Abstract
Introduction
The chemokine CXCL12, designated stromal cell-derived factor-1 (SDF-1), plays a significant role in many cancer metastases. Previous studies have shown that CXCL12-G801A, a single nucleotide polymorphism (SNP) in the 3’ untranslated region, correlates with breast and lung cancer in Iran. The aim of this study was to evaluate the association of the gene variant CXCL12-G801A with colorectal cancer (CRC) in a Taiwanese cohort.
Material and methods
In this study, we used a denaturing high performance liquid chromatography (DHPLC) method to analyze the frequencies of CXCL12-G801A polymorphic variants between CRC patients (n = 258) and healthy controls (n = 300) in Taiwan.
Results
The SNP distribution was higher in CRC patients with TNM stage II (117/258) than healthy controls (52/300). We observed a significant increase in the G/A plus A/A genotype of the CXCL12-G801A polymorphism in CRC patients (45.35%) compared with healthy controls (17.33%). The analysis of allelic frequencies in both groups revealed that CRC patients have a higher frequency of A allele (23.45%) than healthy controls (8.67%). Furthermore, among older CRC patients, the frequency of the CXCL12-G801A genotype was significantly increased (p = 0.0148).
Conclusions
Our observations suggest that the CXCL12-G801A genotype may be associated with some clinical manifestations in CRC patients in Taiwan.
Keywords: chemokine CXCL12, single nucleotide polymorphism, colorectal cancer, genotype
Introduction
Colorectal cancer (CRC) is proven to be the major cause of malignancy-related deaths worldwide, and is the most prevalent cancer in Taiwan. This high mortality is attributed to failure of its early diagnosis, especially in TNM stage II (T2) [1]. However, many of the genetic factors influencing its appearance still remain far from being fully characterized [2]. Chemokines have been proposed to contribute to tumor growth and metastatic spread of CRC [3]. Chemokines are a superfamily of small (8–11 kDa) proteins, which play a pivotal role in the regulation of leukocyte trafficking and extravasation into sites of tissue inflammation [4]. There are currently four subgroups within the chemokine family: CXC, CC, CX3C, and C chemokine ligands (X represents any amino acid) depending on the positioning of the conserved cysteines in the amino terminal part of these small inducible proteins [5]. One of the most extensively studied chemokines in migration is CXCL12 and its receptor, CXCR4. CXCL12, also named stromal cell-derived factor-1 (SDF-1), is primarily produced by stromal cells implicated in migration, proliferation, differentiation and survival of many cell types, including human and murine hematopoietic stem and progenitor cells [6]. CXCL12 is widely expressed in various organs, including heart, liver, brain, kidney, skeletal muscle and lymphoid [7]. The CXCL12/ CXCR4 pathway has been implicated in cancer metastasis of many different neoplasms [8]. Several studies have also demonstrated that CXCL12 can promote CRC cell migration and result in tumor metastasis [9, 10].
Single nucleotide polymorphisms (SNPs) have become increasingly important tools for the study of the structure and history of the human genome. They are also useful polymorphic markers to investigate genetic susceptibility to disease or to pharmacological sensitivity [11, 12]. An SNP at position 801 (G to A) in the 3’-untranslated region (3'UTR) has been shown to possess the ability of up-regulating the expression of CXCL12 [13, 14]. This CXCL12-G801A polymorphism is associated with blast invasion in acute myelogenous leukemia [15]. Furthermore, CXCL12 is also a costimulator for CD4+ T-cells [16]. It has been suggested that a CXCL12-G801A gene variant is associated with delayed progression of both AIDS and early onset of type 1 diabetes [13, 17]. Several studies further found that CXCL12-G801A was associated with an increased likelihood of developing breast cancer, lung cancer and prostate cancer [18–20].
Despite advances in early detection modalities, CRC screening rates remain low. Therefore, the development of screening for the disease is recommended in individuals who are at increased risk [21]. Citing this evidence, we evaluated the frequency of CXCL12-G801A genotypes and alleles in a Taiwanese cohort of CRC patients and healthy controls. Additionally, denaturing high performance liquid chromatography (DHPLC), which has been adopted in many laboratories for the screening of mutations and SNPs [22], was used to measure the gene polymorphisms. We investigated the hypothesis that CXCL12-G801A polymorphism may be an important indicator of CRC patients correlated with clinical characteristics.
Material and methods
Patients and controls
A total of 258 pathologically confirmed CRC patients and 300 healthy controls individuals enrolled at Yongkang Veterans Hospital, Tainan, Taiwan were included in this study (Table I). Genomic DNA was obtained from the peripheral blood of healthy controls and CRC patients with low-risk stage T2. All patients were preoperatively staged by biological and radiological examinations. Disease stage was determined on the basis of the TNM classification system [23]. Healthy controls consisted of random volunteers with no apparent abnormal findings upon medical examination at Yongkang Veterans Hospital. Peripheral blood samples were obtained from the patients and controls after written informed consent was obtained. This study has been cleared by Yongkang Veterans Hospital Institution Ethics Review Board for human studies and that patients have signed informed consent. All patients and healthy controls were native Taiwanese and resided in the Tainan prefecture or adjacent prefectures.
Table I.
Characteristics of CRC patients and healthy controls
| Variable | No. of controls | Age, mean ± SD | No. of CRC patients | Age, mean ± SD |
|---|---|---|---|---|
| Male | 138 | 59.2 ±15.3 | 137 | 63.4 ±12.9 |
| Female | 162 | 57.8 ±16.2 | 121 | 60.8 ±17.6 |
| Total | 300 | 258 |
Genotyping
Polymorphisms were analyzed by polymerase chain reaction-denatured high performance liquid chromatography (PCR-DHPLC). The PCR primers of CXCL12 were 5’-TGA AGG CTT CTC TCT GTG GG-3’ (forward) and 5’-AGC TTT GGT CCT GAG AGT CC-3’ (reverse), respectively. Each PCR reaction was carried out in a total volume of 20 µl consisting of 0.3 µl of a 10 µmol/l solution of each primer, 1.5 mmol/l MgCl2, 0.8 mmol/l deoxynucleotide triphosphate, 0.5 unit RedTaq DNA polymerase (Sigma), 1 µl of genomic DNA (80 ng/µl), and 15.6 µl of H2O using a T-Gradient Thermocycler (Biometra). Polymerase chain reaction products were digested with EcoRI and BamHI (Promega) for CXCL12, respectively. The restricted products were analyzed in a 2% agarose gel containing ethidium bromide, to make sure that only the specific product was amplified and no additional band occurred (data not shown). The PCR products were further used for DHPLC analysis.
Denatured high performance liquid chromatography analysis was performed on a Wave@ DNA Fragment Analysis System (Transgenomic, San Jose, CA, USA). In the first DHPLC, under non-denaturing conditions (50°C), 10 µl of crude PCR products were automatically injected into a temperature-equilibrated DNASep cartridge and eluted at a flow rate of 0.9 ml/min using a linear gradient of acetonitrile in 0.1 M triethylammonium acetate (TEAA), pH 7.0. Samples with G/G genotypes eluted as single peaks, whereas heterozygous G/A samples eluted as double peaks. To distinguish between the G/G and G/A genotypes, 10 µl of crude PCR products of each sample with a single peak pattern in the first DHPLC was mixed with that of known genotype (G/G), denatured at 95°C for 2 min and then slowly re-annealed by decreasing the temperature down to 25°C at a rate of 0.2°C/s to form homo- and/or heteroduplexes and then applied to the DHPLC cartridge under denaturing conditions (63.9°C). The temperature required for successful resolution of heteroduplex molecules was determined using the WaveMaker software (Transgenomic Inc.).
Sequencing
The genotypes revealed by DHPLC analysis were further confirmed by DNA sequencing with an ABI PRISMR 3730 DNA Analyzer Sequencer using the ABI BigDye Terminator kit v3.1 (ABI, Foster City, CA, USA).
Statistical analysis
Differences in the frequencies of the CXCL12 gene polymorphism between CRC patients and healthy control groups were analyzed using the χ2 test. Statistical analysis was performed using the SAS 8.0 for Windows computer package. Results were considered significant at a level of p < 0.05.
Results
Characteristics of CRC patients and controls
To determine the effect of specific CXCL12 polymorphisms in CRC patients, we recruited 258 CRC patients (137 male and 121 female) in stage T2 and 300 healthy controls (138 male and 162 female) for this research. As shown in Table I, the age range of diagnosis of CRC male patients was 51–76 years (63.4 ±12.9 years) (mean ± SD) and that of CRC female patients was 43–78 years (60.8 ±17.6 years) (mean ± SD). None of the patients or healthy controls had smoking or drinking habits.
CXCL12 polymorphism and CRC risk
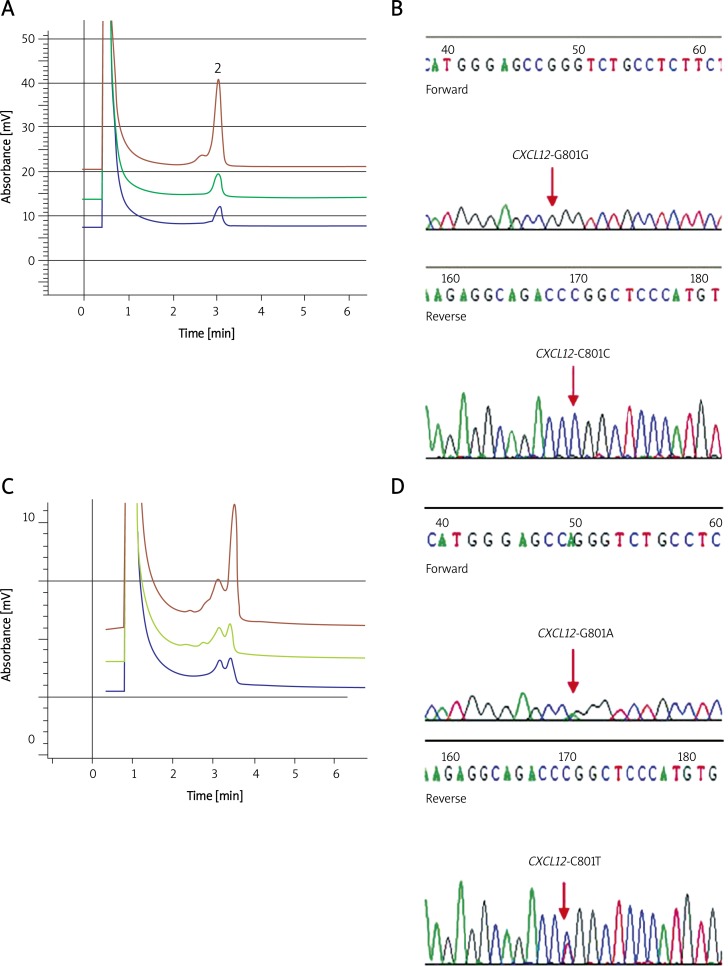
In order to examine the association of CXCL12 gene variants with CRC, we used the PCR-DHPLC system to analyze the prevalence of CXCL12-G801A polymorphism in 258 CRC patients and 300 healthy controls. Figure 1A illustrates the results of three differentiated samples with G/G genotype characterized by a single peak (or homoduplex). Figure 1C illustrates the results of three differentiated samples with G/A genotype characterized by double peaks (or heteroduplex). Figures 1B and 1D show the confirmation of G/G genotype and G/A polymorphism, respectively, by DNA sequencing. Table II shows the genotype distribution of CXCL12-G801A polymorphism in CRC patients and healthy controls. A significant increase in the CXCL12 G/A genotype was observed in CRC patients compared with healthy controls (odds ratio (OR), 0.25; 95% confidence interval (CI), 0.17–0.37; p = 0.0001). The frequency of CXCL12 801 A allele also tended to increase in CRC patients (OR 0.31; 95% CI 0.22-0.44; p = 0.0001; Table III).
Figure 1.
CXCL12-801 on DHPLC elution profile comparison with sequencing. A – CXCL12-801G/G DHPLC elution profile at 63.9°C. B – G/G phenotype revealed by DHPLC was further confirmed by DNA sequencing. C – CXCL12-801G/A DHPLC heteroduplexes profile and sequencing at 63.9°C. D – G/A phenotype revealed by DHPLC was further confirmed by DNA sequencing
Table II.
Distribution of genotypes in percent of CXCL12 polymorphism in CRC patients and healthy controls
| Genotypes | CRC patients | Controls | OR (95% CI) |
|---|---|---|---|
| G/G | 141 (54.65%) | 248 (82.67%) | 1.00 (1.00) |
| G/A | 113 (43.80%) | 52 (17.33%) | 0.25 (0.17–0.37) |
| A/A | 4 (1.55%) | 0 (0) | 0.06 (0.00–1.18) |
Table III.
Allelic frequencies in percent of CXCL12 polymorphism in CRC patients and healthy controls
| Allele | CRC patients | Controls | OR (95% CI) |
|---|---|---|---|
| G | 395 (76.55%) | 548 (91.33%) | 1.00 (1.00) |
| A | 121 (23.45%) | 52 (8.67%) | 0.31 (0.22–0.44) |
Relationship of the CXCL12 polymorphism with clinical parameters in CRC patients
The relationship of the CXCL12-G801A polymorphism with clinicopathologic parameters, including gender and age at diagnosis in CRC patients, was evaluated. As shown in Table IV, there was no signi-ficant difference between male and female CRC patients with CXCL12-G801A SNP (p = 0.0695). Table V shows that among older patients (50 years < age ≤ 70 years and 70 years < age ≤ 90 years), the frequency of the CXCL12-G801A genotype was significantly increased (p = 0.0488 and p = 0.0315). The frequency of CXCL12-G801A genotype of older subjects was significantly higher in CRC patients (n = 22; 68.8%) compared with healthy controls (n = 12; 29.3%; p = 0.0315).
Table IV.
Gender difference of CXCL12 polymorphism in CRC patients and healthy controls
| Variable | Male | Female | Value of p | ||
|---|---|---|---|---|---|
| No. | No. of SNP | No. | No. of SNP | ||
| CRC patients | 137 | 60 | 121 | 57 | 0.0695 |
| Controls | 138 | 33 | 162 | 19 | 0.0797 |
| Value of p | 0.0611 | 0.0203* | |||
Table V.
Difference of distribution of CXCL12-801G/A polymorphism by age in CRC patients and healthy controls
| Variable | 30 < Age ≤ 50 | 50 < Age ≤ 70 | 70 < Age ≤ 90 | Value of p | |||
|---|---|---|---|---|---|---|---|
| No. | No. of SNP | No. | No. of SNP | No. | No. of SNP | ||
| CRC patients | 104 | 32 | 122 | 63 | 32 | 22 | 0.0148* |
| Controls | 127 | 17 | 132 | 23 | 41 | 12 | 0.2353 |
| Value of p | 0.0774 | 0.0488* | 0.0315* | ||||
Discussion
Recent studies have indicated that CXCL12 plays an important role in metastatic cancers [8, 15]. CXCL12-G801A SNP studies have been done in several cancers, including lung, breast, colorectal, and prostate cancer [18–20, 24]. To date, CXCL12-G801A polymorphism in CRC is still the source of some controversy. Previous studies have found that there is no significant difference in genotype distribution and allelic frequencies between CRC patients and healthy controls in Sweden and Spain, using restriction fragment length polymorphism (RFLP) and fluorescence resonance energy transfer (FRET) methods to perform genetic analysis on CXCL12-G801A polymorphism [2, 24]. Recently, Chang et al. found that the frequency of CXCL12 GA/AA genotypes was significantly higher in patients with lymph node metastasis [25]. However, these results need further confirmation, since they do not pass multiple testing corrections, and this feature may also correlate with race, gender, and tumor stage. To our knowledge, this is the first report of DHPLC analysis of CXCL12 polymorphism in CRC.
Denaturing polyacrylamide gel electrophoresis of radiolabelled PCR amplification is time-consuming, laborious, and radioactive. Incomplete enzyme digestion is a problem inherent to the PCR-RFLP technique for genotyping. TaqMan PCR is a cheap, rapid and sensitive method for the detection of SNPs. However, it requires specific equipment and the up front investment of TaqMan PCR instrument is a limiting factor in some areas of the world. In contrast, DHPLC analysis is a cost-effective, rapid, sensitive, and high-throughput technique for CXCL12 G/A genotyping. DHPLC has greater sensitivity at detecting mutations (> 95%), and requires minimal optimization of instrument settings for each SNP studied. This allows high throughput analysis of samples, and the findings are consistent and reproducible [26–28]. These properties should enable a broad screening of CXCL12-G801A polymorphism in routine analysis.
Invasive cancers that are confined within the wall of the colon (TNM stages I (T1) and II (T2)) are often curable with surgery. Over 90% of patients diagnosed at this stage usually will survive the disease beyond 5 years. However, if left untreated, the cancer can spread to regional lymph nodes (T3), and even spread widely around the body (T4). It is usually not curable; approximately 7% of patients diagnosed at this stage survive beyond 5 years [1]. Therefore, the development of an indicator for stage T2 CRC diagnosis is important for reducing the mortality caused by this disease. In this study, we analyzed CXCL12 genotypes in 258 stage T2 CRC patients and 300 healthy controls (Table I). A significant increase in the CXCL12 G/A genotype was observed in CRC patients compared with healthy controls (Table II). The frequency of CXCL12 801 A allele also tended to increase in CRC patients (Table III). Therefore, it is believed that CXCL12-G801A may increase the risk of CRC. The results showed that the CXCL12-G801A polymorphism may be an important tool for indicating and detecting stage T2 CRC.
Furthermore, we explored whether there were any associated age or gender differences in CRC patients and healthy controls. As shown in Table IV, the CXCL12-G801A SNP shows no significant difference between male and female subjects, whether in CRC patients (p = 0.0695) or in healthy controls (p = 0.0797). However, there is a significant difference (p = 0.0203) in CXCL12-G801A of females among CRC patients compared with that among healthy controls. This result is similar to the research performed by Hidalgo-Pascual et al. which indicated a significant difference in female subjects (p = 0.02) [2]. Further, we analyzed the distribution of age in CRC patients with CXCL12-G801A polymorphism. Table V shows that among older CRC patients (50 years < age ≤ 70 years and 70 years < age ≤ 90 years), the frequency of the CXCL12-G801A genotype was significantly increased (p = 0.0488 and p = 0.0315). Our preliminary evidence supported this literature and confirmed that in CRC, the primary risk factor is age, with over 90% of cases diagnosed in people over age 50 [29].
The function of CXCL12-3'UTR-G801A in the transcript and protein expression of CXCL12 is important. Therefore, the influence of CXCL12 genotype CXCL12-G801A on CRC progression may be considered as an important indicator. Colorectal cancer has been seen as a disease with a polygenic background, where oncogenes and tumor suppressor genes and signaling pathways participate [30, 31]. In addition, CXCL12 and its receptor, CXCR4, crosstalk has been found to be crucial for tumor metastasis in colorectal and breast cancer models by inducing chemotactic and invasive responses [32, 33]. Previous studies have noted a trend showing higher CXCR4 expression with higher stages of CRC [3, 34, 35]. However, the detailed mechanism for the involvement of CXCL12/CXCR4 in CRC remains to be elucidated.
In conclusion, this is the first report using the DHPLC method to show a significant association between healthy controls and stage T2 CRC patients with CXCL12-G801A polymorphism in Taiwan. Further studies with larger samples of patients are necessary to determine whether the CXCL12 polymorphism affects the carcinogenesis of CRC, and if the plasma level of CXCL12 could reflect the disease status. The results presented in this study indicated that the CXCL12-G801A polymorphism may be developed as an early diagnostic marker for CRC in the future.
Acknowledgments
Ming-Der Shi and Jing-Hsien Chen contributed equally to this study and therefore share first authorship.
This work was supported by a grant from Yongkang Veterans Hospital Research Program (VHYK-9811), Tainan, Taiwan.
References
- 1.Richards MA. Trends and inequalities in survival for 20 cancers in England and Wales 1986-2001: population-based analyses and clinical commentaries. Foreword. Br J Cancer. 2008;99:S1. doi: 10.1038/sj.bjc.6604570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hidalgo-Pascual M, Galan JJ, Chaves-Conde M, et al. Analysis of CXCL12 3'UTR G > A polymorphism in colorectal cancer. Oncol Rep. 2007;18:1583–7. doi: 10.3892/or.18.6.1583. [DOI] [PubMed] [Google Scholar]
- 3.Ottaiano A, Franco R, Aiello Talamanca A, et al. Overexpression of both CXC chemokine receptor 4 and vascular endothelial growth factor proteins predicts early distant relapse in stage II-III colorectal cancer patients. Clin Cancer Res. 2006;12:2795–803. doi: 10.1158/1078-0432.CCR-05-2142. [DOI] [PubMed] [Google Scholar]
- 4.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–8. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 5.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–7. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 6.Aiuti A, Webb IJ, Bleul C, Springer T, Gutierrez-Ramos JC. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J Exp Med. 1997;185:111–20. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–9. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balkwill F. Chemokine biology in cancer. Semin Immunol. 2003;15:49–55. doi: 10.1016/s1044-5323(02)00127-6. [DOI] [PubMed] [Google Scholar]
- 9.Kollmar O, Rupertus K, Scheuer C, et al. Stromal cell-derived factor-1 promotes cell migration and tumor growth of colorectal metastasis. Neoplasia. 2007;9:862–70. doi: 10.1593/neo.07559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeelenberg IS, Ruuls-Van Stalle L, Roos E. The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res. 2003;63:3833–9. [PubMed] [Google Scholar]
- 11.Brookes AJ. The essence of SNPs. Gen. 1999;234:177–86. doi: 10.1016/s0378-1119(99)00219-x. [DOI] [PubMed] [Google Scholar]
- 12.Hosseini M, Houshmand M, Ebrahimi A. MTHFR polymorphisms and breast cancer risk. Arch Med Sci. 2011;7:134–7. doi: 10.5114/aoms.2011.20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler C, Modi W, Smith MW, et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Science. 1998;279:389–93. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe MA, de Oliveia Cavassin GG, Orellana MD, et al. SDF-1 gene polymorphisms and syncytia induction in Brazilian HIV-1 infected individuals. Microb Pathog. 2003;35:31–4. doi: 10.1016/s0882-4010(03)00088-3. [DOI] [PubMed] [Google Scholar]
- 15.Dommange F, Cartron G, Espanel C, et al. CXCL12 polymorphism and malignant cell dissemination/tissue infiltration in acute myeloid leukemia. FASEB J. 2006;20:1913–5. doi: 10.1096/fj.05-5667fje. [DOI] [PubMed] [Google Scholar]
- 16.Nanki T, Lipsky PE. Cutting edge: stromal-derived factor-1 is a costimulator for CD4+ T-cell activation. J Immunol. 2000;164:5010–4. doi: 10.4049/jimmunol.164.10.5010. [DOI] [PubMed] [Google Scholar]
- 17.Dubois-Laforgue D, Hendel H, Caillat-Zucman S, et al. A common stromal cell-derived factor-1 chemokine gene variant is associated with the early onset of type 1 diabetes. Diabetes. 2001;50:1211–3. doi: 10.2337/diabetes.50.5.1211. [DOI] [PubMed] [Google Scholar]
- 18.Razmkhah M, Talei AR, Doroudchi M, Khalili-Azad T, Ghaderi A. Stromal cell-derived factor-1 (SDF-1) alleles and susceptibility to breast carcinoma. Cancer Lett. 2005;225:261–6. doi: 10.1016/j.canlet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 19.Razmkhah M, Doroudchi M, Ghayumi SM, Erfani N, Ghaderi A. Stromal cell-derived factor-1 (SDF-1) gene and susceptibility of Iranian patients with lung cancer. Lung Cancer. 2005;49:311–5. doi: 10.1016/j.lungcan.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Hirata H, Hinoda Y, Kikuno N, et al. CXCL12 G801A polymorphism is a risk factor for sporadic prostate cancer susceptibility. Clin Cancer Res. 2007;13:5056–62. doi: 10.1158/1078-0432.CCR-07-0859. [DOI] [PubMed] [Google Scholar]
- 21.Whitlock EP, Lin JS, Liles E, Beil TL, Fu R. Screening for colorectal cancer: a targeted, updated systematic review for the U.S. Preventive Services Task Force. Ann Intern Med. 2008;149:638–58. doi: 10.7326/0003-4819-149-9-200811040-00245. [DOI] [PubMed] [Google Scholar]
- 22.Frueh FW, Noyer-Weidner M. The use of denaturing high-performance liquid chromatography (DHPLC) for the analysis of genetic variations: impact for diagnostics and pharmacogenetics. Clin Chem Lab Med. 2003;41:452–61. doi: 10.1515/CCLM.2003.068. [DOI] [PubMed] [Google Scholar]
- 23.Sobin LH, Wittekind C, International union against cancer [UICC] New York: Wiley; 1997. TNM classification of malignant tumors. [Google Scholar]
- 24.Dimberg J, Hugander A, Löfgren S, Wa°gsäter D. Polymorphism and circulating levels of the chemokine CXCL12 in colorectal cancer patients. Int J Mol Med. 2007;19:11–5. [PubMed] [Google Scholar]
- 25.Chang SC, Lin PC, Yang SH, Wang HS, Li AF, Lin JK. SDF-1alpha G801A polymorphism predicts lymph node metastasis in stage T3 colorectal cancer. Ann Surg Onco. 2009;16:2323–30. doi: 10.1245/s10434-009-0501-x. [DOI] [PubMed] [Google Scholar]
- 26.Kuklin A, Munson K, Gjerde D, Haefele R, Taylor P. Detection of single-nucleotide polymorphisms with the WAVE™ DNA Fragment Analysis System. Genet Test. 1998;1:201–6. doi: 10.1089/gte.1997.1.201. [DOI] [PubMed] [Google Scholar]
- 27.Spiegelman JI, Mindrinos MN, Oefner PJ. High-accuracy DNA sequence variation screening by DHPLC. Biotechniques. 2000;29:1084–92. doi: 10.2144/00295rr04. [DOI] [PubMed] [Google Scholar]
- 28.Kuklin A, Davis AP, Hecker KH, Gjerde DT, Taylor PD. A novel technique for rapid automated genotyping of DNA polymorphisms in the mouse. Mol Cell Probes. 1999;13:239–42. doi: 10.1006/mcpr.1999.0239. [DOI] [PubMed] [Google Scholar]
- 29.Potter JD. Colorectal cancer: Molecules and populations. J Natl Cancer Inst. 1999;91:916–32. doi: 10.1093/jnci/91.11.916. [DOI] [PubMed] [Google Scholar]
- 30.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 31.Smith G, Carey FA, Beattie J, et al. Mutations in APC, Kirsten-ras, and p53-alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–8. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 33.Brand S, Dambacher J, Beigel F, et al. CXCR4 and CXCL12 are inversely expressed in colorectal cancer cells and modulate cancer cell migration, invasion and MMP-9 activation. Exp Cell Res. 2005;310:117–30. doi: 10.1016/j.yexcr.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Ottaiano A, di Palma A, Napolitano M, et al. Inhibitory effects of anti-CXCR4 antibodies on human colon cancer cells. Cancer Immunol Immunother. 2005;54:781–91. doi: 10.1007/s00262-004-0636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Takeuchi H, Lam ST, et al. Chemokine receptor CXCR4 expression in colorectal cancer patients increases the risk for recurrence and for poor survival. J Clin Oncol. 2005;23:2744–53. doi: 10.1200/JCO.2005.07.078. [DOI] [PubMed] [Google Scholar]