Abstract
Introduction
Congenital atrial septal defect (ASD) is the second commonest form of cardiac developmental anomaly, responsible for substantial morbidity and mortality in affected individuals. Previous studies have implicated genetic defects in the pathogenesis of ASD. However, ASD is largely a genetically heterogeneous disease and the genetic determinants for ASD in the majority of patients remain to be identified.
Material and methods
The entire coding region of GATA4, a gene encoding a zinc-finger transcription factor essential for normal cardiac morphogenesis, was sequenced in 220 unrelated patients with ASD. The available relatives of the patients harboring the identified mutations and 200 unrelated ethnicity-matched control individuals were genotyped.
Results
Four heterozygous missense GATA4 mutations, p.P36S, p.H190R, p.S262A, and p.V399G, were identified in four unrelated patients with ASD, respectively. These mutations were neither detected in 200 control individuals nor described in the human SNP database. Alignment of multiple GATA4 protein sequences across species indicated that the affected amino acids were highly conserved evolutionarily. Genetic analysis of the available relatives of the mutation carriers showed that in each family the mutation co-segregated with ASD.
Conclusions
The findings expand the spectrum of mutations in GATA4 linked to ASD and provide new insight into the molecular etiology associated with ASD, suggesting the potential implications for the genetic diagnosis and gene-specific therapy for this prevalent cardiovascular abnormality in humans.
Keywords: atrial septal defect, transcription factor, genetics
Introduction
Congenital heart disease (CHD) is the most common form of developmental malformation with an estimated incidence of nearly 1% in live neonates, and is the leading non-infectious cause of infant deaths, with over 29% of infants who die of a birth defect having a cardiovascular abnormality [1]. Clinically CHD comprises at least 25 different types with many additional anatomic variations, of which atrial septal defect (ASD) is the second most prevalent CHD seen in children, accounting for approximately 10% of the overall CHD, and now has become the most frequent CHD found in adults, accounting for as much as 20–40% of all congenital cardiovascular deformities [1, 2]. Congenital ASD may be isolated or associated with other cardiac deformations, such as ventricular septal defect, tetralogy of Fallot, and patent ductus arteriosus. Irrespective of other potential anomalies that accompany ASD, single ASD can result in a spectrum of conditions from no significant cardiac sequelae to cardiac enlargement, reduced exercise capacity, congestive heart failure, pulmonary hypertension, Eisenmenger's syndrome, delayed fetal brain development, arrhythmias, and even sudden cardiac death in the absence of surgical or catheter based repair [3–9]. Despite the high prevalence and the significant association with substantial morbidity and mortality, the fundamental etiology responsible for ASD in the vast majority of cases remains unknown.
Abnormally developed atrial septum is implicated in a heterogeneous, complex pathogenic process with environmental and genetic risk factors playing important roles [10, 11]. There is increasing evidence pointing to a critical role of the zinc finger transcription factor GATA4 in septogenesis [12, 13]. The human GATA4 gene is located on chromosome 8p23.1-p22 and consists of seven exons coding for a protein of 442 amino acids [14]. It is expressed throughout embryonic development and also in the adult heart [12, 13]. Therefore, GATA4 has recently been one of the prime candidate genes in identifying the molecular determinants for structural congenital heart defects. To date, more than 20 germline mutations in the coding exons of the GATA4 gene have been identified in patients with a wide variety of congenital heart defects including ASD, ventricular septal defects, atrioventricular septal defects, tetralogy of Fallot, and endocardial cushion defect [15–32]. Nevertheless, ASD is genetically heterogeneous and the genetic determinants for ASD in most patients are still to be identified.
To investigate the occurrence and prevalence of GATA4 mutations in a newly recruited cohort of 220 unrelated patients with congenital ASD, the coding exons and splice junctions of GATA4 were sequenced in patients with ASD and matched healthy individuals used as controls. As a result, four novel heterozygous missense GATA4 mutations, p.P36S, p.H190R, p.S262A, and p.V399G, were identified in 4 unrelated patients with ASD, respectively, which were absent in a matched control population. Genetic analysis of the available family members of the mutation carriers indicated that the mutation co-segregated with ASD in each family. The findings expand the spectrum of mutations in GATA4 linked to ASD and provide new insight into the molecular rationale involved in the pathogenesis of ASD.
Material and methods
Study population
From January 2010 to August 2011, a cohort of 220 unrelated patients diagnosed with ASD was prospectively recruited among the Chinese Han population. All the subjects were evaluated by individual and familial history, review of the medical records, complete physical examination, 12-lead electrocardiogram (ECG) and two-dimensional transthoracic echocardiography with color flow Doppler. All patients had a classic form of ASD, with a defect diameter of > 5 mm and nearly all patients underwent cardiac catheterization and, if required, cardiac surgery. A total of 200 ethnically matched unrelated healthy individuals who were enrolled from the general population were used as controls to screen for likely mutations in GATA4. Peripheral venous blood specimens from subjects and control individuals were prepared. The study protocol was reviewed and approved by the local institutional ethics committee and written informed consent was obtained from all participants or their guardians prior to investigation.
Genetic studies
Genomic DNA from all participants was extracted from blood lymphocytes with Wizard Genomic DNA Purification Kit (Promega). The candidate gene GATA4 was screened initially in 220 unrelated patients with ASD and genotyping of GATA4 in the available relatives of an index patient carrying an identified mutation and the 200 ethnically matched unrelated healthy control individuals was conducted subsequently. The referential genomic DNA sequence of GATA4 was derived from GenBank (accession No. NC_000008). By the aid of on-line Primer 3 software (http://frodo.wi.mit.edu), the primer pairs used to amplify the coding exons and exon/intron boundaries of GATA4 by polymerase chain reaction (PCR) were designed as shown in Table I. The PCR was carried out using HotStar Taq DNA Polymerase (Qiagen) on a PE 9700 Thermal Cycler (Applied Biosystems), with standard conditions and concentrations of reagents. Amplified products were analyzed on 1% agarose gels stained with ethidium bromide and purified with QIAquick Gel Extraction Kit (Qiagen). Both strands of each PCR product were sequenced with a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) under an ABI PRISM 3130 XL DNA Analyzer (Applied Biosystems). The sequencing primers were the same as previously designed for amplification of specific regions. The DNA sequences were viewed and analyzed with DNA Sequencing Analysis Software v5.1 (Applied Biosystems). The variant was validated by re-sequencing an independent PCR-generated amplicon from the subject and met our quality control thresholds with a call rate > 99%.
Table I.
The intronic primers to amplify the coding exons and exon-intron boundaries of GATA4
| Exon | Forward primer (5’ to 3’) | Reverse primer (5’ to 3’) | Amplicon [bp] |
|---|---|---|---|
| 2-a | GAT, CTT, CGC, GAC, AGT, TCC, TC | GTC, CCC, GGG, AAG, GAG, AAG | 458 |
| 2-b | GCT, GGG, CCT, GTC, CTA, CCT | AAA, AAC, AAG, AGG, CCC, TCG, AC | 554 |
| 3 | GGG, CTG, AAG, TCA, GAG, TGA, GG | GAT, GCA, CAC, CCT, CAA, GTT, CC | 437 |
| 4 | GAG, ATC, TCA, TGC, AGG, GTC, GT | GCC, CCT, TCC, AAA, TCT, AAG, TC | 390 |
| 5 | TCT, TTC, TCG, CTG, AGT, TCC, AG | GGG, ATG, TCC, GAT, GCT, GTC | 379 |
| 6 | GCC, ATC, CCT, GTG, AGA, ACT, GT | GAG, GGT, AGC, TCA, CTG, CTT, GC | 444 |
| 7 | AAG, TGC, TCC, TTG, GTC, CCT, TC | TTC, CCC, TAA, CCA, GAT, TGT, CG | 479 |
Multiple sequence alignments
The multiple GATA4 protein sequences across species were aligned using the online program CLUSTALW (http://www.genome.jp/tools/clustalw/).
Results
Characteristics of the study subjects
A cohort of 220 unrelated patients with ASD was identified and clinically evaluated in contrast to a total of 200 ethnically matched unrelated healthy individuals as controls. None of them had apparent traditional risk factors for ASD. The baseline clinical characteristics of the 220 unrelated patients with ASD are summarized in Table II.
Table II.
Clinical characteristics of the 220 unrelated patients with ASDs
| Variable | Number | Percentage or range |
|---|---|---|
| Male: female | 92: 148 | 42:58:00 |
| Age at the present study [year] | 23 | 1–58 |
| Distribution of different types of ASDs | ||
| Ostium secundum ASD | 180 | 82 |
| Ostium primum ASD | 27 | 12 |
| Sinus venosus ASD | 13 | 6 |
| Prevalence of ASDs with other defects | ||
| Isolated ASD | 185 | 84 |
| ASD and VSD | 12 | 5 |
| ASD and VSD and PDA | 4 | 2 |
| ASD and VSD and PS | 3 | 1 |
| ASD and ASA | 10 | 5 |
| ASD and PDA | 8 | 4 |
| ASD and PS | 5 | 2 |
| Incidence of arrhythmias | ||
| AVB | 8 | 4 |
| AF | 3 | 1 |
| Treatment | ||
| Surgical repair | 135 | 61 |
| Percutaneous closure | 85 | 39 |
ASD – atrial septal defect, VSD – ventricular septal defect, PDA – patent ductus arteriosus, PS – pulmonary stenosis, ASA – atrial septal aneurysm, AVB – atrioventricular block, AF – atrial fibrillation
GATA4 mutations
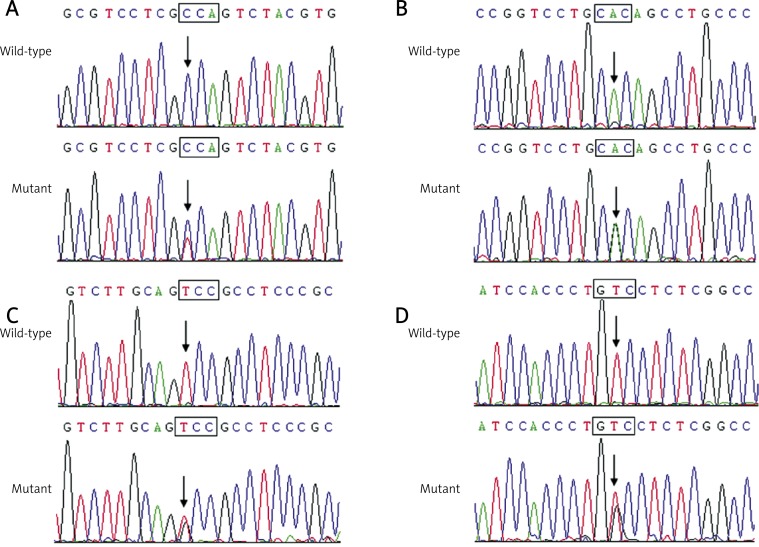
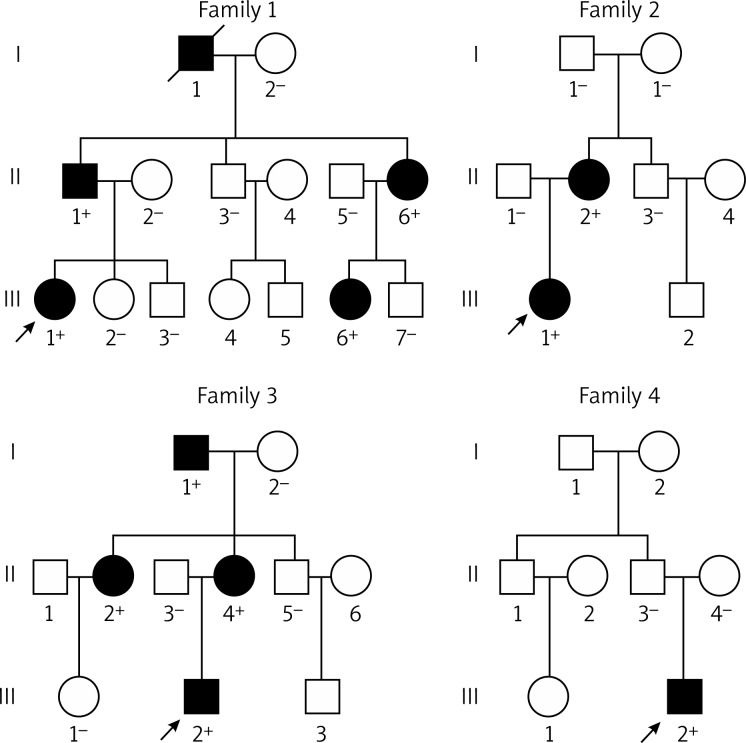
Direct sequencing of the coding exons of the GATA4 gene was conducted after PCR amplification of genomic DNA from the 220 unrelated ASD patients. Four heterozygous missense mutations in GATA4 were identified in 4 out of 220 patients, respectively. The total population prevalence of GATA4 mutations based on the cohort patients was approximately 1.82%. Specifically, a substitution of thymine for cytosine in the first nucleotide of codon 36 of the GATA4 gene (c.106C > T), predicting the transition of proline to serine at amino acid position 36 (p.P36S), was identified in the index patient from family 1. A change of adenine into guanine in the second nucleotide of codon 190 of the GATA4 gene (c.569A > G), corresponding to the transversion of histidine to arginine at amino acid residue 190 (p.H190R), was identified in the proband from family 2. A displacement of thymine by guanine in the first nucleotide of codon 262 of the GATA4 gene (c.784T > G), equivalent to the replacement of serine by alanine at amino acid 262 (p.S262A), was identified in the proband from family 3. A GATA4 variation of c.1196T > G, resulting in the conversion of valine into glycine at amino acid 399 (p.V399G), was identified in the proband from family 4. The sequence chromatograms showing the detected heterozygous GATA4 mutations in comparison to control sequences are shown in Figure 1. The variants were neither detected in 200 control individuals nor described in the human SNP database (http://www.ncbi.nlm.nih.gov/SNP). Genetic scan of the family members available of the mutation carriers revealed that in each family the variant was present in all affected family members alive, but absent in unaffected family members tested. Analysis of the pedigrees indicated that each mutation co-segregated with ASD in the family with complete penetrance. The pedigree structures of the families are illustrated in Figure 2. The phenotypic characteristics and results of genetic screening of the affected pedigree members are listed in Table III.
Figure 1.
Sequence chromatograms of GATA4 in index patients and controls. The arrow indicates the heterozygous nucleotides of C/T (Figure A), A/G (Figure B), T/G (Figure C), and T/G (Figure D), in the probands from families 1, 2 3, and 4, respectively (mutant) or the homozygous nucleotides of C/C (Figure A), A/A (Figure B), T/T (Figure C), and T/T (Figure D), in the corresponding control individuals (wild-type). The square denotes the nucleotides comprising a codon of GATA4
Figure 2.
Pedigree structures of the families with ventricular septal defect. Families are designated as family 1, family 2, family 3, and family 4. Family members are identified by generations and numbers. Squares indicate male family members; circles, female members; closed symbols, affected members; open symbols, unaffected members; symbols with a slash, deceased members; arrow, proband; “ + ”, carriers of the heterozygous mutations; and “–”, non-carriers
Table III.
Phenotypic characteristics and status of the GATA4 mutations in the affected pedigree members
| Identity | Gender | Subject information | Phenotypes | Genotypes Mutations | |||
|---|---|---|---|---|---|---|---|
| Age at time of study [years] | Age at diagnosis of ASD [years] | ASD [mm] | Other structural defects | AVB | |||
| Family 1 | P36S | ||||||
| I-1 | M | 62a | 45 | 12 | VSD, PDA | + | N/A |
| II-1 | M | 34 | 8 | 19 | + | +/– | |
| II-6 | F | 28 | 16 | 8 | VSD | – | +/– |
| III-1 | F | 12 | 2 | 23 | – | +/– | |
| III-6 | F | 4 | 4 | 6 | – | +/– | |
| Family 2 | H190R | ||||||
| II-2 | F | 26 | 22 | 17 | – | +/– | |
| III-1 | F | 3 | 3 | 10 | – | +/– | |
| Family 3 | S262A | ||||||
| I-1 | M | 56 | 40 | 15 | PS | + | +/– |
| II-2 | F | 30 | 10 | 30 | – | +/– | |
| II-4 | F | 27 | 26 | 10 | VSD | + | +/– |
| III-2 | M | 5 | 4 | 21 | – | +/– | |
| Family 4 | V399G | ||||||
| II-2 | M | 2 | 2 | 26 | – | +/– | |
M – male, F – female, ASD – atrial septal defect, N/A – not available or applicable, VSD – ventricular septal defect, PDA – patent ductus arteriosus, PS – pulmonary stenosis, AVB – atrioventricular block; + indicates present and – denotes absent
Age at death
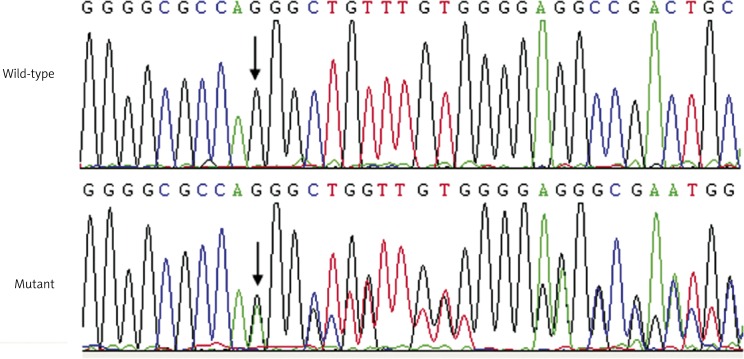
Additionally, a previously reported GATA4 intronic mutation of c.1146 + 24_25insA was identified in a 5-year-old girl with idiopathic ASD, who had a negative family history. The sequence chromatograms showing the heterozygous GATA4 insertion mutation in contrast to a control sequence are shown in Figure 3.
Figure 3.
Sequence chromatograms showing the insertion mutation of GATA4 in contrast to its control. The arrow indicates the heterozygous nucleotides of G/A (mutant) or the homozygous nucleotides of G/G (wild-type)
Multiple alignments of the GATA4 protein sequences across species
A cross-species alignment of GATA4 protein sequences showed that the affected amino acids were highly conserved evolutionarily, as shown in Figure 4, suggesting that these amino acids are functionally important.
Figure 4.
Alignment of multiple GATA4 protein sequences across species. The altered amino acids of P36, H190, S262, and V399 are highly conserved evolutionarily among mammals
Discussion
In the present study, 4 novel heterozygous missense mutations of GATA4 were identified in 4 families with congenital ASD, respectively. In each family, the mutation was present in all the affected family members alive but absent in unaffected relatives tested and 400 normal chromosomes from a matched control population. A cross-species alignment of GATA4 protein sequences demonstrated that the altered amino acids were highly conserved evolutionarily, suggesting the potential pathogenic effect of these novel mutations. Therefore, it is very likely that mutated GATA4 is involved in the pathogenesis of ASD in these families.
In humans, a long list of nonsynonymous GATA4 sequence variants was found to be associated with a great number of cardiac defects including ASD. Garg et al. [15] reported that a heterozygous missense mutation of p.G296S (c.886G > A) and a heterozygous frame-shift mutation of p.E359RfsX44 (c.1075delG) in GATA4 were identified in 2 large unrelated families with ASD, respectively. This is the first report on GATA4 mutation associated with an isolated congenital heart defect. Okubo et al. [16] found a novel GATA4 mutation of p.S358RfsX45 (c.1074delC) in a large Japanese family with ASD. By PCR sequencing, Hirayama-Yamada et al. [18] screened GATA4 in 16 unrelated families with ASD, and a novel mutation of S52F (c.155C > T) and a known mutation of E359fsX45 (c.1075delG) were identified in 2 families, respectively, with a mutation prevalence of 12.5% in the probands with ASD. Tomita-Mitchell et al. [20] investigated the exons and exon-intron boundaries of GATA4 in a large population of 628 unrelated patients with either septal or conotruncal defects and 4 missense sequence variants were identified in 5 patients, of which p.G93A and p.Q316E were observed in 2 out of 122 ASD patients. So far, over 30 nonsynonymous germline mutations in GATA4 have been implicated in congenital cardiovascular abnormalities, of which more than half of mutations have been linked to ASD with or without other defects, showing that although GATA4 mutations are involved in various cardiac malformations, the most frequent phenotype resulting from a mutation in GATA4 is ASD [15–32]. In most of these patients, the ASD-causing mutations are familial, whereas sporadic cases remain relatively infrequent [15–32]. Similar to these findings, the figure of 4/220 mutations (roughly 1.82%) in our cohort of patients suggests that GATA4 mutations could be an uncommon cause of ASD.
Association of compromised GATA4 with increased susceptibility to ASD has been demonstrated in animal experiments. In the embryonic hearts of knock-down chicks generated by using small interfering RNAs targeted to GATA4, the bilateral myocardial rudiments failed to travel to the midline, giving rise to the formation of two separate hearts in lateral positions, an anomaly of cardia bifida [33]. In mice GATA4 is one of the earliest transcription factors expressed in developing cardiac cells and continues to be expressed abundantly in cardiomyocytes throughout the life of mice. Homozygous GATA4-deficient mice died between day 7.0 and 9.5, and analysis of the GATA4-null embryo substantiated the lethal failure to form a linear heart tube [34, 35]. Transgenic mice expressing GATA4 mutants demonstrated a wide variety of cardiac malformations including septal defects, right ventricular hypoplasia, endocardial cushion defect, tetralogy of Fallot, double outlets of the right ventricle, and cardiomyopathy, similar to the anomalies seen in humans [34–36]. Taken together, these results from animal experiments define a critical role for GATA4 in regulating the normal cardiac morphogenesis.
In conclusion, the findings expand the mutation spectrum of GATA4 linked to ASD and provide additional insight into the molecular etiology implicated in the pathogenesis of ASD, suggesting potential implications for genetic diagnosis, early prophylaxis and gene-specific therapy for this common disease in infancy.
Acknowledgments
Yi-Qing Yang and Juan Wang contributed equally to the work.
We are indebted to participants for their dedication to the study. This work was supported in part by grants from the National Basic Research Program of China (2010CB912604), the National Natural Science Fund of China (81070153, 30570768, and 30700776), the Natural Science Fund of Shanghai, China (10ZR1433100, 10ZR1428000), and the Key Program of Shanghai Basic Research (10JC1414000, 10JC1414001, and 10JC1414002).
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics – 2011 update: a report from the American Heart Association. Circulation. 2011;123:e18–209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marelli AJ, Mackie AS, Ionescu-Ittu R, Rahme E, Pilote L. Congenital heart disease in the general population: changing prevalence and age distribution. Circulation. 2007;115:163–72. doi: 10.1161/CIRCULATIONAHA.106.627224. [DOI] [PubMed] [Google Scholar]
- 3.Sommer RJ, Hijazi ZM, Rhodes JF., Jr Pathophysiology of congenital heart disease in the adult: part I: shunt lesions. Circulation. 2008;117:1090–9. doi: 10.1161/CIRCULATIONAHA.107.714402. [DOI] [PubMed] [Google Scholar]
- 4.Trojnarska O, Gwizdała A, Katarzyński S, et al. Evaluation of exercise capacity with cardiopulmonary exercise testing and BNP levels in adult patients with single or systemic right ventricles. Arch Med Sci. 2010;6:192–7. doi: 10.5114/aoms.2010.13893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McQuillen PS, Miller SP. Congenital heart disease and brain development. Ann N Y Acad Sci. 2010;1184:68–86. doi: 10.1111/j.1749-6632.2009.05116.x. [DOI] [PubMed] [Google Scholar]
- 6.Walsh EP, Cecchin F. Arrhythmias in adult patients with congenital heart disease. Circulation. 2007;115:534–45. doi: 10.1161/CIRCULATIONAHA.105.592410. [DOI] [PubMed] [Google Scholar]
- 7.Piechowiak M, Banach M, Ruta J, et al. Risk factors for atrial fibrillation in adult patients in long-term observation following surgical closure of atrial septal defect type II. Thorac Cardiovasc Surg. 2006;54:259–63. doi: 10.1055/s-2006-923955. [DOI] [PubMed] [Google Scholar]
- 8.Trojnarska O, Bręborowicz P, Ochotny O, Cieśliński A. Permanent cardiac pacing in adult patients with congenital heart disease after cardiac surgery. Arch Med Sci. 2005;1:211–7. [Google Scholar]
- 9.Yap SC, Harris L. Sudden cardiac death in adults with congenital heart disease. Expert Rev Cardiovasc Ther. 2009;7:1605–20. doi: 10.1586/erc.09.153. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins KJ, Correa A, Feinstein JA, et al. American Heart Association Council on Cardiovascular Disease in the Young Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- 11.Pierpont ME, Basson CT, Benson DW, Jr, et al. American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:3015–38. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- 12.Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Peterkin T, Gibson A, Loose M, Patient R. The roles of GATA-4, -5 and -6 in vertebrate heart development. Semin Cell Dev Biol. 2005;16:83–94. doi: 10.1016/j.semcdb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 14.White RA, Dowler LL, Pasztor LM, et al. Assignment of the transcription factor GATA4 gene to human chromosome 8 and mouse chromosome 14: Gata4 is a candidate gene for Ds (disorganization) Genomics. 1995;27:20–6. doi: 10.1006/geno.1995.1003. [DOI] [PubMed] [Google Scholar]
- 15.Garg V, Kathiriya IS, Barnes R, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–7. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 16.Okubo A, Miyoshi O, Baba K, et al. A novel GATA4 mutation completely segregated with atrial septal defect in a large Japanese family. J Med Genet. 2004;41:e97. doi: 10.1136/jmg.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkozy A, Conti E, Neri C, et al. Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J Med Genet. 2005;42:e16. doi: 10.1136/jmg.2004.026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirayama-Yamada K, Kamisago M, Akimoto K, et al. Phenotypes with GATA4 or NKX2.5 mutations in familial atrial septal defect. Am J Med Genet A. 2005;135:47–52. doi: 10.1002/ajmg.a.30684. [DOI] [PubMed] [Google Scholar]
- 19.Nemer G, Fadlalah F, Usta J, et al. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat. 2006;27:293–4. doi: 10.1002/humu.9410. [DOI] [PubMed] [Google Scholar]
- 20.Tomita-Mitchell A, Maslen CL, Morris CD, Garg V, Goldmuntz E. GATA4 sequence variants in patients with congenital heart disease. J Med Genet. 2007;44:779–83. doi: 10.1136/jmg.2007.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajagopal SK, Ma Q, Obler D, et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol. 2007;43:677–85. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Posch MG, Perrot A, Schmitt K, et al. Mutations in GATA4, NKX2.5, CRELD1, and BMP4 are infrequently found in patients with congenital cardiac septal defects. Am J Med Genet A. 2008;146A:251–3. doi: 10.1002/ajmg.a.32042. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Li X, Shen A, Jiao W, Guan X, Li Z. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51:527–35. doi: 10.1016/j.ejmg.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Hamanoue H, Rahayuningsih SE, Hirahara Y, et al. Genetic screening of 104 patients with congenitally malformed hearts revealed a fresh mutation of GATA4 in those with atrial septal defects. Cardiol Young. 2009;19:482–5. doi: 10.1017/S1047951109990813. [DOI] [PubMed] [Google Scholar]
- 25.Chen MW, Pang YS, Guo Y, et al. GATA4 mutations in Chinese patients with congenital cardiac septal defects. Pediatr Cardiol. 2010;31:85–9. doi: 10.1007/s00246-009-9576-1. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Han ZQ, Yan WD, et al. A novel mutation in GATA4 gene associated with dominant inherited familial atrial septal defect. J Thorac Cardiovasc Surg. 2010;140:684–7. doi: 10.1016/j.jtcvs.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Mao J, Sun Y, et al. A novel mutation of GATA4 in a familial atrial septal defect. Clin Chim Acta. 2010;411:1741–5. doi: 10.1016/j.cca.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 28.D'Amato E, Giacopelli F, Giannattasio A, et al. Genetic investigation in an Italian child with an unusual association of atrial septal defect, attributable to a new familial GATA4 gene mutation, and neonatal diabetes due to pancreatic agenesis. Diabet Med. 2010;27:1195–200. doi: 10.1111/j.1464-5491.2010.03046.x. [DOI] [PubMed] [Google Scholar]
- 29.Butler TL, Esposito G, Blue GM, et al. GATA4 mutations in 357 unrelated patients with congenital heart malformation. Genet Test Mol Biomarkers. 2010;14:797–802. doi: 10.1089/gtmb.2010.0028. [DOI] [PubMed] [Google Scholar]
- 30.Peng T, Wang L, Zhou SF, Li XT. Mutations of the GATA4 and NKX2.5 genes in Chinese pediatric patients with non-familial congenital heart disease. Genetica. 2010;138:1231–40. doi: 10.1007/s10709-010-9522-4. [DOI] [PubMed] [Google Scholar]
- 31.Liu XY, Wang J, Zheng JH, et al. Involvement of a novel GATA4 mutation in atrial septal defects. Int J Mol Med. 2011;28:17–23. doi: 10.3892/ijmm.2011.638. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Fang M, Liu XY, et al. A novel GATA4 mutation responsible for congenital ventricular septal defects. Int J Mol Med. 2011;28:557–64. doi: 10.3892/ijmm.2011.715. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Toyofuku T, Kamei J, Hori M. GATA-4 regulates cardiac morphogenesis through transactivation of the N-cadherin gene. Biochem Biophys Res Commun. 2003;312:1033–8. doi: 10.1016/j.bbrc.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 34.Kuo CT, Morrisey EE, Anandappa R, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–60. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 35.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–72. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 36.Holtzinger A, Evans T. GATA4 regulates the formation of multiple organs. Development. 2005;132:4005–14. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]