Abstract
Ecdysoneless (Ecd) is an evolutionarily conserved protein whose function is essential for embryonic development in Drosophila and cell growth in yeast. However, its function has remained unknown until recently. Studies in yeast suggested a potential role of Ecd in transcription; however Ecd lacks a DNA binding domain. Using a GAL4-luciferase reporter assay and a GAL4-DNA binding domain (DBD) fusion with Ecd or its mutants, we present evidence that human Ecd has a transactivation activity in its C-terminal region. Importantly, further analyses using point mutants showed that a single amino acid change at either Asp-484 or Leu-489 essentially completely abolishes the transactivation activity of Ecd. We further demonstrate that Ecd interacts with p300, a histone acetyltransferase and the co-expression of Ecd with p300 enhances the Ecd-mediated transactivation activity. Ecd localizes to both nucleus and cytoplasm and shuttles between the nucleus and cytoplasm; however it exhibits strong nuclear export. Based on previous yeast studies and evidence provided here, we suggest that Ecd functions as a transcriptional regulator. This study points out to an important function of human Ecd and provides a basis to explore the transcriptional partners of Ecd.
Keywords: coactivator, Ecdysoneless, hSGT1, transactivation
Introduction
The Ecdysoneless (Ecd) gene was named after the phenotype of a mutant Drosophila that has low levels of ecdysone, an insect steroid hormone responsible for normal embryogenesis, larval molting and metamorphosis. Dysregulation of ecdysone levels during embryonic development leads to the failure of normal development (Garen et al., 1977). Nearly three decades after the initial isolation of the mutant flies, the gene responsible for this mutation was identified (Gaziova et al., 2004). Ecd gene shows a strong evolutionary conservation throughout eukaryotes from fission yeast to humans suggesting a conserved biochemical function.
Human Ecd was first isolated and named as hSGT1 (human suppressor GCR two) by a complementation assay study in Saccharomyces cerevisiae (S. cerevisiae). GCR1 is a core transcription factor that is involved in the expression of glycolytic genes in S. cerevisiae. A gcr1 mutant showed a severe defect in glycolytic gene expression (Uemura et al., 1997). GCR2 is a GCR1-interacting protein and functions as a coactivator of GCR1 in glycolytic gene expression. The GCR2 gene was initially identified through the characterization of a novel mutation that affected glycolytic gene expression in S. cerevisiae. The gcr2 mutant phenotype was similar to that of gcr1 mutant and it was subsequently shown that GCR2 interacts with GCR1 and functions as a coactivator for GCR1-mediated glycolytic gene expression (Zeng et al., 1997). The complementation study of a gcr2 mutant strain with human cDNA library was performed in order to identify human genes that can rescue the gcr2 phenotype. One human cDNA that could reconstitute the GCR2 coactivator function in yeast was named hSGT1. Importantly, the hSGT1/hEcd complementation resulted in a recovery from the cell growth defect seen in the gcr2 mutant apparently by substituting for the coactivator function of GCR2 through hEcd interaction with GCR1. The authors suggested that hSGT1 may be a functional analog of GCR2. Notably, there is no sequence similarity between hSGT1/hEcd and GCR2 (Sato et al., 1999).
A recent study in Schizosaccharomyces pombe (S.pombe) showed that S. pombe Ecd (called as spSGT1 in the reported study) is required for cell survival and regulates gene expression involved in carbohydrate metabolism, amino acid metabolism and energy pathways (Kainou et al., 2006). In addition, we previously showed that hEcd interacts with and stabilizes p53 and its overexpression in mammalian cells increases the transcription of p53 target genes (Zhang et al., 2006).
Given the paucity of knowledge on the structure and function of this protein, we hypothesized that Ecd may have a role in transcriptional regulation based on several lines of evidence: i) in S. cerevisiae, human Ecd is able to bind to GCR1 and act as a coactivator by substituting for GCR2 (Sato et al., 1999), ii) in S. pombe, Ecd was shown to be important in cell survival and loss of its expression suggested a possible role as a transcription regulator (Kainou et al., 2006), iii) human Ecd binds to p53, and increases p53-mediated transcription (Zhang et al., 2006) and iv) hEcd also binds to Rb and regulates Rb/E2F pathway (Kim et al., In press).
Here, using a GAL4-DBD fusion protein of Ecd and GAL4-reporter luciferase assays, we demonstrate that Ecd has an intrinsic transactivation activity. Significantly, a point mutation in the C-terminal region completely abolished the transactivation activity of Ecd. Ecd binds to p300 and cooperates with p300 to increase the transactivation activity. Moreover, Ecd shuttles between the nucleus and cytoplasm, and its cytoplasmic localization depends on active CRM1-mediated nuclear export. These results suggest that mammalian Ecd may function as a transcriptional regulator and this function appears to be conserved through evolution.
Results
Human Ecd has an intrinsic transactivation activity
There is no identifiable DNA binding-domain on hEcd and no potential primary transcription factor for hEcd. Our previous studies showed that Ecd interacts with p53 and increases p53-mediated transcription (Zhang et al., 2006). However, Ecd may have a function in stabilizing p53 protein level rather than increasing p53 transcription via associating with p53 on the p53 target promoter since chromatin immunoprecipitation experiment failed to show that Ecd associate with p53 on the p21 promoter (data not shown). Regarding the role of Ecd in Rb/Ecd pathway, Ecd is not likely to associate with E2F target promoter through the interaction with Rb because i) Rb can associate with E2F target promoter only through the interaction with E2F transcription factor and ii) Ecd competes with E2F for Rb binding. Chromatin immunoprecipitation experiment also failed to show that Ecd associates with E2F target gene promoters, such as B-myb, cdc2, and cyclin B1 (data not shown).
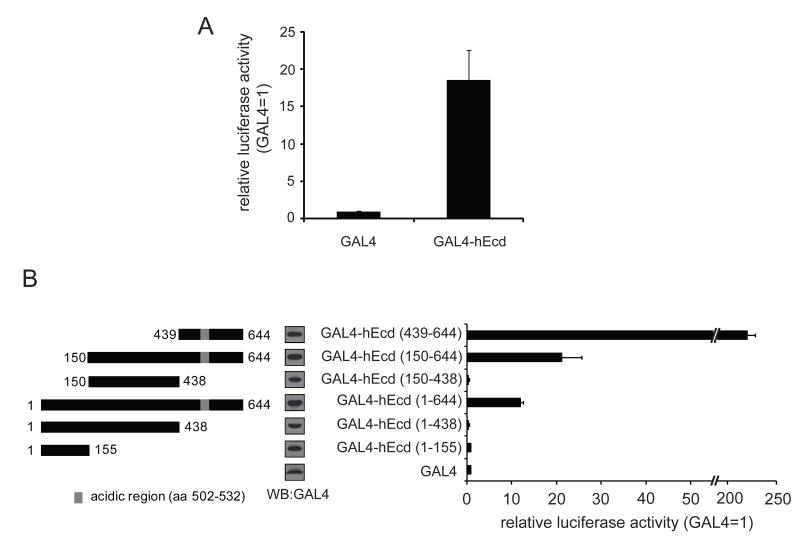
Therefore, in order to examine whether Ecd has a transactivation activity, a GAL4-reporter assay was performed. The GAL4-reporter assay is commonly used to study transactivation functions of transcription factors or transcriptional coactivators (Bratton et al., 2009; Kim et al., 2003). Full-length Ecd was fused to GAL4-DBD (DNA binding domain) and introduced into cells together with pG5/luciferase reporter containing 5X GAL4 DNA binding element upstream of the luciferase gene. The expression of luciferase reporter gene is regulated by transcription factors that associate with the upstream GAL4 DNA binding element. In order to examine if mammalian Ecd has a transactivation activity, GAL4-DBD-fused full length human Ecd was introduced into U2OS cells together with pG5/luciferase reporter plasmid (SV40-driven Renilla luciferase cloned within the GAL4-DBD vector served as a transfection efficiency control). As shown in Figure 1A, human Ecd was able to enhance the luciferase gene expression compared to GAL4-DBD control, which suggests that mammalian Ecd has an intrinsic transactivation activity. These results are consistent with the possible transactivation function of Ecd previously shown in yeast (Sato et al., 1999).
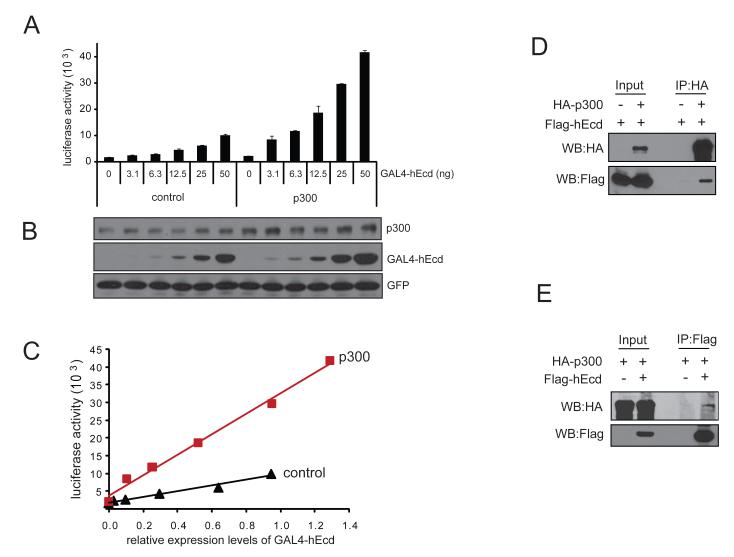
Figure 1. Mammalian Ecd has an intrinsic transactivation activity.
(A) To test if mammalian Ecd has transactivation activity, GAL4-fused human Ecd was transfected into U2OS cells together with GAL4-reporter plasmid. Twenty hours after transfection, cells were lysed and luciferase activities were measured as described in Materials and Methods. (B) Five deletion mutants of GAL4-hEcd were tested by GAL4-reporter assay. GAL4-fused hEcd constructs and pG5/luciferase plasmid were co-transfected into U2OS cells and luciferase activities were measured 20 hours after transfection using Dual-Luciferase Reporter Assay System. Luciferase activities were normalized with control Renilla luciferase values and relative activity is shown compared to GAL4 control. Error bars represent standard deviation from the means of triplicate samples. Cell lysates from the luciferase activity measurement were subjected to western blotting using anti-GAL4 antibody to show the expression levels of different GAL4-hEcd forms.
The transactivation activity of Ecd resides in the C-terminal region
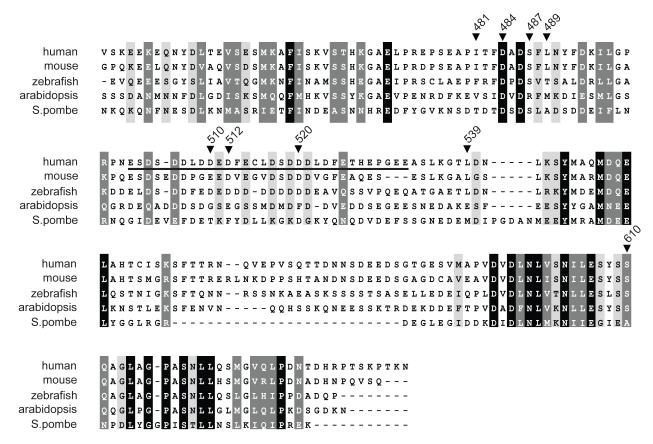
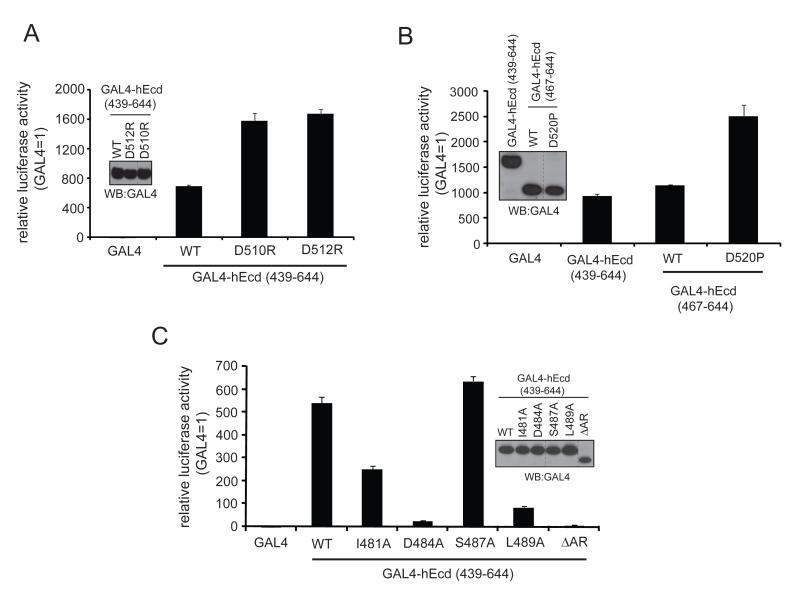
Given the lack of identifiable domains in Ecd, five GAL4-fused deletion mutants (aa 1-155, 1-438, 150-438, 150-644, and 439-644) were generated based on the secondary structure prediction (Jpred software) In order to define the region(s) in Ecd required for transactivation. these GAL4-fused truncated forms of hEcd were then tested for transactivation activity. Notably, the C-terminal region (aa 439-644) was required for strong transactivation activity. A small fragment of the C-terminus (aa 439-644) showed even stronger activity when compared to the full-length hEcd (Figure 1B). The N-terminal region (aa 1-438) of Ecd protein may have a inhibitory function for the autonomous transactivation activity of its C-terminal region by changing the protein intramolecular folding structure like other transcription factors (Dennig et al., 1996; Lillycrop et al., 1994; Park et al., 2008; Zhao et al., 2002). As shown in Figure 2, the C-terminal region (aa 439-644) is well conserved in other species. Notably, Ecd contains a putative acidic region (aa 502-532) located in the C-terminal fragment (Figure 2). Acidic domains in other proteins are often associated with a transactivation function (Ma and Ptashne, 1987). In order to examine if the acidic region is involved in the transactivation activity, several point mutants (D510R, D512R, or D520P) of the acidic region were generated in the context of the hEcd C-terminal fragment and analyzed using the GAL4 reporter assay. The mutations D510R or D512R in aa 439-644 fragment increased Ecd transactivation by two-fold (Figure 3A). A shorter fragment (aa 467-644) exhibits transactivation activity comparable to that of the 439-644 fragment (Figure 3B); mutation of D520P in this shorter fragment also increased the transactivation activity (Figure 3B). These mutational analyses suggest that the structural changes in this acidic region introduced by single amino acid substitutions affect the transactivation activity of hEcd protein.
Figure 2. Amino acid sequence alignment of C-terminal region of human Ecd with other species.
Amino acid sequence alignment of the human Ecd protein with mouse, zebrafish, Arabidopsis thaliana, and Schizosaccharomyces pombe was done using ClustalW2. Black color indicates identical or conserved residues in all sequences, and dark gray and gray colors indicate conserved substitutions and semi- conserved substitutions, respectively. Underline (aa 502-532, human Ecd) indicates acidic region, and the location of mutations which were used in this studies are marked.
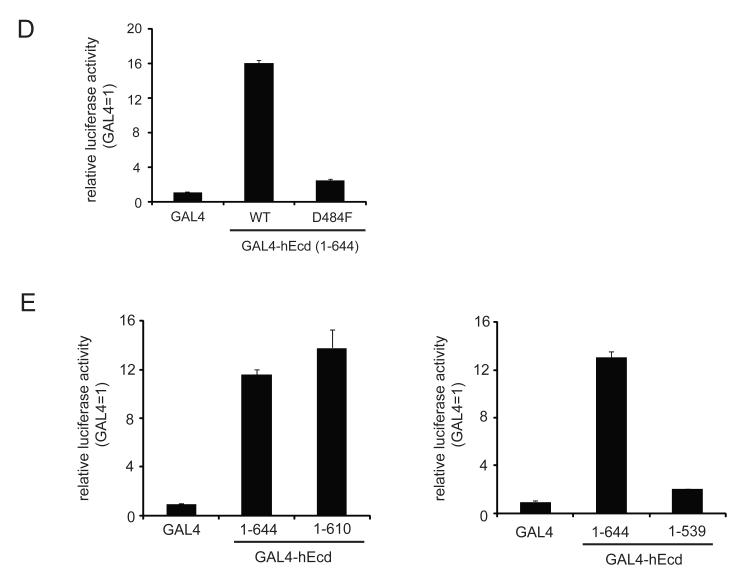
Figure 3. Mutational analyses of human Ecd in a transactivation assay.
(A) Two point mutations (D510R, or D512R) were introduced in GAL4-hEcd (aa 439-644) and these plasmids were transfected into U2OS cells. Relative activities are shown compared to GAL4 control. (B) D520P mutation was introduced in GAL4-hEcd (aa 467-644) and tested for transactivation activity. (C) Several indicated mutants (I481A, D484A, S487A, L489A and ΔAR) in GAL4-hEcd (aa 439-644) were tested for their effect on transactivation activity. (D) Full-length wild type and D484F mutant GAL4-hEcd were tested for transactivation activity. (E) Two C-terminally truncated mutants of GAL4-hEcd (aa 1-539 and 1-610) were used for reporter gene assay. All luciferase assays were performed as described in Materials and Methods. The relative activities of GAL4-hEcd are shown compared to GAL4 control. Error bars represent standard deviation from the means of triplicate samples. Western blotting using anti-GAL4 antibody shows relative expression levels of GAL4-hEcd.
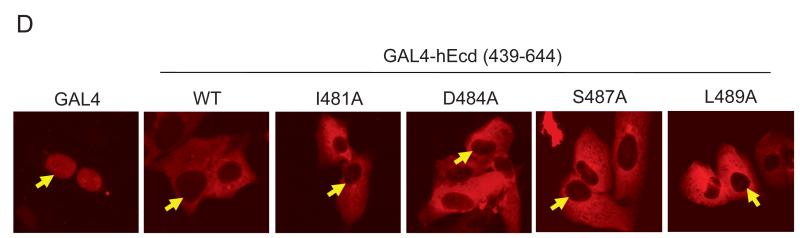
Given that the mutations in the acidic region affects transactivation activity, further mutational analyses in the upstream sequences of the acidic region were tested in the GAL4-reporter assay. The I481A mutation led to about 50% decrease in transactivation activity compared to wild type, whereas the S487A mutation had no effect (Figure 3C). Interestingly, a single point mutation D484A in the context of 439-644 fragment essentially abrogated all the transactivation activity (Figure 3C). Moreover, another mutation L489A clearly eliminated the transactivation activity of hEcd (Figure 3C). The same effect was confirmed in full-length hEcd when a single point mutation (D484F) was introduced (Figure 3D). Thus, Asp-484 and Leu-489 appear to be critical residues for hEcd transactivation activity and substitution of either of these amino acids can disrupt the transactivation activity of hEcd.
The C-terminus part of the acidic region is also well conserved between species (Figure 2). Therefore, we tested two C-terminally truncated forms (aa 1-610 and 1-539) of hEcd fragment for transactivation function. The fragment aa 1-610 demonstrated similar transactivation activity as full length Ecd (Figure 3E, left), which suggests that deletion of aa 611-644 does not affect transactivation activity. However, the truncated fragment that spans from aa 1-539 was defective in transactivation (Figure 3E, right). The acidic region (aa 502-532) may play a role in exerting potential transactivation activity together with proper functional structure of adjacent sequences.
p300, a global transcriptional coactivator interacts with Ecd and increases the transactivation activity of Ecd
The p300 protein is a global transcriptional coactivator which binds to a number of transcription factors and associates with other coactivators, thereby increasing the expression of target genes. This function involves the recruitment of the basal transcription machinery to the promoter and promotion of acetylation of transcription factors and the histone proteins (Blobel, 2000; Chan and La Thangue, 2001; Goodman and Smolik, 2000; Imhof et al., 1997; Sterner and Berger, 2000; Vo and Goodman, 2001). To test whether p300 plays a role in Ecd-mediated transactivation, p300 was co-expressed with GAL4-hEcd in the GAL4-reporter assay. Transfection of increasing amounts of GAL4-hEcd plasmid led to a dose-dependent transactivation activity in both the absence and presence of p300. Notably, coexpression of p300 increased the transactivation activity of GAL4-hEcd compared to that of control plasmid expression (Figure 4A). GFP plasmid was co-transfected in all samples to verify comparable transfection efficiency and the expression levels of p300, GAL4-hEcd, and GFP were shown by western blotting (Figure 4B). Endogenous p300 was detected in control plasmid transfected cells and about two-fold increase in the total level of p300 is observed in the samples where ectopic p300 was co-expressed (Figure 4B). Comparison of luciferase activity with the expression level of GAL4-hEcd in the absence or presence of p300 co-expression is shown in Figure 4C. These data demonstrate that the increased expression of p300 enhances the transactivation activity of Ecd.
Figure 4. Co-expression of p300 with Ecd increases Ecd transactivation activity and hEcd interacts with p300.
(A) 293T cells seeded in 24 well plate were transfected with different amounts of GAL4 control plasmid or full length GAL4-hEcd plasmid together with 100 ng of pG5/luciferase, 50 ng of GFP (as a transfection control) and 100 ng of HA-p300 plasmids. Total luciferase activity was measured from cell lysates of transfected cells, which were cultured for additional 24 hours post-transfection. (B) The expression of GAL4-hEcd and p300 is shown by western blotting using GAL4 and p300 antibody, respectively. GFP expression is shown as a transfection control. Immunoblotting with p300 antibody detects both endogenous and exogenous p300. (C) The graph represents the comparison of luciferase activity versus the expression level of GAL4-hEcd in the absence or presence of p300 co-expression. The expression of GAL4-hEcd and GFP was quantified using ImageJ software and the relative expression level of GAL4-hEcd (signal ratio of GAL4-hEcd to GFP) is shown. (D) 293T cells were transected with Flag-hEcd plasmid along with or without HA-p300 plasmid. Cell extracts were immunoprecipitated using anti-HA antibody and Flag-hEcd associated with HA-p300 was detected by western blotting using anti-Flag antibody. (E) Extracts of 293T cells expressing HA-p300 together with or without Flag-hEcd were immunoprecipitated with anti-Flag antibody followed by western blotting using HA antibody.
Given the effect of p300 on Ecd-mediated transactivation, we examined whether Ecd directly interacts with p300 by performing immunoprecipitation followed by western blotting. For immunoprecipitation, Flag-hEcd was co-expressed in 293T cells in the absence or presence of HA-tagged p300. Cell extracts were immunoprecipitated using anti-HA antibody and Flag-hEcd that is associated with p300 was detected by western blotting using anti-Flag antibody. A clear association of Flag-hEcd with p300 was detected in the immunoprecipitates from HA-p300 co-expressed cell extracts (Figure 4D). This interaction was also confirmed by the reverse approach where HA-p300 was expressed in 293T cells in the absence or presence of Flag-tagged hEcd followed by immunoprecipitation using anti-Flag antibody and western blotting using HA antibody (Figure 4E). Collectively, these results provide evidence for the potential role of Ecd as a transcriptional regulator.
Subcellular localization of Ecd protein
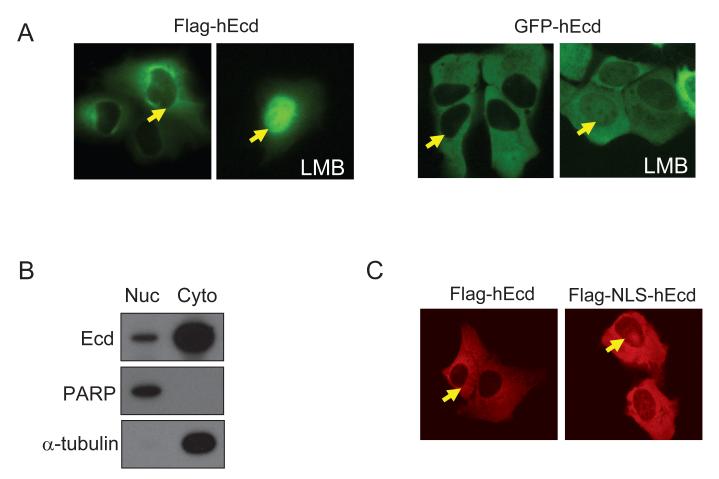
One of the criteria for a protein to be a transcription factor or a transcriptional regulator is its ability to localize to the nucleus. In order to examine the subcellular localization of human Ecd, Flag or GFP-fused full length hEcd proteins were expressed in U2OS cells. The majority of Flag-hEcd and GFP-hEcd expressions were found in the cytoplasm (Figure 5A). However, biochemical fractionation of nuclear and cytoplasmic extracts showed the presence of nuclear as well as cytoplasmic Ecd (Figure 5B). The integrity of the fractionation was confirmed by immunoblotting against PARP and α-tubulin which are markers for nuclear and cytoplasmic extracts, respectively. These results suggest that Ecd localizes in both the nucleus and cytoplasm and the localization may be dynamic. In order to test whether Ecd shuttles between the nucleus and cytoplasm, Flag- or GFP-tagged hEcd were expressed in U2OS cells and the localization of Ecd was detected with or without treatment with leptomycin B (LMB), a CRM1-mediated nuclear export inhibitor. A significant amount of Ecd accumulated in the nucleus upon leptomycin B-induced inhibition of nuclear export (Figure 5A). These results suggest that Ecd shuttles between the nucleus and cytoplasm, and that Ecd may be rapidly exported to the cytoplasm.
Figure 5. Ecd shuttles between nucleus and cytoplasm.
(A) Flag-hEcd was transfected into U2OS cells. Twenty-four hours after transfection, the localization of Flag-hEcd was shown by fluorescence immunostaining using anti-Flag antibody. GFP-hEcd was also analyzed 24 hours after transfection. Twenty hours after transfection, cells were treated with 5 ng/ml of leptomycin B (LMB) for 4 hours. (B) Biochemical fractionation of U2OS cell extract is shown. Nuclear (Nuc) and cytoplasmic (Cyto) extracts were prepared as described in Materials and Methods. To confirm purity of fractionation, extracts were probed against PARP (nuclear protein marker) and α-tubulin (cytoplasmic protein marker). (C) Chimera of Flag-hEcd fused with SV40 large T antigen NLS (PKKKRKV) was generated by inserting the NLS motif between Flag and hEcd sequences. Flag-NLS-hEcd plasmid was transfected into U2OS cells. Twenty hours after transfection, Flag-NLS hEcd signal was visualized by immunostaining using anti-Flag antibody. (D) Wild type and mutant (I481A, D484A, S487A, and L489A) of GAL4-hEcd (aa 439-644) plasmids were transfected into U2OS cells and twenty hours after transfection, GAL4-hEcd signal was visualized by immunostaining using anti-GAL4 antibody. Fluorescence immunostaining was performed as described in Materials and Methods. Arrow heads indicate nucleus-cytoplasm boundary.
To further characterize the nuclear localization and export of Ecd, a chimera of Flag-hEcd fused with SV40 large T antigen nuclear localization signal (NLS; PKKKRKV) was generated. As shown in Figure 5C, although there was a moderate increase in the nuclear Ecd pool, a majority of Flag-NLS-hEcd was again localized in the cytoplasm. Additionally, we analyzed the subcellular localization of GAL4-hEcd. Since GAL4-DBD possesses an intrinsic NLS and GAL4-hEcd showed transactivation activity in the GAL4-reporter assay, we expected GAL4-hEcd to localize to the nucleus. As expected GAL4 expression alone was in the nucleus, however the GAL4-hEcd (aa 439-644) chimera was predominantly present in the cytoplasm albeit it showed very strong transactivation activity in the GAL4-reporter assay (Figure 5D). Next, several mutants of GAL4-hEcd (aa 439-644) containing single amino acid substitution (I481A, D484A, S487A and L489A) that showed different transactivation activity were tested for localization. Notably, none of these mutants showed any difference in the subcellular localization (Figure 5D), suggesting that the transactivation defective phenotypes of D484A and L489A mutation were not due to different subcellular localization of these mutant forms of Ecd. These results support the idea that transient nuclear localization is sufficient for Ecd to exhibit transactivation activity. These results also suggest that Ecd has a strong nuclear export signal (NES) and its nuclear localization is transient.
The deletion of aa 481-497 region disrupts the cytoplasmic localization of Ecd
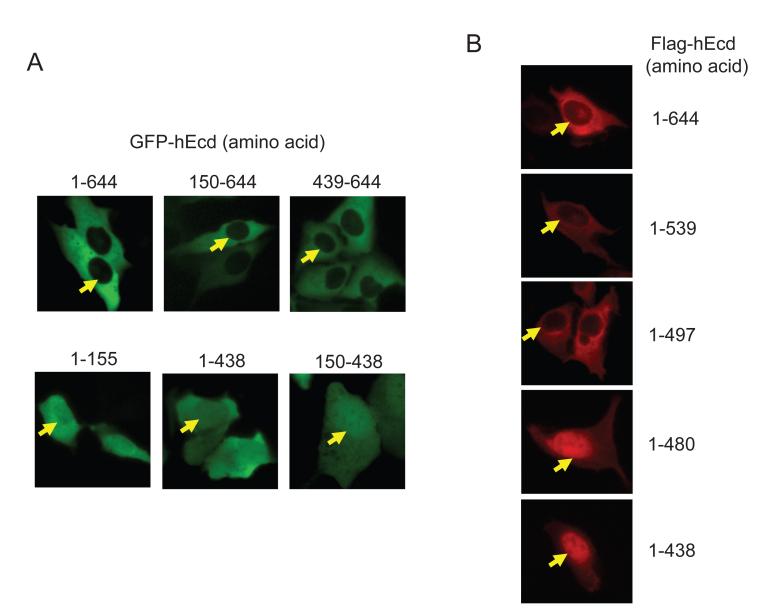
To characterize the regions of Ecd required for predominant cytoplasmic localization, five GFP-tagged deletion constructs (aa 1-155, 1-438, 150-438, 150-644, and 439-644) were transiently expressed in U2OS cells and the localization of these different fragments was examined. While the full length Ecd or its aa 150-644 and 439-644 fragments were mostly detected in cytoplasm, three fragments of Ecd (aa 1-155, 1-438, and 150-438) were detected in both the nucleus and cytoplasm (Figure 6A). These results suggest that the aa 439-644 region is responsible for the cytoplasmic localization of Ecd.
Figure 6. The deletion of aa 481-497 region disrupts the cytoplasmic localization of Ecd.
(A) GFP-tagged full length or truncated forms of hEcd (aa 150-644, 439-644, 1-155, 1-438, 150-438) were transfected into U2OS cells. After 24 hours post-transfection, GFP signal was observed under fluorescent microscope. (B) Flag-tagged full length or C-terminal truncated forms of hEcd (aa 1-539, 1-497, 1-480, and 1-438) were transfected into U2OS cells. Twenty four hours after transfection, Flag-hEcd signal was detected by fluorescence immunostaining using anti-Flag antibody as described in Materials and Methods. Arrowheads indicate nucleus-cytoplasm boundary.
To further examine the subcellular localization, we analyzed additional truncated hEcd fragments. A series of Flag-tagged C-terminal truncated fragments (aa 1-539, 1-497, 1-480 and 1-438) were expressed in U2OS cells and tested for the localization by immunostaining using an anti-Flag antibody. The aa 1-539 and 1-497 fragments localized in the cytoplasm similar to the full length Ecd; however aa 1-480 fragment was localized in the nucleus (Figure 6B). This observation leads us to conclude that the short segment 481-497 seems necessary for Ecd cytoplasmic localization. Although, this region does not have a canonical nuclear export sequence (NES) (la Cour et al., 2003), it may function as a potential nuclear export signal. It is also possible that the deletion of 481-497 region may disrupt the motif for the cytoplasmic localization that resides in the vicinity of this segment. Taken together, these results demonstrate that Ecd localizes to both nucleus and cytoplasm and shuttles between the nucleus and cytoplasm with a rapid nuclear export.
Discussion
The Ecd protein family is highly conserved from yeast to man, indicative of a conserved function of sequences retained through evolution. At present, however, no specific structural domains or motifs linked to Ecd function have been identified in any species. The first potential role of Ecd was suggested in yeast where human Ecd (called hSGT1) was able to substitute for the coactivator function of GCR2 in S. cerevisiae (Sato et al., 1999) and deletion of Ecd in S.pombe (spSGT1) showed that Ecd is important for cell growth and regulates the expression of many genes involved in cellular processes including various metabolic pathways (Kainou et al., 2006). Furthermore S. pombe Ecd appears to be localized to the nucleus and to play an important role in the transcription of various genes. These observations led us to explore whether human Ecd possesses properties of a transcriptional regulator.
Results of a GAL4-reporter gene assay using GAL4-DBD-fusion of Ecd show that human Ecd possesses transactivation activity. This is the first evidence that mammalian Ecd may play a role in transcriptional regulation. Due to lack of known DNA binding motif, Ecd is expected to function as a transcriptional regulator rather than a transcription factor, which is consistent with its ability to substitute for coactivator GCR2 in yeast. Human Ecd contains 644 amino acids. Several regions in Ecd are highly conserved from yeast to humans, while other regions are conserved among higher organisms (Drosophila and higher). Initial mutational analyses presented here show that the C-terminal region of Ecd is crucial for its function as a transcriptional regulator. By a series of mutational analysis, we showed that Asp-484 and Leu-489 are critical for transactivation, as each single point mutation was able to abolish the transactivation activity. We opted to examine this region considering its strict conservation (Asp-484) among different species.
At present, it is difficult to speculate how the point mutations in the acidic region (D510R, D512R, or D520P) increase transactivation activity since the structure of Ecd protein is not known. These point mutations in acidic region may cause a slight structural change in activation domain, which might impart increase transactivation activity of these mutants. In our analyses, the two mutant fragments 1-610 and 1-539 showed different transactivation abilities although both mutants contain acidic activation region (aa 502-532). Although not experimentally proven, we speculate that 1-539 fragment may disrupt proper structure of activation domain due to its close proximity to the acidic region.
Further evidence to support the observation that Ecd functions as a transcriptional regulator is provided by its cooperativity with p300, a histone acetyltransferase (HAT) for its transcriptional function. It is well documented that transcriptional factors or other transcriptional regulators directly interact with HAT proteins, such as p300 to perform a role in transcriptional regulation. Using in vivo assays, we present evidence for direct interaction of Ecd with p300 and cooperatively of p300 to augment the transactivation activity of Ecd. These results support the notion that mammalian Ecd has the biochemical properties of a transcriptional regulator.
Ecd derives its name from Drosophila studies where Ecd mutants show the defective phenotypes in embryo development that may be caused by the insufficient ecdysone hormone production (Garen et al., 1977). However, several lines of evidence suggest other functions of Ecd in Drosophila and these defects cannot be fully explained by the low level of ecdysone in these mutant flies. To date, the precise biochemical function of Ecd in ecdysone pathway, such as an enzyme function or other regulatory role, has not been elucidated. It is also important to note that the execution of ecdysone pathway in Drosophila constitutes transcription of various genes during embryonic development and together with the information presented here, it is reasonable to suggest that Ecd may play a role in transcription in flies.
Transcriptional regulator function requires localization in the nucleus; however there is a discrepancy of Ecd subcellular localization observed in S.pombe vs. Drosophila. The majority of Ecd localizes to the cytoplasm in Drosophila (Gaziova et al., 2004); however, S.pombe Ecd localizes to the nucleus (Kainou et al., 2006). We analyzed the localization of human Ecd in mammalian cells in detail. Although the steady state distribution of Ecd is mainly cytoplasmic, we demonstrate that mammalian Ecd shuttles between the nucleus and cytoplasm and inhibition of its nuclear export by a CRM1 inhibitor led to the accumulation of Ecd in the nucleus. Thus, our data suggests that the localization of Ecd may be regulated under certain physiological conditions, which remains to be studied.
Through mutational studies, the region of Ecd responsible for nuclear export was determined. Dramatic change in subcellular localization of Ecd between truncated mutants aa 1-497 and aa 1-480 suggests that the region between aa 480-497 harbors a potential nuclear export signal (NES) sequence. Although the NES of Ecd does not precisely match the canonical NES sequences (Ф-X(2-3)-Ф-X(2-3)-Ф-X-Ф), the sequence ILNYFDKIL in the aa 488-496 region of Ecd represents a potential NES sequence.
Our extensive efforts to direct Ecd to the nucleus by fusing nuclear localization signals (NLS) of SV40 large T antigen or GAL4-DNA binding domain with Ecd did not significantly alter Ecd localization, suggesting that Ecd is actively exported out of the nucleus. Furthermore cytoplasmic localization of GAL4-hEcd suggests that Ecd localizes in the nucleus transiently, but this transient nuclear localization is apparently sufficient for Ecd to execute its role in transcriptional regulation as shown by the GAL4-reporter assay.
In conclusion, we have presented the biochemical characteristics of Ecd protein that support its function as a transcriptional regulator. At present, it is not known which transcription factor(s) associates with Ecd to exert its biological functions. Future studies will be focused on the identification of primary transcription factors that directly associate with Ecd on the promoter. Such studies, together with identification of Ecd target genes will shed light on the biological function of Ecd as a transcriptional regulator.
Materials and Methods
Cell culture
293T cells were maintained in Dulbecco’s Modified Eagle Medium (Invitrogen) supplemented with 10% Fetal Calf Serum (HyClone), 0.1 mM non-essential amino acids (Invitrogen), 2 mM L-glutamine (Invitrogen), 1 mM sodium pyruvate (Invitrogen) and 10 mM HEPES (Invitrogen). U2OS cells were grown in Alpha-Minimum Essential Medium (Invitrogen) supplemented with 10% Fetal Calf Serum, 0.1 mM MEM non-essential amino acids, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, and 1μg/ml insulin.
Plasmids
For generating N-terminally tagged GAL4-DBD full length or truncated forms (with amino acid numbers shown in parentheses) of human Ecd, different primer sets (shown below) were used to amplify by PCR the region of interest using Flag-hEcd plasmid (Zhang et al., 2006) as the template. Full length mouse Ecd was amplified by RT-PCR using total RNA extracted from mouse embryonic fibroblast cells. Amplified PCR fragments were subcloned into SalI and NotI sites of pBIND vector (Promega). GAL4-hEcd (467-644) was generated from self-ligated GAL4-hEcd (439-644) construct followed by digestions with SalI and XhoI. For expression of N-terminal Flag-tagged Ecd constructs, different Ecd fragments were recovered from the corresponding GAL4-Ecd constructs with EcoRV and NotI digestions and subcloned into EcoRV and NotI sites of modified pcDNA3.1(+)-Flag1(Invitrogen) in which Flag sequence was added between NheI and HindIII sites by inserting annealed double-stranded oligonucleotides (5′-CTAGCATGGACTACAAGGACGACGATGACAAGA-3′ and 5′-AGCTTCTTGTCATCGTCGTCCTTGTAGTCCATG-3′). Flag-hEcd (1-497) and Flag-hEcd (1-480) were generated by PCR cloning. PCR primers and cloning sites are shown in below. For EGFP-tagged hEcd constructs, PCR fragments of hEcd were cloned into pEGFP-C1 (Clontech). EGFP-hEcd (1-155) plasmid was constructed by subcloning the fragment (KpnI/Xba) of pcDNA3.1(+)-Flag-hEcd (1-155) into the KpnI and XbaI sites of pEGFP-C1 vector. EGFP-hEcd (150-438) and EGFP-hEcd (439-644) were also constructed by subcloning the fragment (EcoRI/Xba) of corresponding pcDNA3.1(+)-Flag-hEcd plasmids into the EcoRI and XbaI sites of pEGFP-C1 vector. Nuclear localization signals (NLS) of SV40 large T antigen was tagged by inserting annealed double-stranded oligonucleotides (5′-TCGACAAGATATCCCAAAAAAGAAGAGAAAGGTA-3′ and 5′-TACCTTTCTCTTCTTTTTTGGGATATCTTG-3′) into GAL4-hEcd (1-644) constructs digested with SalI and EcoRV. The fragment of NLS-hEcd (1-644), then recovered from GAL4-NLS-hEcd (1-644) by digesting with EcoRV and NotI were subcloned into the EcoRV and NotI sites of modified pcDNA3.1(+)-Flag1, thereby generating Flag-NLS-hEcd (1-644).
Site-directed mutagenesis
PCR-based site-directed mutagenesis method was used to generate various Ecd mutants. About 5-50 ng of plasmid DNA was used as templates for PCR reaction (12-18 cycles, 1 min/Kb of plasmid length for elongation times at 68 °C) using Pfu turbo DNA polymerase (Stratagene) and specific primers. PCR products were treated with DpnI for 30 min to remove parental DNA and then used for transformation into E.coli. The mutations were verified by sequencing. The primers for mutagenesis are shown below.
GAL4-reporter gene assay
For reporter gene assay, the cells were plated in twenty four-well plates for 24 hours and were then transfected with indicated pBIND fusion plasmid, and pG5/luciferase plasmid (Promega) using Lipofectamine 2000 (Invitrogen) or Fugene 6 (Roche) transfection reagent according to the manufacturer’s instructions. GAL4-fused human Ecd (200 ng) constructs were transfected in U2OS cells in 24-well plate together with 200 ng of GAL4-reporter plasmid (pG5/luciferase). Twenty hours after transfection, cells were lysed in 100 μl of passive lysis buffer (Promega). Luciferase activitiy , in 2-10 μl of lysates, was measured using a luminometer and Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol.
Immunofluorescence staining
Immunofluorescence staining of Flag-hEcd or GAL4-hEcd was performed using anti-Flag M2 mouse monoclonal antibody (Sigma) or anti-GAL4 mouse monoclonal antibody (Santa Cruz Biotechnology). Transfected cells on coverslips were fixed for 20 min in 1% paraformaldehyde, and then subjected to permeabilization (in PBS containing 0.5% Triton X-100, for 15 min at RT) and blocking (in PBS containing 1% BSA, for 30 min at RT). After blocking, cells were incubated for 1 hour with anti-Flag antibody (1:1000) and then with anti-mouse IgG Alexa Fluor 488 or 564 (1:1000) (Molecular Probes) in PBS containing 0.1% Tween-20. After washing in PBS containing 0.1% Tween-20, cells on coverslips were mounted and visualized under fluorescence microscope. To block nuclear export of Ecd proteins, cells were treated with 5 ng/ml of leptomycin B (LMB) for 4 hours.
Western blotting and immunoprecipitation
Western blotting was performed using a primary antibodies against Ecd (generated at the Monoclonal Antibody Facility at the Lurie Cancer Center, Northwestern University, Chicago, IL), GAL4 (Santa Cruz Biotechnology), Flag (Sigma), HA (Sigma), PARP (Zymed), p300 (Santa Cruz Biotechnology), α-tubulin (Sigma), and GFP (Santa Cruz Biotechnology). For immunoprecipitations, cell extracts were prepared in lysis buffer (20 mM Tris-HCl [pH7.5], 150 mM NaCl, 0.5 % NP-40 and a protease inhibitor cocktail from Roche) and immunoprecipitated using 2 μg of antibodies for overnight at 4°C and the immunocomplexes were pulled down with protein A/G agarose (Santa Cruz Biotechnology) for an additional 2 hours. 2X sample loading buffer was added to the purified immunocomplexes, boiled for 5 min and then loaded on SDS-PAGE. For biochemical fractionation of nuclear/cytoplasmic extracts, cells were lysed in hypotonic buffer (20 mM HEPES pH7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5% NP-40, 0.5 mM DTT, protease inhibitors). Nuclei were isolated by centrifugation of lysates at 1000g for 5 min and supernatant was saved as cytosolic fraction. The cytosolic fraction was cleared by centrifugation at 15000 rpm for 20 min. For nuclear fraction, nuclei were washed once with hypotonic buffer without NP-40, and lysed with high salt buffer containing 400 mM NaCl for 30 min on ice. Nuclear lysates were cleared by centrifugation at 15000 rpm.
Primer sequences used in cloning experiments
| Plasmid (amino acids) |
Cloning sites (5′/3′) |
Primers (forward) (reverse) |
|---|---|---|
| GAL4-hEcd (1-644) | SalI/NotI | 5′-AAGTCGACAAGATATCATGGAAGAAACCATGAAGCT–3′ |
| 5′–AAAGCGGCCGCTTAATTTTTTGTTGGCTTACT–3′ | ||
| GAL4-hEcd (1-610) | SalI/NotI | 5′–AAGTCGACAAGATATCATGGAAGAAACCATGAAGCT–3′ |
| 5′–ACCGCGGCCGCTCAGGAGCTATAGGATTCCAA–3′ | ||
| GAL4-hEcd (1-539) | SalI/NotI | 5′–AAGTCGACAAGATATCATGGAAGAAACCATGAAGCT–3′ |
| 5′–AATGCGGCCGCAAGTGTTCCTTTCAGGGAAGC–3′ | ||
| GAL4-hEcd (1-155) | SalI/NotI | 5′–AAGTCGACAAGATATCATGGAAGAAACCATGAAGCT–3′ |
| 5′–ACCGCGGCCGCTCATCCAGATTTTCTTGGTGC–3′ | ||
| GAL4-hEcd (1-438) | SalI/NotI | 5′–AAGTCGACAAGATATCATGGAAGAAACCATGAAGCT–3′ |
| 5′–ACCGCGGCCGCTCAAGACTCGGATTCTTAAAAC–3′ | ||
| GAL4-hEcd (150-438) | SalI/NotI | 5′–AAGTCGACAAGATATCGCACCAAGAAAATCTGGA–3′ |
| 5′–ACCGCGGCCGCTCAAGACTCGGATTCTTAAAAC–3′ | ||
| GAL4-hEcd (150-644) | SalI/NotI | 5′–AAGTCGACAAGATATCGCACCAAGAAAATCTGGA–3 |
| 5′–AAAGCGGCCGCTTAATTTTTTGTTGGCTTACT–3′ | ||
| GAL4-hEcd (439-644) | SalI/NotI | 5′–AAGTCGACAAGATATCGTTTCCAAGGAGGAGAAGGAGC- 3′ |
| 5′–AAAGCGGCCGCTTAATTTTTTGTTGGCTTACT–3′ | ||
| Flag-hEcd (1-497) | EcoRV/NotI | 5′–AAGTCGACAAGATATCATGGAAGAAACCATGAAGCT–3′ |
| 5′–ACCGCGGCCGCTTACCCTAAAATCTTATCAAAATA–3′ | ||
| Flag-hEcd (1-480) | EcoRV/NotI | 5′–AAGTCGACAAGATATCATGGAAGAAACCATGAAGCT–3′ |
| 5′–ACCGCGGCCGCTCATGGAGCCTCAGAAGGTTCTCG–3′ | ||
| EGFP-hEcd (1-644) | SacI/SacII | 5′–GAGGAGCTCACATGGAAGAAACCATGAAGCTTGC–3′ |
| 5′–AGACCGCGGATTTTTTGTTGGCTTACTTGTTGGTCTG–3′ | ||
| EGFP-hEcd (1-438) | BspE1/SalI | 5′–GCTCCGGAGAAGAAACCATGAAGCTTG–3′ |
| 5′–CGGTCGACTTAAGACTCGGATTCTTTTTTGCC–3′ | ||
| EGFP-hEcd (150-644) | BspE1/SalI | 5′–GCTCCGGAGCACCAAGAAAATCTGGAGC–3′ |
| 5′–CGGTCGACTTAATTTTTTGTTGGCTTACTTGTTGG–3′ |
Primer sequences used in site-directed mutagenesis
| Mutation site (human Ecd) | Primers |
|---|---|
| I481A | 5′–CCTTCTGAGGCTCCAGCTACTTTTGATGCAGAT–3′ |
| 5′–ATCTGCATCAAAAGTAGCTGGAGCCTCAGAAGG–3′ | |
| D484A | 5′–GCTCCAATCACTTTTGCTGCAGATTCTTTTCT–3′ |
| 5′–AGAAAAGAATCTGCAGCAAAAGTGATTGGAGC–3′ | |
| D484F | 5′–CCAATCACTTTTTTTGCAGATTCTTTT–3′ |
| 5′–AAAAGAATCTGCAAAAAAAGTGATTGG–3′ | |
| S487A | 5′–CACTTTTGATGCAGATGCTTTTCTTAATTATTTTG–3′ |
| 5′–CAAAATAATTAAGAAAAGCATCTGCATCAAAAGTG–3′ | |
| L489A | 5′–GATGCAGATTCTTTTGCTAATTATTTTGATAAG–3′ |
| 5′–CTTATCAAAATAATTAGCAAAAGAATCTGCATC–3′ | |
| D510R | 5′–TCTGATGATCTGGATCGGGAAGACTTTGAATGT–3′ |
| 5′–ACATTCAAAGTCTTCCCGATCCAGATCATCAGA–3′ | |
| D512R | 5′–GATCTGGATGATGAACGCTTTGAATGTTTAGAT–3′ |
| 5′–ATCTAAACATTCAAAGCGTTCATCATCCAGATC–3′ | |
| D520P | 5′–GTTTAGATAGTGATCCGGACTTGGACTTTG–3′ |
| 5′–CAAAGTCCAAGTCCGGATCACTATCTAAAC–3′ | |
| ΔAR (502-532) | 5′–ATTAGGCCTTGGCCCTAAAAT–3′ |
| 5′–GCTTCCCTGAAAGGAACACTT–3′ |
Acknowledgements
We thank Dr. Xiangshan Zhao and other members of Band laboratory for helpful discussions and suggestions. This work was supported by the NIH Grant R01 CA96844-06, R01 CA94143 and Department of Defense grant W81XWH-07-1-0351 to V.B; NIH CA 87986, CA 105489, CA 99900, CA99163 and CA116552 to H.B. YZ is a Carol and Marvin Gollob Fellow. JHK is supported by a Korea Science and Engineering Foundation Grant (M06-2004-000-10547-0) and the Department of Defense Pre-doctoral Traineeship grant # W81XWH-08-1-0366. VB acknowledges the support of the Duckworth family through the Duckworth Family Chair for Breast Cancer Research. HB acknowledges the support from the Jean Ruggles-Romoser Chair for Cancer Research.
The work presented here was initiated while the investigators were at the Department of Medicine, Evanston Northwestern Healthcare Research Institute, Northwestern University, Evanston, IL
References
- Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- Bratton MR, Frigo DE, Vigh-Conrad KA, Fan D, Wadsworth S, McLachlan JA, Burow ME. Organochlorine-mediated potentiation of the general coactivator p300 through p38 mitogen-activated protein kinase. Carcinogenesis. 2009;30:106–113. doi: 10.1093/carcin/bgn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Dennig J, Beato M, Suske G. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J. 1996;15:5659–5667. [PMC free article] [PubMed] [Google Scholar]
- Garen A, Kauvar L, Lepesant JA. Roles of ecdysone in Drosophila development. Proc Natl Acad Sci U S A. 1977;74:5099–5103. doi: 10.1073/pnas.74.11.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziova I, Bonnette PC, Henrich VC, Jindra M. Cell-autonomous roles of the ecdysoneless gene in Drosophila development and oogenesis. Development. 2004;131:2715–2725. doi: 10.1242/dev.01143. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- Kainou T, Shinzato T, Sasaki K, Mitsui Y, Giga-Hama Y, Kumagai H, Uemura H. Spsgt1, a new essential gene of Schizosaccharomyces pombe, is involved in carbohydrate metabolism. Yeast. 2006;23:35–53. doi: 10.1002/yea.1336. [DOI] [PubMed] [Google Scholar]
- Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12:1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillycrop KA, Dawson SJ, Estridge JK, Gerster T, Matthias P, Latchman DS. Repression of a herpes simplex virus immediate-early promoter by the Oct-2 transcription factor is dependent on an inhibitory region at the N terminus of the protein. Mol Cell Biol. 1994;14:7633–7642. doi: 10.1128/mcb.14.11.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Park HJ, Wang Z, Costa RH, Tyner A, Lau LF, Raychaudhuri P. An N-terminal inhibitory domain modulates activity of FoxM1 during cell cycle. Oncogene. 2008;27:1696–1704. doi: 10.1038/sj.onc.1210814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Jigami Y, Suzuki T, Uemura H. A human gene, hSGT1, can substitute for GCR2, which encodes a general regulatory factor of glycolytic gene expression in Saccharomyces cerevisiae. Mol Gen Genet. 1999;260:535–540. doi: 10.1007/s004380050926. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura H, Koshio M, Inoue Y, Lopez MC, Baker HV. The role of Gcr1p in the transcriptional activation of glycolytic genes in yeast Saccharomyces cerevisiae. Genetics. 1997;147:521–532. doi: 10.1093/genetics/147.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Zeng X, Deminoff SJ, Santangelo GM. Specialized Rap1p/Gcr1p transcriptional activation through Gcr1p DNA contacts requires Gcr2p, as does hyperphosphorylation of Gcr1p. Genetics. 1997;147:493–505. doi: 10.1093/genetics/147.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen J, Gurumurthy CB, Kim J, Bhat I, Gao Q, Dimri G, Lee SW, Band H, Band V. The human orthologue of Drosophila ecdysoneless protein interacts with p53 and regulates its function. Cancer Res. 2006;66:7167–7175. doi: 10.1158/0008-5472.CAN-06-0722. [DOI] [PubMed] [Google Scholar]
- Zhao C, York A, Yang F, Forsthoefel DJ, Dave V, Fu D, Zhang D, Corado MS, Small S, Seeger MA, Ma J. The activity of the Drosophila morphogenetic protein Bicoid is inhibited by a domain located outside its homeodomain. Development. 2002;129:1669–1680. doi: 10.1242/dev.129.7.1669. [DOI] [PubMed] [Google Scholar]