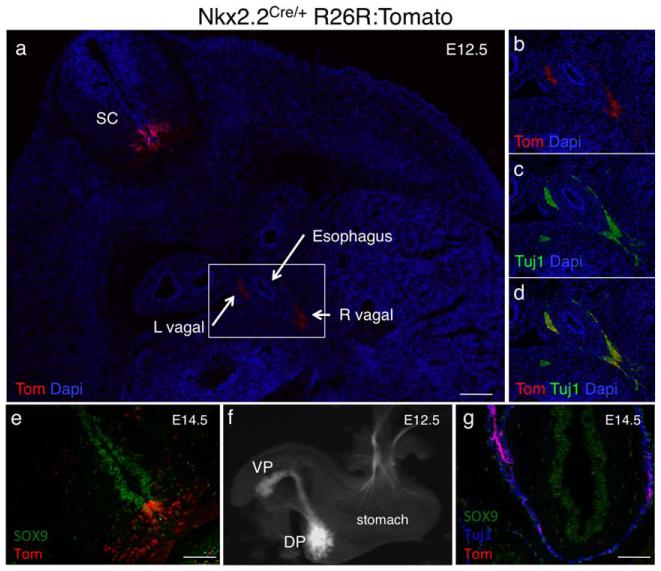
As evident in whole mount embryos (Fig. 2), Nkx2.2-Cre is active along the A/P axis of the CNS. In transverse sections at the level of the thoracic spinal cord, we could detect tomato reporter expression adjacent to the floor plate in the midline Sox9+ progenitor domain and extended laterally into the V3 interneurons that are derived from the Nkx2.2-expressing P3 population (Fig. 6a, e). Nkx2.2-Cre remains active in the Olig2+/Sox10+ oligodendrocyte precursor cells (OPCs), which develop from the P3 domain at later stages (data not shown). Furthermore, consistent with endogenous Nkx2.2 expression in ventral hindbrain progenitors that give rise to the visceral motor neurons (vMN), tomato expression could be detected in the axons of the vMNs that project along the vagus (Xth cranial) nerve and co-express the neuronal marker Tuj1 (Ericson et al., 1997b; Fig. 6b-d). Tomato expression was also maintained in efferent neurons extending into the stomach, face and ear (Fig. 6f-g, Supporting Information Fig. 2).
FIG. 6.
Nkx2.2-Cre activity in the central nervous system. Analysis of tomato expression in the CNS of Nkx2.2CreEGFP/+; R26R:Tomato embryos. (a) Transverse section through the thoracic region of an e14.5 embryo. The tomato reporter is expressed in the ventral spinal cord in the P3 progenitor domain adjacent to the floor plate and in the more lateral V3 interneuron domain. Tomato expression can also be detected adjacent to the esophagus in the left (L) and right (R) vagal nerves. Dapi (blue) labels cell nuclei. Magnification 5×; Scale bar = 500 μm. The rectangle represents the region shown at higher magnification in b-d. (b-d) The tomato reporter is co-expressed with Tuj1 in motor neuron axons extending from Nkx2.2-derived visceral motor neurons in the hindbrain. Magnification 20×; Scale bar = 100 μm. (e) Transverse section of an e14.5 thoracic spinal cord showing co-expression of Tomato and Sox9 (red) in the ventricular zone containing neural precursors. Tomato expressing cells are also present in the more lateral domain containing differentiated interneuron populations. Magnification 20×; Scale bar = 100 μm. (f) Whole mount of dissected e12.5 visceral tissue from a Nkx2.2CreEGFP/+; R26R:Tomato embryo. Tomato is expressed in the dorsal (DP) and ventral (VP) and in a subset of axons extending into the stomach. (g) Section through the distal stomach of an e14.5 Nkx2.2CreEGFP/+; R26R:Tomato embryo. Tomato+ axons extend into the Tuj1 expressing neurons innervating the stomach. Magnification 20×; Scale bar = 100 μm.
In summary, we have generated and characterized a novel Nkx2.2:CreEGFP knockin mouse line. We have demonstrated that Cre activity is highly efficient and restricted to the known Nkx2.2 expression domains in the CNS, pancreas and intestine. Although Cre and EGFP are expressed as a fusion protein, EGFP signal is almost undetectable and cannot be used to label Nkx2.2-expressing cells in vivo. This may indicate Cre-EGFP expression from the Nkx2.2 regulatory domain is generally low, but produces sufficient Cre recombinase to catalyze recombination of the R26R:tomato reporter. Furthermore, our data showing a paucity of tomato expression in the e12.5 ventral pancreatic bud may also suggest that Nkx2.2 is expressed at even lower levels within the ventral pancreas resulting in sub-threshold Cre activity. This unexpected feature of the Nkx2.2:CreEGFP allele may prove to be useful in studies wishing to delete pancreatic genes primarily from the early dorsal pancreas primordia. We also demonstrate the usefulness of the Nkx2.2:CreEGFP allele to perform lineage analyses of the neuronal populations derived from the Nkx2.2-expressing progenitor domains in the ventral spinal cord and hindbrain.
MATERIALS AND METHODS
Generation of Nkx2.2:Cre-EGFP Mice
Nkx2.2:Cre-EGFP mice were generated using the RMCE protocol as described previously (Arnes et al., 2012; Chen et al., 2011). Schematic representation of the strategy is shown in Figure 1. The Nkx2.2:Cre-eGFP allele was created using recombineering to insert the CreEGFP fusion gene into the Nkx2.2 basal exchange vector pNkx2.2.Ex1.H (Arnes et al., 2012). The CreEGFP gene was fused in frame with the Nkx2.2 start ATG and replaced all but 25 base pairs of the Nkx2.2 first coding exon (Fig. 1 “targeting vector”). The modified Nkx2.2:CreEGFP targeting vector was sequence verified. For RMCE, the targeting vector was co-injected with a Cre-expressing vector (Sauer, 1998) into ES cells carrying the Nkx2.2LCA allele (4H9). Cells were screened for loss of the selection cassette in the Nkx2.2LCA allele and for hygromycin resistance, as described previously (Arnes et al., 2012). Surviving clones were PCR verified for the correct insertion. The 5′ primers (P1 and P2) verified integration of the hygromycin cassette within the correct genomic context (606 bp PCR product). The 3′ primers (P3 and P4) verified incorporation of the 3′ Lox2272 site (479 bp wild-type PCR product versus 556 bp Nkx2.2CreEGFP allele), as described previously (Arnes et al., 2012; Fig. 1A,B). Correctly exchanged ES clone 4H9/1E4 was injected into C57/B16/J1 blastocysts and four resulting >95% chimeric male mice were mated to C57/B16/J1 mice (Jackson Labs). Germline passage of the allele from two chimeric males was confirmed by PCR using primers P5 (oDB1) (5′-CAGTCACAGACTACATTTCT-3′) and P6 (ODB128) 5′-TCATCACTCGTTGCATCGA-3′). Removal of the FRT-flanked Hygromycin cassette was accomplished by breeding with ACTB:FLPe mice (Jackson Labs). The Nkx2.2:Cre-EGFP mice will be available to the research community upon request.
Animals
All mice were maintained on a C57/B16/J1 background and housed and treated according to the Columbia University Institutional Animal Care and Use Committee approval protocol AAAC0259 and AAAG3206. Nkx2.2-Cre activity was assessed using the R26R:Tomato reporter strain: R26-TOM (B6.Cg-Gt(RO-SA)26Sortm14(CAG-tdTomato)Hze/J) obtained from the Jackson Laboratory.
Immunofluorescence
Tissue processing and immunofluorescence was performed as previously described (Mastracci et al., 2011). The primary antibodies used were: rabbit α-amylase (1:500; Sigma-Aldrich), goat α-ghrelin (1:500; Santa Cruz Biotechnologies), guinea pig α-glucagon (1:1000; Linco) rabbit α-glucagon (1:1000; DAKO), guinea pig α-insulin (1:1000; Linco), rabbit α-insulin (1:1000; Cell Signaling Technology), rabbit α-pancreatic polypeptide (1:200; Zymed), rabbit α-somatostatin (1:200; Phoenix Pharmaceuticals), rabbit α-Pdx1 (1:1000; Millipore), rabbit α-Sox9 (1:500; Santa Cruz), rabbit α-Neurog3 (1:500; BCBC Antibody Core), rabbit α-Tuj1(1:200; Covance). Sections were incubated with appropriate secondary antibodies conjugated to DyLight-488, DyLight-549 and DyLight-649, Alexa-488, Alexa-647 (Jackson Immunoresearch). Fluorescein-Dolichos Biflorus Agglutinin (DBA, Vector Labs, Burlingame, CA) was added with the primary antibodies. DAPI (1:1000; Invitrogen) was applied for 30 min following secondary antibody incubation. Tomato signal was detected by direct fluorescence of the protein. Images were acquired with either an epifluorescence (Leica DM5500) or a confocal (Zeiss LSM710) microscope.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chang Qing Zhang for performing the RMCE. We acknowledged the Columbia University Cancer Center Transgenic Core and Columbia University DERC Histology Core (P30 DK63608) for expert technical support. We thank members of the Sussel laboratory for critical reading of the manuscript.
Contract grant sponsor: National Institutes of Health (NIH) Beta Cell Biology Consortium (BCBC), Contract grant number: U01 DK089523 (to LS and MM); Contract grant sponsor: NIH, Contract grant number: R01 DK082590 (to LS)
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Arnes L, Leclerc K, Friel JM, Hipkens SB, Magnuson MA, Sussel L. Generation of Nkx2.2:lacZ mice using recombination-mediated cassette exchange technology. Genesis. 2012 doi: 10.1002/dvg.22037. DOI:10.1002/dvg.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O’Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeo-box gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- Chen SX, Osipovich AB, Ustione A, Potter LA, Hipkens S, Gangula R, Yuan W, Piston DW, Magnuson MA. Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis Model Mech. 2011 doi: 10.1242/dmm.006569. DOI:dmm.006569 [pii] 10.1242/dmm.006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S, Loomis Z, Pugh-Bernard A, Schrunk J, Doyle MJ, Minic A, McCoy E, Sussel L. Nkx2.2 regulates cell fate choice in the enteroendocrine cell lineages of the intestine. Dev Biol. 2008;313:58–66. doi: 10.1016/j.ydbio.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Briscoe J, Rashbass P, van Heyningen V, Jessell TM. Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harbor Symp Quant Biol. 1997a;62:451–466. [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997b;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Gu G, Brown JR, Melton DA. Direct lineage tracing reveals the ontogeny of pancreatic cell fates during mouse embryogenesis. Mech Dev. 2003;120:35–43. doi: 10.1016/s0925-4773(02)00330-1. [DOI] [PubMed] [Google Scholar]
- Harno E, Cottrell EC, White A. Metabolic pitfalls of CNS cre-based technology. Cell Metab. 2013;18:21–28. doi: 10.1016/j.cmet.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- Lei Q, Jeong Y, Misra K, Li S, Zelman AK, Epstein DJ, Matise MP. Wnt signaling inhibitors regulate the transcriptional response to morphogenetic Shh-Gli signaling in the neural tube. Dev Cell. 2006;11:325–337. doi: 10.1016/j.devcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson MA, Osipovich AB. Pancreas-specific cre driver lines and considerations for their prudent use. Cell metabolism. 2013;18:9–20. doi: 10.1016/j.cmet.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci TL, Wilcox CL, Arnes L, Panea C, Golden JA, May CL, Sussel L. Nkx2.2 and Arx genetically interact to regulate pancreatic endocrine cell development and endocrine hormone expression. Dev Biol. 2011;359:1–11. doi: 10.1016/j.ydbio.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Samad OA, Krumlauf R, Rijli FM, Brunet JF, Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003a;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Sander M, Ericson J. Complementary roles for Nkx6 and Nkx2 class proteins in the establishment of motoneuron identity in the hindbrain. Development. 2003b;130:4149–4159. doi: 10.1242/dev.00641. [DOI] [PubMed] [Google Scholar]
- Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci USA. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Smedley D, Salimova E, Rosenthal N. Cre recombinase resources for conditional mouse mutagenesis. Methods. 2011;53:411–416. doi: 10.1016/j.ymeth.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- Wang H, Lei Q, Oosterveen T, Ericson J, Matise MP. Tcf/Lef repressors differentially regulate Shh-Gli target gene activation thresholds to generate progenitor patterning in the developing CNS. Development. 2011;138:3711–3721. doi: 10.1242/dev.068270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.