Abstract
The transposase of IS911, a member of the IS3 family of bacterial insertion sequences, is composed of a catalytic domain located at its C-terminal end and a DNA binding domain located at its N-terminal end. Analysis of the transposases of over 60 members of the IS3 family revealed the presence of a helix–turn–helix (HTH) motif within the N-terminal region. Alignment of these potential secondary structures further revealed a completely conserved tryptophan residue similar to that found in the HTH motifs of certain homeodomain proteins. The analysis also uncovered a similarity between the IS3 family HTH and that of members of the LysR family of bacterial transcription factors. This information was used to design site-directed mutations permitting an assessment of its role in transposase function. A series of in vivo and in vitro tests demonstrated that the HTH domain is important in directing the transposase to bind the terminal inverted repeats of IS911.
INTRODUCTION
Bacterial insertion sequences (ISs) are small (<2.5 kb), generally phenotypically cryptic, segments of DNA capable of inserting at multiple sites in a target DNA molecule. Typical ISs have a simple genetic organization. They include a single or two open reading frames encoding an enzyme (the transposase) that promotes displacement of the IS, and sometimes also encode proteins implicated in regulation. ISs are generally bordered by two short terminal inverted repeat sequences (IR). Transposase specifically binds to the IRs where it catalyses the DNA strand cleavages and transfer reactions necessary for insertion of the IS into a target site (for reviews see 1,2). Recognition and binding to the IR is therefore a key feature of transposase activity.
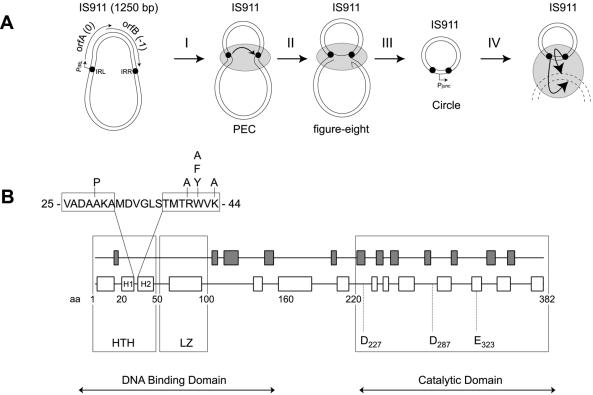
The insertion sequence IS911, originally isolated from Shigella dysenteriae (3), is a representative member of perhaps the largest group of ISs, the IS3 family. It has become one of the paradigms of IS transposition (for a review see 4). IS911 is 1250 bp long with imperfect 36 bp terminal inverted repeats (IRL and IRR) (Fig. 1A) terminating with a 5′-CA-3′ dinucleotide characteristic of the majority of IS3 family members and implicated in strand cleavage and transfer. Previous evidence showed that IS911 transposes using a circular intermediate (5,6) where the two IRs are covalently linked. This is capable of integrating into a target DNA molecule (6) (Fig. 1A, I–IV). Transposon circles are generated by a process in which the transposase assembles both IS ends into a synaptic or paired end complex (PEC) (Fig. 1A, I), introduces a single-strand break at one end to generate a 3′-OH group and transfers the cleaved strand to the second, target, end, creating a single-strand bridge between the two ends (Fig. 1A, II). This generates a molecule resembling a figure-of-eight. Transposon circles (Fig. 1A, III) are generated from the figure-of-eight forms probably by a replicative pathway (5; G. Duval-Valentin, in preparation). Circular transposon forms have been observed for at least three other IS3 family elements (IS2, IS3 and IS150) and this two-step transposition mechanism is thought to be conserved throughout the family (for reviews see 1,4).
Figure 1.
The IS911 transposition cycle and organization of its transposase. (A) This scheme describes the four steps of the transposition of IS911 as presently understood (5). Irs, black circles; orfs and promoters, arrows; transposase OrfAB, grey ellipse. The strand cleavage and transfer reactions are also shown as arrows. Note that the stoichiometry of the transposase in these complexes is not known. PEC (paired end complex), figure-of-eight and circle transposition intermediates are described in the text. (B) This scheme represents the known organization of the transposase (OrfAB) of IS911. White boxes, predicted α-helices; grey boxes, predicted β-strands. The DNA binding domain is composed of a helix–turn–helix motif (HTH; H1, helix 1; H2, helix 2) and a leucine zipper (LZ). The catalytic domain carries a classical DDE motif. The amino acid sequence of the HTH motif is represented together with the mutants constructed for this work using the single letter amino acid code.
Like most IS3 family members, IS911 carries two consecutive and partially overlapping open reading frames (ORFs), orfA and orfB. The transposase, OrfAB (Fig. 1A), is a fusion protein produced from both ORFs by programmed –1 translational frameshifting while the product of the upstream ORF, OrfA, plays a regulatory role. The N-terminal region of OrfAB, also partially contained in orfA, includes the DNA binding domain (DBD) (Fig. 1B). This region specifically recognizes the terminal IRs (7–9). The C-terminal region of OrfAB, encoded by orfB, carries the highly conserved DD(35)E catalytic motif and includes the active site. On its own this domain is capable of catalysing sequence-specific strand cleavage in a process known as disintegration (10). Frameshifting thus serves to assemble two distinct functional domains, a DNA binding domain and a catalytic domain, into the single OrfAB polypeptide.
To initiate a transposition reaction the transposase must bind to the left and right terminal IRs (IRL and IRR, defined with respect to the orientation of the transposase gene). Previous computer analyses of the N-terminal DBD revealed two distinct and well-defined motifs. The first, a leucine zipper (LZ) located between codons 63 and 94 (Fig. 1B), is involved in DNA binding and ensures protein–protein interactions which are required for OrfAB binding to either of the two IRs. Mutations in the LZ can eliminate both mutltimerization and IR binding (7). Although the full-length transposase binds poorly to the ends in vitro, binding by a truncated form of the transposase which carries the N-terminal region (OrfAB[1–149]) generates a PEC in which the protein bridges two IRs (7). These observations therefore strongly suggest that OrfAB binds DNA as a multimer (7–9). The second motif is a helix–turn–helix (HTH) (codons 25–44, Fig. 1B) thought to be responsible for the sequence specificity of DNA binding (3). However, no experimental data are yet available to assess this potential role of the HTH in DNA recognition.
We have addressed this question here by a combination of computer-aided alignment of transposases of the entire available collection of IS3 family members and site-directed mutagenesis. The alignments demonstrated that an HTH motif is strongly conserved in an equivalent position in all the proteins. Interest ingly, this analysis revealed the presence of a tryptophan residue conserved in all family members for which we propose a role in helix packing. Moreover, more extensive analysis revealed that the IS911 HTH motif shared sequence characteristics of the LysR protein family (11). Based on these alignments, site-directed mutagenesis of the HTH motif of IS911 confirmed that this region is essential for transposase activity.
MATERIALS AND METHODS
Bacterial strains and media
The Escherichia coli strain used in this study was a derivative of MC1061 (recA). Cells were grown in LB medium, supplemented where necessary with ampicillin (100 µg/ml) and kanamycin (25 µg/ml).
Sequence alignment
Amino acid sequence alignments were carried out using DNAMAN®. The results obtained were assessed and modified manually. The following IS transposase amino acid sequences used for sequence alignment were extracted from the general IS database (http://www-is.biotoul.fr) (origin of the gene and corresponding SWIS-PROT ID given in parentheses): CysB (Haemophilus influenzae, P45105), LysR (Escherichia coli, P03030) and CbnR (Ralstonia eutropha, Q9WXC7).
Oligonucleotides
Pairs of complementary oligonucleotides (AB04/AB05 and AB06/AB07) were designed for PCR mutagenesis of orfAB (bold characters represent mutated positions): ONhe1, GTTTTTTTGGGCTAGCGAATTCGAGC; OSph1, GAAGCTTGCATGCCTGCAGG; AB149His, CTTGCATGCTAGTG GTGGTGGTGGTGGTGTAGCTCAAGTACCTGACTGCG; AB04, GAACTACACGGTGGCAGATGCCGCCAAAGCTATGG; AB04A29P, GAACTACACGGTGGCAGATGCCCCCAAAGCTATGG; AB05, CCATAGCTTTGGCGGCA TCTGCCACCGTGTAGTTC; AB05A29P, CCATAGCTTTGGGGGCATCTGCCACCGTGTAGTTC; AB06, CCACAATGACAAGATGGGTCAAACAACTGCG; AB06R41A, CCACAATGACAGCATGGGTCAAACAACTGCG; AB06 K44A, CCACAATGACAAGATGGGTCGCACAACTGCG; AB06W42A, CCACAATGACAAGAGCGGTCAAACAACTGCG; AB06W42F, CCACAATGACAAGATTTGTCAAACAACTGCG; AB06W42Y, CCACAATGACAAGATATGTCAAACAACTGCG; AB07, CGCAGTTGTTTGACCCATCTTGTCATTGTGG; AB07R41A, CGCAGTTGTTTGA CCCATGCTGTCATTGTGG; AB07K44A, CGCAGTTGTGCGACCCATCTTGTCATTGTGG; AB07W42A, CGCAGTTGTTTGACCGCTCTTGTCATTGTGG; AB07W42F, CGCAGTTGTTTGACAAATCTTGTCATTGTGG; AB07 W42Y, CGCAGTTGTTTGACATATCTTGTCATTGTGG.
Plasmids and directed mutagenesis
The plasmids used are described in Table 1. Mutant derivatives of orfAB were constructed using a series of consecutive PCR steps as described (12). PCRs were carried out with Pfu polymerase (Promega) (20 cycles of 30 s at 94°C, 30 s at 55°C and 30 s at 72°C). The final PCR product was digested with NheI and SphI (New England Biolabs®) and ligated to similarly digested pAPT153 (5) to give plasmids pRP06, pRP10, pRP11, pRP12, pRP14 and pRP15 (Table 1). This places the ORF under control of a pAra promoter. The DNA sequence of the different clones obtained was determined on both strands. To transfer the mutations into orfAB[1–149], PCRs were carried out on these plasmids with oligonucleotides ONhe1 and AB149His, which allow amplification of the coding sequence of the first 149 amino acids of OrfAB containing the mutation, fused at its C-terminal end to a six histidine tag followed by a termination codon and a SphI site. After NheI and SphI digestion the fragment was inserted into similarly digested pAPT153 to give pRP13, pRP16, pRP17, pRP18, pRP19, pRP22 and pRP23 (Table 1). For the in vivo pJunc repression experiments, a PmeI–SphI fragment carrying the mutated ORFs was inserted downstream of the pLacUV5 promoter in pAPT158 (13) to give plasmids pRP33–pRP3939 (Table 1).
Table 1. Description of plasmids used in or constructed for this work.
| Plasmid name | Characteristics | Source | Oligonucleotides used |
|---|---|---|---|
| pAPT153 | pAra::orfAB | (5) | |
| pAPT158 | PLacUV5::orfAB | (13) | |
| pBST2 | pJunc::orfA-lacZ | (6) | |
| pLH37 | IS911 orfA-LacZ omegon | (14) | |
| pRP06 | pAra::orfAB-A29P | This work | AB04A29P + AB05A29P |
| pRP10 | pAra::orfAB-R41A | This work | AB05R41A + AB06R41A |
| pRP11 | pAra::orfAB-W42A | This work | AB05W42A + AB06W42A |
| pRP12 | pAra::orfAB-K44A | This work | AB05K44A + AB06K44A |
| pRP14 | pAra::orfAB-W42F | This work | AB05W42F + AB06W42F |
| pRP15 | pAra::orfAB-W42Y | This work | AB05W42Y + AB06W42Y |
| pRP13 | pAra::orfAB[1–149]-A29P::His | This work | |
| pRP16 | pAra::orfAB[1–149]-R41A::His | This work | |
| pRP17 | pAra::orfAB[1–149]-K44A::His | This work | |
| pRP18 | pAra::orfAB[1–149]-W42A::His | This work | |
| pRP19 | pAra::orfAB[1–149]::His | This work | |
| pRP22 | pAra::orfAB[1–149]-W42F::His | This work | |
| pRP23 | pAra::orfAB[1–149]-W42Y::His | This work | |
| pRP33 | PLacUV5::orfAB[1–149]-A29P::His | This work | |
| pRP34 | PLacUV5::orfAB[1–149]-R41A::His | This work | |
| pRP35 | PLacUV5::orfAB[1–149]-K44A::His | This work | |
| pRP36 | PLacUV5::orfAB[1–149]-W42A::His | This work | |
| pRP37 | PLacUV5::orfAB[1–149]::His | This work | |
| pRP38 | PLacUV5::orfAB[1–149]-W42Y::His | This work | |
| pRP39 | PLacUV5::orfAB[1–149]-W42F::His | This work |
Pairs of oligonucleotides used for mutation introduction by PCR are specified.
Protein purification and in vitro activity assay
OrfAB and derivatives were partially purified as described (5). OrfAB[1–149] derivatives were partially purified using the C-terminal histidine tag as indicated by the supplier (Qiagen®).
Electrophoretic mobility shift assays (EMSA)
IRR fragments (153 bp) were generated by PCR using pAPT166 as a template (9). In a standard EMSA (8), 7 nM DNA fragment (5000 c.p.m. Cerenkov) was incubated with OrfAB[1–149], in a final volume of 16 µl. An aliquot of 8 µg of protein extract was used in the reaction. Comparable amounts of OrfAB[1–149] in each extract were checked by western blot as described (14). Complexes were separated in a 5% polyacrylamide gel (8).
Figure-of-eight formation in vitro
Figure-of-eight formation was performed at 30°C for 30 min in a final volume of 40 µl containing 2.5 µg plasmid DNA substrate (pAPT166) and 4 µg protein extract (OrfAB), in 20 mM HEPES pH 7.5, 5 mM DTT, 200 mM KCl, 10 mM MgCl2 and 10% glycerol. The reaction was stopped by treatment with 0.5 mg/ml (final concentration) proteinase K for 1 h at 50°C and purified with QiaQuick® purification minicolumns as described by the supplier (Qiagen®). DNA was digested with EcoRV and analysed on 0.8% agarose gels.
Figure-of-eight and circle formation in vivo
Overnight cultures at 42°C (to inactivate residual transposase activity) of MC1061 (recA) containing the constructions to be tested (wild-type or mutated orfAB on one plasmid and an artificial IS911-based transposon on the other, pLH37) were diluted in fresh LB medium, supplemented with appropriate antibiotics, at 42°C at an OD600 of 0.05. After ∼90 min of growth (OD600 = 0.3–0.4), cultures were shifted to 30°C and after 5 min incubation (to equilibrate the temperature) arabinose was added (0.01%) to induce transposase expression. Cultures were grown for a further 30 min at 30°C and stopped by centrifugation at 4°C. DNA was then extracted by the cleared lysate method, digested with EcoRV and loaded on a 0.8% agarose gel (9).
Alkaline and cleared lysates
Alkaline lysates were prepared using a QiaPrep® kit as described by the supplier (Qiagen®). Cleared lysates were prepared as described (9).
β-Galactosidase assays
Cultures of MC1061(recA) transformed with pBST2 and with the different constructions to be tested (pRP33–pRP39, Table 1) were grown overnight at 37°C in LB medium supplemented with ampicillin and kanamycin and then diluted into fresh medium at OD600 = 0.05. IPTG (1 mM final concentration) was added to induce transposase expression under control of the placUV5 promoter. After ∼2 h of growth (OD600 = 0.3–0.4), β-galactosidase was measured according to the method of Miller (15).
RESULTS
Computer-aided identification of the HTH motif of transposases of the IS3 family
The presence of an HTH motif in the N-terminal region of the transposase of certain members of the IS3 family had been reported previously (3,16). To obtain a more global view of this secondary structure feature within the IS3 family, the N-terminal region (residues 1–60) of more than 60 members registered in the IS database (http://www-is.biotoul.fr) were aligned using DNAMAN® software (data not shown). Unlike the C-terminal region, which includes the catalytic site (DDE), the N-terminal regions are poorly conserved at the amino acid sequence level. This is presumably due to the fact that different IS elements of this family carry terminal IRs with different sequences and therefore necessitate different arrays of amino acid residues for recognition. On the other hand, examination of the N-terminal region of each transposase for its potential to form HTH and α-helices (http://npsa-pbil.ibcp.fr/) (17) predicted that each possessed this motif.
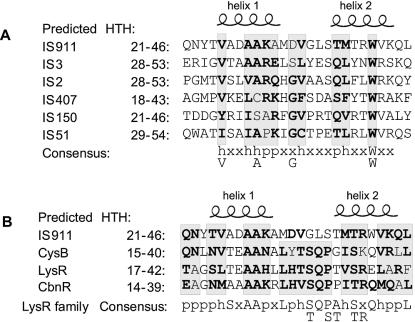
The IS3 family can be conveniently divided into distinct subgroups (the IS2, IS3, IS51, IS150 and IS407 subgroups) based on features such as similarities in transposase sequences, the nucleotide sequence of their terminal IRs and the number of target base pairs which are duplicated on insertion (1). To simplify analysis of secondary structure predictions, a single representative of each subgroup was chosen and the predicted HTH motifs were aligned. The results are presented in Figure 2A. Inspection of this alignment reveals some degree of sequence conservation, most of which is localized in the first helix (helix 1). Three of the five conserved amino acids of the amphiphatic helix 1 are hydrophobic (V25, A28 and A29) and would be located on the same face of the helix. This is consistent with the fact that, in other well-characterized HTH structures (18), this helix ensures the maintenance of the structure of the DNA binding motif. The conserved hydrophobic residues are presumably implicated in the hydrophobic face which contacts the second helix (helix 2). Helix 2 is generally involved in DNA sequence discrimination (18). Less conservation is observed in this second helix. This is consistent with a role in DNA recognition since the position of polar residues in this helix would be dictated by the sequence of different IRs. Furthermore, this second helix is more basic than the first, an observation which is also consistent with a DNA binding activity. Interestingly, helix 2 of the HTH motifs of all IS3 family members contains a tryptophan (W) residue. Together with those of the DDE catalytic motif (Fig. 1B), this is the most highly conserved residue in the transposases of the entire IS3 family, suggesting that it fulfils an important function other than in sequence discrimination.
Figure 2.
Sequence alignments of the HTH motif. (A) Alignment of the predicted HTH motif of the transposase of the five defining members of subgroups within the IS3 family with that of IS911. Grey boxes represent identical or similar residues; bold characters represent residues that fit the consensus. The consensus sequence is presented using the following code: h (hydrophobic: A, V, I, L, M, Y, F, C); p (hydrophilic: T, S, N, Q, D, E, K, R, H); x (any). The most frequently found residue (at least 4/6) is specified below this consensus. These codes include only those residues observed and do not include the entire set of hydrophobic or hydrophilic residues. (B) Alignment of the predicted HTH motif of IS911 transposase with HTH motifs of LysR (E.coli), CysB (H.influenzae) and CbnR (R.eutropha) proteins. The consensus sequence is presented using the following code, which is that proposed for the LysR family (11): h (hydrophobic: V, I, L, M); p (hydrophilic: T, S, N, Q, D, E, K, R, H); x (any). The amino acids TSTTR specified below are the frequently observed alternatives in this consensus (11).
To determine the relationship between the HTH motif of the IS3 transposases with those found in other DNA binding proteins, a BlastP analysis was performed with the non-redundant protein SWISSPROT database (http://www.ncbi.nlm.nih.gov) using the first 60 amino acids of the IS911 transposase (data not shown). This analysis revealed several identities with the H.influenzae CysB protein. CysB is a member of the large LysR family of transcriptional repressor/activator proteins (11). An alignment of the OrfAB, CysB, LysR and CbnR sequences (Fig. 2B) revealed good conservation in the region predicted to form the HTH motif. The proposed IS911 HTH motif fits well with the consensus sequence previously described for the LysR family HTH motif (11) (Fig. 2B).
Directed mutagenesis
Although no crystal structure was available for a LysR family member at the beginning of this work, many mutational studies defining key amino acid residues had been reported in the literature (11,19). This information was used as a guide for targeting mutations to specific residues designed to affect the DNA binding activity of the IS911 HTH motif. The following mutations were chosen (Fig. 1B): A29P (Ala29→Pro), R41A (Arg41→Ala), K44A (Lys44→Ala), W42A (Trp42→Ala), W42F (Trp42→Phe) and W42Y (Trp42→Tyr). Mutation of the central alanine of helix 1 (A29P) would be expected to disrupt the helix. Mutations R41A and K44A concern charged residues located in helix 2 and would be expected to interfere with specific DNA contacts. Finally, the three mutations at W42 (A, F and Y) should provide information concerning the role of this conserved residue: the requirement for an aromatic residue (W42A) or for a non-polar (F) or polar (Y) hydrophobic aromatic residue.
The HTH mutant proteins no longer repress the IR–IR junction promoter, pjunc
Formation of the IR–IR junction in the circular transposition intermediate generates a strong promoter, pjunc, by assembling a –35 promoter element located within the IRR with a –10 element located in the IRL (13) (Fig. 1A). This promoter drives transposase expression from the circularized IS and strongly stimulates integration. The full-length transposase, which weakly binds the IRs in vitro, was found to only slightly repress pjunc. However, a transposase derivative truncated for the catalytic C-terminal region and carrying the first 149 amino acids of OrfAB binds strongly to the IRs in vitro and is capable of generating a synaptic complex between the terminal IRS (8,9). This derivative was previously found to repress pjunc more strongly in vivo (20). It was therefore of interest to test whether derivatives of OrfAB[1–149], with the mutated HTH motif retained the capacity of wild-type OrfAB[1-149] to repress pjunc. For this, the HTH mutations were introduced separately into a copy of orfAB[1–149] under control of the placUV5 promoter. A set of strains was then constructed, each carrying the pBST2 plasmid, in which pjunc drives expression of a lacZ reporter gene, and a second plasmid to supply each of the mutant OrfAB[1–149] derivatives in trans under control of the inducible placUV5 promoter. The levels of expression from pjunc, measured as expression of the β-galactosidase reporter gene, are presented in Table 2. All mutants showed some reduction in their ability to repress pjunc but only two, A29P and W42A, were completely inactive. W42F showed only 18% repression (compared with 90% with the wild-type) and W42Y exhibited an intermediate activity (40% repression). R41A also exhibited reduced activity (69% repression) while K44A appeared as active as the wild-type (88% repression). These in vivo results strongly suggest that modification of the HTH motif influences the IR binding properties of the IS911 transposase (or at least of derivatives containing the DBD of IS911 transposase). The results also suggest an important role for W42, since all three mutants exhibit a strong decrease in their capacity to repress pjunc. The A29 position also seems to be important for DNA binding activity. Modifications of the charged residues R41 and K44 have only a slight effect, suggesting that, alone, they are not able to abolish DNA binding activity.
Table 2. Measurement of pjunc activity.
| Substrate pBST2 | Protein supplied in trans | pjunc activity (β-Gal) |
|---|---|---|
| pjunc::lacz | None | 100% |
| pjunc::lacz | OrfAB[1–149] | 10% |
| pjunc::lacz | OrfAB[1–149]-A29P | 100% |
| pjunc::lacz | OrfAB[1–149]-R41A | 31% |
| pjunc::lacz | OrfAB[1–149]-K44A | 12% |
| pjunc::lacz | OrfAB[1–149]-W42A | 100% |
| pjunc::lacz | OrfAB[1–149]-W42F | 82% |
| pjunc::lacz | OrfAB[1–149]-W42Y | 60% |
β-Galactosidase activities (third column, percentage relative to pjunc activity in the absence of any protein in trans, 100% = 6000 Miller units) were measured in strains containing a plasmid (pBST2, first column) in which pjunc drives lacZ expression and a plasmid that produces wild-type (wt) or variant forms of OrfAB[1–149] (second column). These results are the average of three independent experiments with a standard error <15%.
DNA binding activity of HTH mutants
To confirm these results, the effect of these mutations on binding to the IS911 IRs was tested in vitro. Previous studies showed that full-length OrfAB binds poorly in vitro to the terminal IR sequences. However, as for pjunc repression, binding can be significantly increased using transposase derivatives truncated for the C-terminal region (8,9). OrfAB[1–149] was therefore also used in this experiment. The mutations were introduced into a copy of orfAB[1–149] carrying a C-terminal His6 tag (orfAB[1–149]::6His) under control of the arabinose promoter (para). The partially purified proteins were used in EMSAs with radiolabelled DNA fragments carrying IRR. Comparable amounts of OrfAB[1–149], as judged by western blot analysis, were used in these assays. The results of gel electrophoresis of the resulting complexes are presented in Figure 3. Preparations of wild-type OrfAB[1–149] generate different complexes when bound to the IRR (Fig. 3, right panel schemes) (8,9). Complex I had previously been identified as a synaptic complex including two IR-carrying DNA fragments paired by a proposed oligomeric form of OrfAB[1–149]. Complex II appeared at higher protein concentrations and was proposed to result from the interaction of a single IR-carrying DNA fragment with the same oligomeric form of OrfAB[1–149]. Complex III was proposed to represent a complex containing OrfAB[1–149] and the IRR in another, but as yet undefined, configuration. This third complex was observed when OrfAB[1–149] was mixed with the regulatory protein OrfA or with an equivalent protein produced by proteolysis of OrfAB[1–149] in partially purified protein extracts (8,9). To simplify the interpretation of the results, experimental conditions were used in which the partially purified protein extracts of OrfAB[1–149] gave rise only to complexes I and III (Fig. 3, lane 3). Each mutant was found to modify these complexes quantitatively or qualitatively. As expected, the A29P mutant (Fig. 3, lane 4) completely abolished DNA binding. Removal of the basic residues in the recognition helix 2 (R41A and K44A) did not abolish DNA binding. However, these mutants exhibit patterns of complex formation that are clearly different from the wild-type protein (Fig. 3, compare lanes 5 and 6 with lane 3). Both produce more of complexes II and III and less of complex I than the wild-type protein and, in addition, the K44A mutant shows a qualitative difference in the migration of complex II. However, the higher levels of complex III, which are correlated with a reduction in the level of free DNA, might also be explained by there being more of the truncated forms shorter than OrfAB[1–149] in the extract (9). Results obtained with mutants of the conserved tryptophan within helix 2 (W42) were also interesting. While only traces of DNA–protein complexes were detected with W42A and W42F (Fig. 3, lanes 7 and 8), the DNA binding activity of the W42Y mutant was only slightly affected compared with the wild-type protein (Fig. 3; compare lanes 9 and 3). This suggests that an aromatic, polar and hydrophobic residue is needed at this position in helix 2.
Figure 3.
Electrophoretic mobility shift assay (EMSA) of wild-type and mutant OrfAB[1–149] derivatives. Protein extracts were incubated with 5′-32P- labelled IRR-containing DNA fragments as described (8,9). Schemes on the right represent the structures of the different DNA–protein complexes as proposed (8,9). Lane 1, no protein; lane 2, protein extracts from a strain expressing no IS911 proteins (V); lane 3, protein extracts from a strain expressing wild-type (WT) orfAB[1–149]; lanes 4–9, protein extracts from strains expressing the mutated orfAB[1–149].
These results are in agreement with those obtained in the pjunc repression assay (Table 2). Furthermore, there is an excellent correlation between the generation of complex I in EMSA and repression in the pjunc repression assay. This reinforces the idea that the HTH motif mediates correct recognition of IRs. Residues A29 and W42 are clearly important for DNA binding activity of the IS911 transposase. Whether this is a direct effect on DNA recognition or a more indirect effect on protein structure remains to be determined (see Discussion).
In vitro activity of HTH mutant transposases
The results presented above indicate that the HTH motif is required for binding of the truncated form of the transposase to a single IRR both in vitro or to pjunc in vivo. It was therefore of interest to determine whether these mutants were capable of sustaining transposase-mediated recombination. To examine this, they were first transferred into a full-length copy of orfAB under control of para. Note that in these constructions the OrfAB protein was produced by artificially fusing the orfA and orfB reading frames to generate the orfAB gene without changing the OrfAB amino acid sequence (mutation from ATTA6G to ACTCA6G) (5). After partial purification of these modified proteins, their in vitro activity in formation of the figure-of-eight intermediates was examined in a standard reaction (5). Following incubation of a plasmid carrying an IS911-based transposon, pAPT166, with each of the partially purified mutant transposases, the products were digested with EcoRV and figure-of-eight and parental plasmid forms were separated by agarose gel electrophoresis. The results are presented in Figure 4. Since the parent plasmid carries two EcoRV sites, one within the transposon and the other within the donor backbone, EcoRV-digested figure-of-eight transposition intermediates (8) appear as a χ DNA form migrating high in the gel while the parental plasmid simply generates two distinguishable DNA fragments (marked * in Fig. 4). The persistence of linearized parental plasmid (marked ** in Fig. 4) is due to the difficulty of obtaining complete EcoRV digestion after treatment with the protein extract.
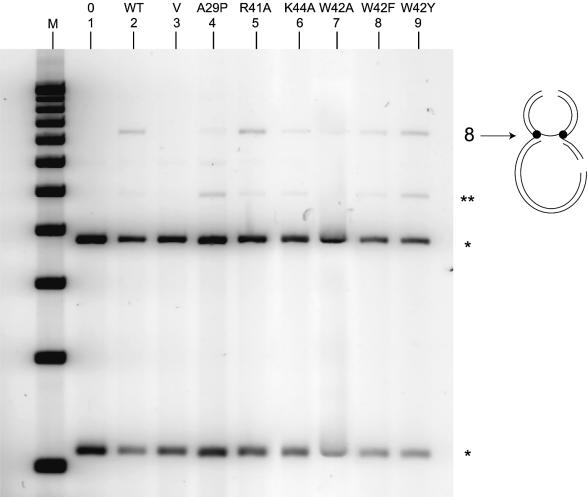
Figure 4.
In vitro figure-of-eight formation assay. An IS911-containing plasmid was incubated with purified wild-type or mutated transposases to form a figure-of-eight transposition intermediate. After EcoRV restriction, singly (**) or doubly cut (*) plasmids from figure-of-eight intermediates migrating as a χ were separated by agarose gel electrophoresis as described (5). The scheme on the right represents the digested figure-of-eight intermediate (see Fig. 1). Lane M, standard 1 kb ladder (New England Biolabs®); lane 1, no protein; lane 2, protein extracts from a strain expressing wild-type (WT) orfAB; lane 3, protein extracts from a strain expressing no IS911 proteins (V); lanes 4–9, protein extracts from strains expressing the mutated orfAB.
In this assay, mutants A29P and W42A, as expected, showed low activity in figure-of-eight formation (Fig. 4, lanes 4 and 7) compared with wild-type OrfAB (Fig. 4, lane 2). Mutants K44A and W42F (Fig. 4, lanes 6 and 8) exhibited reduced activity, whereas the R41A and W42Y mutants (Fig. 4, lanes 5 and 9) generated quantities of the figure-of-eight intermediate comparable to those obtained with the wild-type protein. Globally, these results are consistent with those obtained with the OrfAB[1–149] variants in the in vitro and in vivo DNA binding assays.
In vivo activity of HTH mutant transposases
As a further test, the activity of the mutant OrfAB derivatives was assessed in vivo using a transposase-mediated recombination assay. In contrast to the in vitro assay, in which IS911 transposition is arrested at the figure-of-eight step, both figure-of-eight and transposon circle formation can be followed with this procedure (21). A two plasmid system was used. One plasmid carried each of the orfAB derivatives under the control of para while the other, pLH37 (14), carried an IS911-derived transposon with appropriately oriented IRs but without the transposase gene. After 30 min of induced OrfAB expression, DNA was isolated from strains using a cleared lysate procedure and the products were analysed by agarose gel electrophoresis following EcoRV digestion. The results are shown in Figure 5. Again, as in the other assays described above, the mutants A29P and W42A exhibited a clearly reduced level of both figure-of-eight (8) and circular transposon forms (circle) compared with the wild-type OrfAB (Fig. 5, compare lane 3 with lanes 4 and 7). The K44A mutant appeared to be less affected in its activity than either of these (Fig. 5, lane 6), while the other mutant derivatives did not appear to be affected at all in their capacity to generate either of these transposition intermediates.
Figure 5.
In vivo figure-of-eight and circle formation assay. An IS911-containing plasmid was co-transformed together with a plasmid expressing wild-type or mutated orfAB under the control of para promoter in a MC1061 (recA) strain to form figure-of-eight and circle transposition intermediates. DNA was purified 30 min after orfAB induction by arabinose, digested by EcoRV and analysed by agarose gel electrophoresis. The two fragments obtained with the IS911-containing plasmid (* and ***) and the linearized transposase expression vector (**) are indicated. The control vector plasmid (V) is smaller due to absence of the orfAB gene. The figure-of-eight intermediate (8) which migrates as a χ and the circle intermediate migrating as linear DNA species as described (21) are also indicated. Lane 1 (M), a standard 1 kb ladder (New England Biolabs®); lane 2 (V), DNA from the strain expressing no IS911 gene; lane 3 (WT), DNA purified from the strain expressing wild-type orfAB; lanes 4–9, DNA isolated from strains expressing the various mutated orfAB derivatives.
In contrast to the DNA binding activities of the equivalent OrfAB[1–149] derivatives (Fig. 3), all mutants, except for A29P, retained at least some activity both in vivo (Fig. 5) and in vitro (Fig. 4). This difference may be due to the presence of the C-terminal catalytic domain in the full-length OrfAB mutants. Since this domain must also interact with the tip of the IR (9) to catalyse strand cleavage and transfer, this interaction may contribute to the overall binding capacity of the protein.
Taken together, these results demonstrate that mutations within the predicted HTH motif affect transposase activity.
DISCUSSION
Previous results had located a HTH motif in the N-terminal end of the IS911 transposase, OrfAB, between residues 25 and 44 (3), and this had been proposed to play a central role in recognition and binding to the terminal IRs of this insertion sequence. This idea was supported by the observation that an OrfAB derivative including only the first 149 residues retains the capacity to bind these ends and to assemble the two ends into a synaptic or paired end complex (7).
Conservation of the HTH motif sequence
Since IS911 is a member of one of the largest IS families, the IS3 family, in which all members have presumably adopted a similar transposition mechanism, the importance of this HTH motif should be reflected in its conservation throughout the family. Computer-aided analysis indeed detected a HTH motif at a similar position in the transposases of the entire IS3 family (>60 members, http://www-is.biotoul.fr/is.html). Although there was only limited sequence conservation between the different members, most of this was concentrated in the first helix of the motif (Fig. 2, consensus hxxhhppx), considered to be a structural helix (18). Moreover, the conservation includes the hydrophobic residues of this helix which could be involved in helix–helix interactions. We interpret this as reflecting a conservation of folding of the HTH motif within the family. The fact that less conservation is observed in the second helix, generally considered to be the DNA recognition helix, is consistent with the fact that these transposases interact with different DNA sequences (IRs). Remarkably, the second helix exhibits a highly conserved tryptophan residue (W42 in the case of IS911) which, apart from those located in the catalytic active site, is the most conserved residue in the entire IS3 family of transposases. This conservation must reflect an important function unrelated to DNA sequence discrimination. A similarly conserved tryptophan has been reported in the recognition helix in a family of DNA binding proteins, the homedomain family (22). Although not explicit in Wintjens and Rooman (22), visualization of the structure of these domains in several members of the homeodomain family (PdbViewer, www.expasy.org/spdbv) revealed that the W is involved in hydrophobic contacts between helix 2 (recognition) and helix 1. This suggests a structural role for this residue.
Further analysis of the IS911 HTH motif showed it to be related to those of the LysR family of transcriptional activators and repressors. This conservation is particularly notable between OrfAB and CysB. Once again the most important conservation is observed in the first helix, where both the transposase and LysR families exhibit a clear consensus (Fig. 2A and B, consensus hxxAAp).
OrfAB derivatives with mutations in the HTH motif exhibit modified DNA binding properties
All HTH mutations of OrfAB[1–149], based on the alignment presented in Figure 2, exhibited modified DNA binding properties as judged by EMSA in vitro or by repression of pjunc in vivo. Three mutants, A29P, W42A and W42F, were particularly affected in their capacity to bind: no complexes could be detected with A29P, whereas W42A and W42F generated only trace levels. The other mutants exhibited less clear-cut effects. W42Y appeared to be only slightly inhibited, whereas R41A and K44A exhibited differences in the pattern of complex formation. For example, complex II generated by R41A migrated as does that generated by the wild-type OrfAB[1–149], whereas that generated by K44A migrated significantly higher in the gel. This shows that these two residues are not essential for DNA binding although their substitution modifies DNA binding. More detailed examination of the formation of these complexes is required to determine the exact molecular consequences of the mutations.
Globally these results clearly show that point mutations of the predicted HTH motif of IS911 modify the DNA binding properties of this truncated form of the transposase. The two clear-cut effects are obtained for A29P and W42A, two mutations that are thought to strongly destabilize the HTH structure: A29P should disrupt the first helix and, based on homeodomain family structures (22), W42A would be expected to destabilize the cohesion between helices. The fact that tryptophan is partially complemented in vitro by tyrosine (polar, hydrophobic and aromatic) but neither by alanine (small hydrophobic) nor phenylalanine (hydrophobic and aromatic) suggests that tryptophan could form a polar and hydrophobic arm allowing interaction between the helices. A recent structure obtained for CbnR from Ralstonia eutropha, a LysR-type transcriptional regulator protein, revealed that the A22 residue (corresponding to A29 in OrfAB) is exposed and may interact directly with DNA (23). This suggests that OrfAB A29 residue could have more than a structural role. Residues R41 and K44 are equivalent to R34 and Q37 in CbnR (Fig 2B) (23), which, like A22, are also clearly exposed in the structure. Based on modelling studies, these residues were not proposed to interact with DNA, rather the authors suggested A22 and P30 (equivalent to OrfAB A29 and S37) (23). Moreover, the ‘HTH’ motif within the CbnR structure is composed of three rather than two helices. This is also true for the ‘HTH’ motifs of the homeodomain family which carry a conserved W residue (22).
Furthermore, although few transposases have been characterized for their DNA binding activity [e.g. those of IS50, phage Mu, Tc3 and Sleeping Beauty (24–27)], all carry an HTH-like structure with more than two helices in the motif. Structural studies have shown that the transposase of IS50 carries four helices (24), while those of phage Mu and Tc3 include three (25,26). Although these present no homology with IS3 family transposases either in primary sequence or in the organization of the DNA domain, making it difficult to make direct comparisons, OrfAB also contains a third predicted helix located N-terminal to helices 1 and 2 (Fig. 1B). This raises the possibility that the DNA binding domain we have targeted here may carry structural features in addition to the simple HTH motif.
Inactivation of transposase activity
In the context of the full-length OrfAB protein, mutants A29P and W42A, which strongly alter the HTH motif, exhibited dramatic effects on transposase activity both in vivo and in vitro. The mutant K44A, which exhibited a slightly modified DNA binding activity, showed partial activity in both in vivo and in vitro transposase activity tests, while W42Y and R41A, which exhibited only slight changes in DNA binding activity, appeared to be as active as the wild-type protein in catalysing strand cleavage and transfer. Thus there is an excellent correlation between DNA binding and transposition activity.
The case of the W42F mutation is more difficult to interpret. This mutant is relatively inactive in the DNA binding assay (Fig. 3 and Table 1), partially active in the in vitro recombination assay (Fig. 4) and active in the in vivo recombination assay (Fig. 5). We note that all the effects observed in the context of the full-size transposase are less marked than those obtained with truncated forms tested in the DNA binding assay. These differences may be due to the DNA binding properties of the C-terminal part of the transposase carrying the catalytic site (DDE). The DDE domain must clearly recognize the outside tip of the IR since it catalyses sequence-specific strand cleavage and transfer at the conserved 5′-CA-3′ termini of the IS. It is thus possible that this domain contributes to DNA binding and can to some extent ‘complement’ the effect of some of the HTH mutations, particularly that of W42F. This type of effect would accord with the present model for transposase–IR interactions, where the transposase DNA binding domain (HTH plus LZ) serves as an anchor by binding to an internal region of the IR in order to correctly position the catalytic domain (DDE) on the termini (9). Furthermore, the known structure of DNA bound transposases (Tn5, phage Mu and retroviral IN) suggests that the protein–DNA contacts appear along almost the entire length of the transposase (28).
In summary, the results presented in this article indicate that the N-terminal HTH motif carried by the IS911 transposase and, by extension, by all other members of IS3 family, plays an essential role in transposase activity at the level of recognition of and binding to the terminal IRs. However, comparison with the LysR family member CbnR, homeodomain proteins and other transposases whose structures have been solved raises the possibility that additional structural elements, such the predicted N-terminal helix, may also be associated with this DNA binding motif.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank members of the Mobile Genetic Elements group (C. Loot, B. Marty, Z. Nagy, N. Pouget, P. Siguier, B. Ton-Hoang and C. Turlan) for discussions. This work was supported by grants from the CNRS (UMR5100), the Programme Microbiologie (MENRST), the Association pour la Recherche sur le Cancer, the Groupement de Recherche (GDR 2157) and the Institut Fédératif de Recherche (IFR 109). E.G. was supported by a grant from the Ministère de l’Education Nationale.
REFERENCES
- 1.Mahillon J. and Chandler,M. (1998) Insertion sequences. Microbiol. Mol. Biol. Rev., 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandler M. and Mahillon,J. (2002) Insertion sequences revisited. In Craig,N.L. et al. (eds), Mobile DNA II. ASM Press, Washington, DC, pp. 305–366. [Google Scholar]
- 3.Prere M.F., Chandler,M. and Fayet,O. (1990) Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J. Bacteriol., 172, 4090–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rousseau P., Normand,C., Loot,C., Turlan,C., Alazard,R., Duval-Valentin,G. and Chandler,M. (2002) Transposition of IS911. In Craig,N.L. et al. (eds), Mobile DNA II. ASM Press, Washington, DC, pp. 367–383. [Google Scholar]
- 5.Polard P., Ton-Hoang,B., Haren,L., Betermier,M., Walczak,R. and Chandler,M. (1996) IS911-mediated transpositional recombination in vitro. J. Mol. Biol., 264, 68–81. [DOI] [PubMed] [Google Scholar]
- 6.Ton-Hoang B., Polard,P. and Chandler,M. (1998) Efficient transposition of IS911 circles in vitro. EMBO J., 17, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haren L., Polard,P., Ton-Hoang,B. and Chandler,M. (1998) Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. J. Mol. Biol., 283, 29–41. [DOI] [PubMed] [Google Scholar]
- 8.Haren L., Normand,C., Polard,P., Alazard,R. and Chandler,M. (2000) IS911 transposition is regulated by protein–protein interactions via a leucine zipper motif. J. Mol. Biol., 296, 757–768. [DOI] [PubMed] [Google Scholar]
- 9.Normand C., Duval-Valentin,G., Haren,L. and Chandler,M. (2001) The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. J. Mol. Biol., 308, 853–871. [DOI] [PubMed] [Google Scholar]
- 10.Polard P. and Chandler,M. (1995) Bacterial transposases and retroviral integrases. Mol. Microbiol., 15, 13–23. [DOI] [PubMed] [Google Scholar]
- 11.Schell M.A. (1993) Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol., 47, 597–626. [DOI] [PubMed] [Google Scholar]
- 12.Schanke J.T., Quam,L.M. and Van Ness,B.G. (1994) Flip-PCR for DNA sequence motif inversion. Biotechniques, 16, 414–416. [PubMed] [Google Scholar]
- 13.Ton-Hoang B., Betermier,M., Polard,P. and Chandler,M. (1997) Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J., 16, 3357–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haren L. (1998) PhD Thesis: Relation structure-fonction des facteurs protéiques exprimés par l‘élément transposable bactérien IS911. Université Paul Sabatier, Toulouse, France.
- 15.Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 16.Fu R. and Voordouw,G. (1998) ISD1, an insertion element from the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough: structure, transposition and distribution. Appl. Environ. Microbiol., 64, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodd I.B. and Egan,J.B. (1990) Improved detection of helix–turn–helix DNA-binding motifs in protein sequences. Nucleic Acids Res., 18, 5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison S.C. and Aggarwal,A.K. (1990) DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem., 59, 933–969. [DOI] [PubMed] [Google Scholar]
- 19.Lochowska A., Iwanicka-Nowicka,R., Plochocka,D. and Hryniewicz,M.M. (2001) Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization and positive control. J. Biol. Chem., 276, 2098–2107. [DOI] [PubMed] [Google Scholar]
- 20.Duval-Valentin G., Normand,C., Khemici,V., Marty,B. and Chandler,M. (2001) Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. EMBO J., 20, 5802–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polard P. and Chandler,M. (1995) An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev., 9, 2846–2858. [DOI] [PubMed] [Google Scholar]
- 22.Wintjens R. and Rooman,M. (1996) Structural classification of HTH DNA-binding domains and protein–DNA interaction modes. J. Mol. Biol., 262, 294–313. [DOI] [PubMed] [Google Scholar]
- 23.Muraoka S., Okumura,R., Ogawa,N., Nonaka,T., Miyashita,K. and Senda,T. (2003) Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol., 328, 555–566. [DOI] [PubMed] [Google Scholar]
- 24.Davies D.R., Goryshin,I.Y., Reznikoff,W.S. and Rayment,I. (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science, 289, 77–85. [DOI] [PubMed] [Google Scholar]
- 25.Schumacher S., Clubb,R.T., Cai,M., Mizuuchi,K., Clore,G.M. and Gronenborn,A.M. (1997) Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains. EMBO J., 16, 7532–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Pouderoyen G., Ketting,R.F., Perrakis,A., Plasterk,R.H. and Sixma,T.K. (1997) Crystal structure of the specific DNA-binding domain of Tc3 transposase of C.elegans in complex with transposon DNA. EMBO J., 16, 6044–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izsvak Z., Khare,D., Behlke,J., Heinemann,U., Plasterk,R.H. and Ivics,Z. (2002) Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem., 277, 34581–34588. [DOI] [PubMed] [Google Scholar]
- 28.Rice P.A. and Baker,T.A. (2001) Comparative architecture of transposase and integrase complexes. Nature Struct. Biol., 8, 302–307. [DOI] [PubMed] [Google Scholar]