Abstract
Sporadic Burkitt lymphoma (sBL) can be delineated from diffuse large B-cell lymphoma (DLBCL) by a very homogeneous mRNA expression signature. However, it remained unclear whether all three BL variantsFsBL, endemic BL (eBL) and human immunodeficiency virus-associated BL (HIV-BL)F represent a uniform biological entity despite their differences in geographical occurrence, association with immunodeficiency and/or incidence of Epstein-Barr virus (EBV) infection. To address this issue, we generated micro RNA (miRNA) profiles from 18 eBL, 31 sBL and 15 HIV-BL cases. In addition, we analyzed the miRNA expression of 86 DLBCL to determine whether miRNA profiles recapitulate the molecular differences between BL and DLBCL evidenced by mRNA profiling. A signature of 38 miRNAs containing MYC regulated and nuclear factor-kB pathway-associated miRNAs was obtained that differentiated BL from DLBCL. The miRNA profiles of sBL and eBL displayed only six differentially expressed miRNAs, whereas HIV and EBV infection had no impact on the miRNA profile of BL. In conclusion, miRNA profiling confirms that BL and DLBCL represent distinct lymphoma categories and demonstrates that the three BL variants are representatives of the same biological entity with only marginal miRNA expression differences between eBL and sBL.
Keywords: Burkitt lymphoma, epidemiological variants, diffuse large B-cell lymphoma, micro RNA profiling
Introduction
Burkitt lymphoma (BL) is a mature aggressive B-cell neoplasm, which histologically presents with sheets of monomorphic medium-sized round tumor cells. They show an exceptionally large growth fraction commonly close to 100% and a high rate of apoptosis. The apoptotic cells are phagocytosed by macrophages with a light cytoplasm leading to the characteristic ‘starry sky’ pattern.1 Owing to the consistent expression of CD10 and BCL6 and the mutational status of the immunoglobulin genes BL is regarded as a neoplastic outgrowth of germinal center (GC) B cells.2-4 A further characteristic feature of BL is a translocation of the MYC gene to one of the immunoglobulin gene loci, predominantly the IGH locus,5 resulting in constitutive MYC protein expression.
Based on epidemiological features BL is subdivided into three variants.1 Endemic BL (eBL) occurs in humid areas of equatorial Africa and Papua New Guinea at a high frequency, affects mainly children and is Epstein-Barr virus (EBV) infected in nearly all instances. The most common sites of involvement are jaw and abdomen. In addition, malaria, arbovirus infection as well as plant tumor promoters are considered cofactors of lymphomagenesis.6 In the rest of the world, BL is 100 to 200 times less frequent, having led to the term sporadic BL (sBL). sBL is predominantly found in patients with a higher age than eBL and only a minority of sBL (approximately 30%) is EBV infected.1 The third variant, namely the human immunodeficiency virus-associated BL (HIV-BL) most commonly arises in HIV-infected patients where its occurrence is often one of the first signs of the HIV-induced disease.1
Only few studies looked for molecular differences in the three BL variants. One distinction was found with respect to the t(8;14)(q24;q32). In eBL the MYC gene is translocated to the IGH joining region whereas in sBL the MYC gene is juxtaposed to the IGH MU SWITCH (S-MU) region.7 Also the breakpoint region of the MYC gene differs between both variants and there is a higher incidence of mutations in the 5′-region of the MYC gene in eBL than in sBL.7 Another study revealed a higher number of somatic mutations in the IG genes as well as a higher frequency of features attributed to antigen selection in eBL and HIV-BL compared with sBL.8 These results lead to the hypothesis that the cell of origin of eBL and HIV-BL is more closely related to late GC B cells or post-GC memory B cells and that the cell of origin of sBL is more closely related to early centroblasts.8
On the basis of the current WHO (World Health Organization) diagnostic criteria, BL cannot reliably be distinguished from other types of mature aggressive B-cell lymphoma, especially from diffuse large B-cell lymphoma (DLBCL) with MYC breaks. Therefore messenger RNA (mRNA) signatures of BL fulfilling all criteria of the WHO classification 2001 were established and used for the delineation of BL from DLBCL.9,10 It was found that (i) the morphological spectrum of BL is broader than previously assumed, ranging from typical BL morphology to centroblast-like DLBCL morphology, (ii) expression of CD10 and BCL6 in the absence or in combination with weak expression of BCL2 and a growth fraction >95% are consistent features of BL, (iii) a MYC break is not detectable in 11% of the cases assigned as molecular BL and (iv) there are intermediate cases with overlapping features between BL and DLBCL.
A promising tool to further explore the molecular features of BL is a recently discovered class of molecules, namely micro RNAs (miRNAs). MiRNAs are small non-coding, single-stranded RNA molecules that have been shown to bind to complementary sequences in the 3′-untranslated regions of their target mRNAs.11,12 This leads to inhibition of translation or the degradation of the coding mRNA and consequently to a reduced level of the corresponding protein.13 By these means miRNAs influence important cellular processes like differentiation,14 proliferation15 and apoptosis,16 which also applies to the cells of the hematopoietic lineage.17-20 Meanwhile deregulation of miRNA expression has been found in numerous types of cancer, including lymphoma.21-24 MiRNA expression profiling has also successfully been employed to distinguish tumor subgroups, for example, GCB- and non-GCB-type DLBCL.24-26 To elucidate whether miRNA expression patterns can differentiate the three BL variants and distinguish these variants from DLBCL we generated miRNA profiles from biopsies of 18 eBL, 31 sBL, 15 HIV-BL and 86 DLBCL.
Patients and methods
Patient samples
Formalin-fixed and paraffin-embedded (FFPE) tumor specimens of 86 DLBCL and 36 BL patients were obtained from the Institute of Pathology, Campus Benjamin Franklin, Charité-Universitätsmedizin Berlin, Germany. Additional 28 BL FFPE cases were retrieved from the Department of Human Pathology and Oncology, University of Siena, Italy and the Department of Human Pathology, University of Nairobi, Kenya. The DLBCL samples have previously been reviewed by a panel of expert hematopathologists and their clinical data were published.27 The diagnosis of BL cases was confirmed by histopathology review according to the WHO classification criteria.
DNA isolation
Genomic DNA was isolated from FFPE sections by the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols. Quantity and purity of the DNA was assessed by photometric measurement (Nanodrop ND-1000, Thermo Scientific, Schwerte, Germany).
RNA isolation
Total RNA was isolated from FFPE tissue sections with the RecoverALL Kit (Ambion, Austin, TX, USA) according to the manufacturer’s recommendations and quantified using the Nanodrop ND-1000 UV-vis spectrophotometer (Thermo Scientific).
PCR for detection of HIV infection
A fragment of the HIV-1 DNA was amplified by nested PCR using the lentivirus universal primer pair UNIPOL1/2 as outer primers (25 cycles) and the degenerate primers UNIPOL3 (5′-GAAACAGGAMRRGAGACAGC-3′) and UNIPOL4 (5′-TTCA TDGMTTCCACTACTCCTTG-3′) as inner primers (30 cycles).28 This nested primer set, when used at low-stringency annealing, specifically amplifies all HIV-1, HIV-2 and HIV-0 pol sequences known to date. PCR products were visualized on agarose gels and the specificity of the products was confirmed by direct sequencing.
Tissue microarrays
Tissue cores (1 mm in diameter) were removed from representative areas of the FFPE blocks of all samples and from two tonsils and inserted into a recipient paraffin block. Sections were cut from the newly assembled tissue microarray and used for immunohistochemical stainings and fluorescence in situ hybridization analysis.
Immunohistochemistry
Sections from FFPE blocks or from tissue microarrays were deparaffinized and antigenicity was retrieved by cooking in citrate buffer (100 mm) for 5 min (BCL6 and MYC antibody) or 2 min (all other antibodies). Stainings were performed using mouse monoclonal antibodies against BCL2 (1:25; Dako, Glostrup, Denmark), BCL6 (1:50; Clone PG-B6p, Dako), CD10 (1:25; Novocastra, Leica Microsystems, Berlin, Germany), CD20 (1:50; clone L26, Dako), Ki-67 (1:2000; clone Mib1, Dianova, Hamburg, Germany), IRF4 (1:25; clone Mum1p, kindly provided by B Falini) and MYC (1:300; clone Y69, Epitomics, Burlingame, CA, USA). Binding was visualized with the APAAP Mouse REAL Detection System (Dako) for antibodies against BCL2, CD10, CD20 and Ki-67 and REAL Detection System, Alkaline Phosphatase/RED (Dako) for detection of anti-BCL6, anti-IRF4 and anti-MYC antibodies.
Analysis of MYC protein expression levels
To account for the proportion of MYC-positive tumor cells as well as the intensity of the staining we applied the semiquantitative immunoreactivity score (IRS) as described by Noske et al.29 Briefly, the percentage of stained tumor cells (score 0–4) was multiplied with the staining intensity (score 0–3) to give the IRS of each case (score 0–12). The statistical difference of the expression level between BL and DLBCL was then evaluated by t-test.
Fluorescence in situ hybridization
Interphase fluorescence in situ hybridization was performed on FFPE tissues as described previously.9,30 Assays comprised LSI MYC break-apart and LSI MYC/IGH double-color double-fusion assays (all from Abbott/Vysis, Champaign, IL, USA). BLs with a LSI MYC signal pattern indicating a breakpoint in the MYC locus but lacking IGH-MYC fusion were further investigated with probes for the detection of IGK-MYC and IGL-MYC fusions.9,30
EBER in situ hybridization
Chromogenic EBER in situ hybridization for the detection of the EBER transcripts of the EBV was carried out with DIG-labeled sense and antisense run-off transcripts (DIG RNA Labeling Kit, Roche Diagnostics, Mannheim, Germany) as described before.31 After application of a DIG-specific antibody labeled with Alkaline Phosphatase (Roche Diagnostics), bound probes were visualized by REAL Detection System, Alkaline Phosphatase/RED (Dako).
MiRNA microarray hybridization
Sample preparation for and hybridization to miRNA microarrays was performed as described before.32 Briefly, 5 μg of total RNA were reverse transcribed using Biotin-labeled random octamers. The labeled targets were then hybridized to Ohio State University custom miRNA microarray chips, which contain probes for 602 human miRNAs spotted in duplicate. The hybridized chips were washed, stained with streptavidin-Alexa647 conjugate (Invitrogen, Carlsbad, CA, USA) and the fluorescence intensity was assessed with an Axon GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA, USA).
MiRNA microarray data analysis
Microarray images were analyzed using GenePix Pro 6.0 software (Molecular Devices). Average values of the replicate spots of each miRNA were background subtracted, thresholded to 75 and normalized using the quantiles method as implemented in the Bioconductor package/function Affymetrix/ normalization.33 MiRNAs showing low expression variation among all samples were excluded from further analysis. Statistical analysis including correction for multiple testing (analysis of variance, false discovery rate (q-value) cut-off 0.05) and hierarchical clustering (Euclidean distance, complete linkage, standardized intensities by mean) were performed using Partek Genomics Suite 6.5 beta (Partek Inc., St Louis, MO, USA).
The microarray data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus repository (GSE22420, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=fxalnmququoicle&acc=GSE22420).
Real-time reverse transcriptase (RT)-PCR
Eleven miRNAs (hsa-miRs-9, -23a, -23b, -26a, -29b, -30d, -146a, -146b-5p, -155, -221 and -222) were selected from our BL/DLBCL microarray signature for real-time RT-PCR validation. TaqMan assays for these miRNAs and for small nuclear RNAs (RNUs) 43, 44 and 48, serving as endogenous controls, were plated onto custom 96-well fast plates (Applied Biosystems, Darmstadt, Germany). RT-reactions of 22 BL and 31 DLBCL samples for which RNA was still available were carried out with 20 ng of RNA, respectively, and a primer pool consisting of the stem loop primers for all miRNAs and RNUs. Undiluted RT-reaction of each case was then combined with TaqMan Universal Master Mix II, no UNG (Applied Biosystems), pipetted to the respective wells of the assay plate and amplified (7900HT Fast Real-Time PCR System, Applied Biosystems). Baseline and thresholds were analyzed using RQ Manager 1.2.1 (Applied Biosystems). Exported result files were then loaded into Data Assist Software v2.0 (Applied Biosystems) for statistical analysis. On the basis of the implemented geNorm algorithm, RNU48 showed the most stable expression (mean expression levels: group BL: 24.8 Ct, SD 1.33; group DLBCL 24.4 Ct, SD 1.2) and was thus used as endogenous control for the calculation of 2^-dCt values. A t-test was then performed to compare the expression of each miRNA in the analyzed group of BL and DLBCL cases, respectively.
Results
Classification of BL and DLBCL samples
On the basis of their geographical origin, 31 BL samples were designated as sBL and 18 samples as eBL. Fifteen BL cases were diagnosed as HIV-BL. Of the 18 eBL, 14 cases were EBV-positive (87.5%), 2 samples were EBV-negative (12.5%) and for 2 eBL cases the EBV status was not evaluable. Of the sBL samples 26 were EBV-negative (86.7%), 4 cases were EBV-positive (13.3%) and for 1 case the EBV status was not evaluable. Among the HIV-BL 5 (33.3%) were EBV-positive, whereas 10 (66.7%) were EBV-negative. Fluorescence in situ hybridization analysis detected a MYC translocation in almost all BL (92.6%), whereas this was only the case for 6% of DLBCL. The subtyping of the DLBCL based on the immunohistochemical classifier by Hans et al.34 revealed that 48.8% had a GCB and 45.4% had a non-GCB phenotype. An overview of all patient characteristics is given in Table 1.
Table 1.
Clinical and histopathological/biological characteristics of BL and DLBCL patients
| Sporadic BL | Endemic BL | HIV-BL | DLBCL | |
|---|---|---|---|---|
| Number of patients | 31 | 18 | 15 | 86 |
| Age at time of diagnosis (years) | ||||
| Mean | 45.1 | 29.8 | 39.6 | 68.6a |
| Range | 7–84 | 3–47 | 21–67 | 61–80 |
| Sex (%) | ||||
| Female | 6 (20.7) | 8 (47.1) | 2 (13.3) | 36 (41.9) |
| Male | 23 (79.3) | 9 (52.9) | 13 (86.7) | 50 (58.1) |
| NA (lack of patient data) | 2 | 1 | 0 | 0 |
| EBV infection (%) | ||||
| Positive | 4 (13.3) | 14 (87.5) | 5 (33.3) | Not done |
| Negative | 26 (86.7) | 2 (12.5) | 10 (66.7) | Not done |
| NA | 1 | 2 | 0 | Not done |
| MYC translocation (%) | ||||
| IGH–MYC fusion | 19 (65.5) | 5 (45.4) | 10 (71.5) | Not done |
| IGL/IGK–MYC fusion | 6 (20.7) | 4 (36.4) | 2 (14.3) | Not done |
| MYC breakpoint (other than to IG-loci) | 1 (3.4) | 2 (18.2) | 1 (7.1) | 4 (6.0) |
| Negative | 3 (10.3) | 0 | 1 (7.1) | 63 (94.0) |
| NA | 2 | 7 | 1 | 19 |
| IHC classification (Hans et al. 34 ) (%) | ||||
| GCB-like | Not done | Not done | Not done | 42 (48.8) |
| Non-GCB-like | Not done | Not done | Not done | 39 (45.4) |
| NA | Not done | Not done | Not done | 5 (5.8) |
Abbreviations: BL, Burkitt lymphoma; DLBCL, diffuse large B-cell lymphoma; EBV, Epstein–Barr virus; GC, germinal center; HIV-BL, human immunodeficiency virus-associated BL; IHC, immunohistochemistry; NA, not evaluable.
Patients were enrolled in the RiCOVER-60 study that demanded a minimum age of 60 at diagnosis.
MYC protein expression levels in BL and DLBCL
The MYC protein expression was determined by immunohistochemistry and staining intensity was assessed by the IRS. The mean IRS for MYC of the BL samples analyzed (n = 56) was 10.05 whereas the DLBCL samples (n = 46) displayed a mean IRS of 7.13 (P = 5.6 × 10−6). Correlation of the IRS with the MYC status showed a high MYC IRS in cases with MYC translocation irrespective of their morphological diagnosis (P = 1.9 × 10−8). This includes also MYC-break negative BLs, which had a low MYC-IRS (7.0) indistinguishable from MYC-break negative DLBCL cases (IRS of 7.1).
Unsupervised data analysis
By unsupervised cluster analysis employing all 150 lymphoma cases a clear distinction of BL and DLBCL was possible, however, without subdivision into the BL subgroups (Supplementary Figure 1). In contrast, within the DLBCL cases a heterogeneous miRNA expression pattern was present supporting their molecular heterogeneity as previously demonstrated. As unsupervised approaches are unable to identify subtle molecular differences we next performed statistical analyses to clarify if miRNAs are able to separate the three BL subgroups.
MiRNA expression differences between BL and DLBCL
Statistical analysis revealed a differential expression of 38 mature miRNAs (false discovery rate (q-value) cut-off 0.05) between BL and DLBCL cases (Table 2). In all, 17 miRNAs were downregulated in BL compared with DLBCL and 21 miRNAs showed a higher expression in BL than in DLBCL. Six of the miRNAs differentially expressed between BL and DLBCL (hsa-miRs-9, -221, -222, -146a, -146b and -155) have been shown to be associated with the transcription factor nuclear factor (NF)-kB system.24-26,35-37 In addition, for several miRNAs expressed at lower level in BL than in DLBCL in our data set (hsa-miRs-23a, -23b, -26a, -29b, -30d and -146a) a repression by the MYC protein has been demonstrated.38-40 Of the MYC upregulated miRNAs,41,42 the miR-17-92 cluster showed, however, no differential expression between BL and DLBCL, whereas hsa-miR-9 was higher expressed in the BL cases than in the DLBCL cases. Finally, miR-328, which targets CD4443 commonly downregulated in BL,9,10 was also found to be higher expressed in the BL cases than in DLBCL cases.
Table 2.
Differentially expressed miRNAs between BL and DLBCL based on ANOVA analysis (FDR (q-value) <0.05)
| MiRNA name | Fold change BL/DLBCL | Description |
|---|---|---|
| hsa-miR-221 | −15.55 | BL down vs DLBCL |
| hsa-miR-155 | −14.65 | BL down vs DLBCL |
| hsa-miR-146a | −11.57 | BL down vs DLBCL |
| hsa-miR-146b-5p | −6.66 | BL down vs DLBCL |
| hsa-miR-26b | −6.44 | BL down vs DLBCL |
| hsa-miR-23a | −6.28 | BL down vs DLBCL |
| hsa-miR-30d | −5.18 | BL down vs DLBCL |
| hsa-miR-107 | −4.00 | BL down vs DLBCL |
| hsa-miR-103 | −3.99 | BL down vs DLBCL |
| hsa-miR-222 | −3.90 | BL down vs DLBCL |
| hsa-miR-26a | −3.18 | BL down vs DLBCL |
| hsa-miR-30a | −2.55 | BL down vs DLBCL |
| hsa-miR-142-5p | −2.31 | BL down vs DLBCL |
| hsa-miR-23b | −1.92 | BL down vs DLBCL |
| hsa-miR-342-3p | −1.80 | BL down vs DLBCL |
| hsa-miR-29b | −1.73 | BL down vs DLBCL |
| hsa-miR-34b | −1.25 | BL down vs DLBCL |
| hsa-miR-371-5p | 1.43 | BL up vs DLBCL |
| hsa-miR-185 | 1.44 | BL up vs DLBCL |
| hsa-miR-93* | 1.50 | BL up vs DLBCL |
| hsa-miR-326 | 1.51 | BL up vs DLBCL |
| hsa-miR-497 | 1.68 | BL up vs DLBCL |
| hsa-miR-26b* | 1.70 | BL up vs DLBCL |
| hsa-miR-339-5p | 1.75 | BL up vs DLBCL |
| hsa-miR-485-3p | 1.79 | BL up vs DLBCL |
| hsa-miR-9 | 1.86 | BL up vs DLBCL |
| hsa-miR-193a-5p | 1.87 | BL up vs DLBCL |
| hsa-miR-448 | 1.88 | BL up vs DLBCL |
| hsa-miR-202* | 1.92 | BL up vs DLBCL |
| hsa-miR-483-3p | 2.03 | BL up vs DLBCL |
| hsa-miR-26a-1* | 2.05 | BL up vs DLBCL |
| hsa-miR-328 | 2.23 | BL up vs DLBCL |
| hsa-miR-192 | 2.23 | BL up vs DLBCL |
| hsa-miR-429 | 2.27 | BL up vs DLBCL |
| hsa-miR-324-5p | 2.27 | BL up vs DLBCL |
| hsa-miR-340 | 2.33 | BL up vs DLBCL |
| hsa-miR-105* | 2.63 | BL up vs DLBCL |
| hsa-miR-124* | 2.64 | BL up vs DLBCL |
Abbreviations: ANOVA, analysis of variance; BL, Burkitt lymphoma; DLBCL, diffuse large B-cell lymphoma; FDR, false discovery rate; miRNA, micro RNA.
Validation of miRNAs differentially expressed between BL and DLBCL by real-time RT-PCR
In all, 11 miRNAs were chosen from the 38 miRNA signature differentiating between BL and DLBCL based on our microarray data for validation employing 22 BL and 31 DLBCL cases. As the results for hsa-miR-222 demonstrated reproducible signals in the no template control assay, this miRNA was omitted from further analysis. Thus, 10 miRNAs were used for the statistical calculations. The t-test confirmed that the eight hsa-miRs-23a, -26a, -29b, -30d, -146a, -146b-5p, -155 and -221 are statistically significantly higher expressed in DLBCL than in BL samples (Table 4). For hsa-miR-23b also an increased expression in DLBCL was calculated in consistency with the array data but statistical significance was not reached (P-value 0.07). The differential expression of hsa-miR-9 could not be confirmed by means of real-time RT-PCR.
Table 4.
Real-time RT-PCR-based validation of miRNAs differentially expressed between BL and DLBCL from microarray experiments
| Assay |
Fold change
BL/DLBCL |
P-value | Description |
Concordance with
microarray data |
|---|---|---|---|---|
| hsa-miR-155 | −27.99 | 0.0002 | BL down vs DLBCL | Yes |
| hsa-miR-146a | −11.12 | 0.0 | BL down vs DLBCL | Yes |
| hsa-miR-146b-5p | −5.42 | 0.0001 | BL down vs DLBCL | Yes |
| hsa-miR-221 | −4.34 | 0.0001 | BL down vs DLBCL | Yes |
| hsa-miR-30d | −4.32 | 0.0 | BL down vs DLBCL | Yes |
| hsa-miR-26a | −4.14 | 0.0 | BL down vs DLBCL | Yes |
| hsa-miR-23a | −3.97 | 0.0001 | BL down vs DLBCL | Yes |
| hsa-miR-29b | −3.13 | 0.0372 | BL down vs DLBCL | Yes |
| hsa-miR-9 | −3.04 | 0.0125 | BL down vs DLBCL | No |
| hsa-miR-23b | −2.17 | 0.0727 (NS) | BL down vs DLBCL | Yes |
Abbreviations: BL, Burkitt lymphoma; DLBCL, diffuse large B-cell lymphoma; miRNA, micro RNA; NS: not significant; RT, reverse transcriptase. Fold change and P-value were calculated by Data Assist Software based on 2^-dCt values using RNU48 as endogenous control.
Hierarchical clustering of BL and DLBCL samples
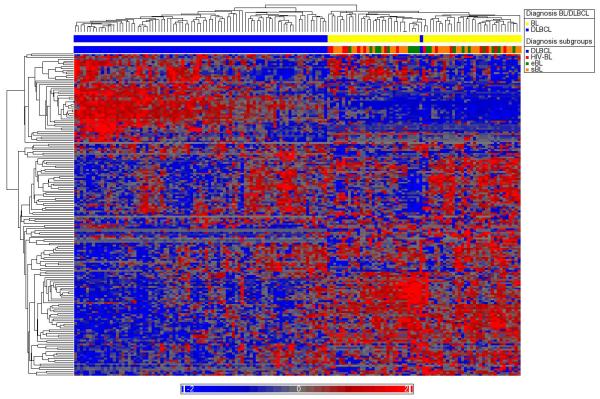
On the basis of the 38 miRNAs differentially expressed between BL and DLBCL, a hierarchical cluster analysis was performed (Figure 1). This revealed that the group of miRNA-defined BLs showed a homogeneous expression pattern with respect to the 38 miRNAs. Interestingly, two DLBCL cases also clustered within this group. MiRNA-defined DLBCLs (miR-DLBCL) were also homogeneous in their miRNA expression for the majority of cases. However, a group of 23 miR-DLBCL cases displayed overlapping miRNA expression features of miR-DLBCL and miRNA-defined BL. Four histological BL cases, all EBV-negative sBL, fell within this sub-cluster. Two of these BL cases showed a translocation of the MYC gene, whereas the other two were negative in this respect. The DLBCL cases present in this subgroup displayed no common characteristic regarding the MYC translocation or the GCB and non-GCB phenotype based on the immunohistochemical classifier developed by Hans et al.34 Furthermore, they could not be delineated from the remaining DLBCL by these means.
Figure 1.
Hierarchical clustering of all samples based on the 38 miRNAs differentially expressed between BL and DLBCL. MiRNA-defined BL denotes the miRNA-defined group of BL cases, miR-DLBCL the group of miR-DLBCL cases. The hatched portion of the miR-DLBCL bar indicates the cases with intermediate miRNA expression features. MYC translocation: results from fluorescence in situ hybridization analysis (NA, not evaluable because of low tissue quality; negative, no translocation detectable with the probes described in the method section; positive, MYC translocation to either of the IG loci detectable). Diagnosis: according to the WHO criteria as described in the Patients and methods section.
Differential miRNA expression patterns within the BL variants
Analysis of variance (false discovery rate (q-value) cut-off 0.05) was applied to detect differences in the miRNA expression between the three BL variants, that is, eBL, sBL and HIV-BL. There were either no or only minor miRNA expression differences between the BL variants. Merely six miRNAs showed marginal expression differences between eBL and sBL (Table 3). One of the differentially expressed miRNAs (miR-10b) is embedded in a HOX gene cluster44 and targets HOXD10.45 There were no statistically significant miRNA expression differences between the HIV-negative and the HIV-positive BL cases.
Table 3.
Differentially expressed miRNAs between eBL and sBL BL based on ANOVA analysis (FDR (q-value) <0.05)
| MiRNA name | Fold change eBL/sBL | Description |
|---|---|---|
| hsa-miR-191 | −2.53 | eBL down vs sBL |
| hsa-miR-374a | −1.95 | eBL down vs sBL |
| hsa-miR-193a-5p | −1.92 | eBL down vs sBL |
| hsa-miR-10b | 1.73 | eBL up vs sBL |
| hsa-miR-216b | 1.74 | eBL up vs sBL |
| hsa-miR-499-3p | 2.51 | eBL up vs sBL |
Abbreviations: ANOVA, analysis of variance; BL, Burkitt lymphoma; eBL, endemic BL; FDR, false discovery rate; miRNA, micro RNA; sBL, sporadic BL.
Comparison of miRNA expression in EBV-positive and -negative BL
There were no miRNA expression differences between EBV-positive and EBV-negative BL cases.
Discussion
Molecular profiling of neoplasms is gaining increasing significance for the definition and characterization of established tumor entities and for the identification of new biological disease groups or subgroups. One of the first highlights in this respect was the identification of GCB- and ABC-like subgroups within the DLBCL by expression profiling of mRNAs. These subgroups were not recognizable by standard diagnostic methods.46,47 In addition, by combining mRNA expression profiling and genetic analyses a biologic definition of BL was established that revealed a broader morphological spectrum, a consistent immunophenotype and lack of a MYC break in approximately 11% of the cases. Furthermore, cases with intermediate features between BL and DLBCL were identified.1,9,10
More recently miRNA genes were introduced into array expression profiling and this method was also efficiently employed to characterize and classify different types of cancer.21,22,26,48 For the classification of poorly differentiated tumors, it was even more informative than mRNA profiling.49 Furthermore, miRNA profiling can be applied to highly degraded RNA derived from FFPE tissue specimens because of the short length of the target molecules.
We have therefore used miRNA profiling to gain further insights into the molecular pathology of BL with respect to its distinction from DLBCL and with respect to the differences between its epidemiological variants (eBL, sBL and HIV-BL). In addition, the impact of EBV infection on miRNA expression in BL was addressed by comparing EBV-positive and -negative cases.
Unsupervised analysis by clustering the miRNA profiles of all cases revealed a clear distinction of BL from DLBCL cases but no further subdivision of BL cases was identifiable. In order to elucidate which miRNAs are responsible for the distinction between BL and DLBCL, the miRNA profiles of 64 BL were compared with those of 86 DLBCL samples by analysis of variance. Thirty-eight miRNAs were found to be differentially expressed between the two lymphoma types (Table 2). Of the miRNAs downregulated in the BL cases, miR-155, has previously been reported to be lower expressed in BL than in DLBCL,50,51 which is in keeping with our results.
Validation of our microarray data was performed by real-time RT-PCR analysis with 11 of the 38 miRNAs that differentiates BL and DLBCL using about one third of the cases (22 BL and 31 DLBCL). These 11 miRNAs (hsa-miRs-9, -23a, -23b, -26a, -29b, -30d, -146a, -146b-5p, -155, -221 and -222) were chosen because of their involvement in MYC or NF-kB signalling and their accordant importance for the biology of BL and DLBCL. Hsa-miR-222 showed reproducible signals in the no template control assay, thus the results of 10 miRNAs were eligible for statistical analysis. With the exception of hsa-miR-9, which displayed an inconsistent expression pattern among the BL and DLBCL cases analyzed, all other miRNAs examined by real-time RT-PCR were able to confirm the microarray data (Table 4).
MiR-155 and four further miRNAs (hsa-miRs-221, -222, -146a and -146b), which we found to be downregulated in our BL cases have also been shown to be lower expressed in the GCB-DLBCL subgroup when compared with the non-GCB-DLBCL subgroup.24-26 The GCB-DLBCL subgroup is known to differ from the non-GCB-DLBCL subgroup by a decreased activity of the NF-kB system, which is also a hallmark of BL.9,10,47 Interestingly, the expression of three of the miRNAs downregulated in BL (hsa-miRs-146a, -146b and -155) is induced via the NF-kB signalling.36,37 These findings point toward the NF-kB system as a major distinctive pathogenetic pathway between BL and DLBCL, including the GCB-DLBCL subgroup. Thus, our observations at the miRNA level are in agreement with the molecular definition of BL based on gene expression studies.9,10
The 38-miRNA signature seems also to be shaped by the influence of MYC. MYC is commonly upregulated in BL because of its chromosomal translocation to one of the immunoglobulin gene loci and is considered a key player of BL pathogenesis.52 It was shown that MYC is able to reduce as well as to induce the expression of miRNAs at a large scale.38,39,41,42 Six of the miRNAs, which have been identified to be repressed by MYC (hsa-miRs-23a, -23b, -26a, -29b, -30d and -146a) also show reduced levels in our BL cases. MiRNAs described to be transcriptionally activated by MYC in in vitro systems are the miR-17-92 cluster and hsa-miR-9. MiR-17-92 cluster is not part of the 38-miRNA signature that differentiates BL and DLBCL and differential expression of hsa-miR-9 could not be confirmed by real-time RT-PCR. The inability to identify miRNAs known to be activated by MYC can most likely be explained by the fact that a substantial number of DLBCL cases is also associated with elevated MYC protein expression and, potentially, with higher activity of MYC. Furthermore, it was shown that high expression levels of the miR-17-92 cluster are associated with amplification of the corresponding genomic locus.53,54 In fact it was demonstrated that 12.5% of GCB-like DLBCL harbor an amplification of C13orf25, the locus that encodes the miR-17-92 cluster.55 DLBCL may also feature alterations of the MYC locus like amplifications or translocations, which was true for four cases of our DLBCL series. These and other possibly MYC-independent mechanisms might be responsible for the activation of the miR-17-92 cluster in DLBCL cases and might thereby explain why the miR-17-92 cluster was not detected as differentially expressed between BL and DLBCL.
By comparing a small series of six high MYC-expressing BL samples with a variety of other, mostly MYC low expressing B-cell non-Hodgkin lymphomas (mantle cell lymphoma, follicular lymphoma and chronic lymphocytic leukemia) Robertus et al.54 identified a MYC-induced miRNA profile for BL. Several of these MYC-associated miRNAs are also found in our BL signature (hsa-miRs-23a, -23b, -26a, -29b and -146a). However, since in our study BL was differentiated from DLBCL that, like BL, express MYC at quite high levels in about ¼ of cases, the influence of MYC in our BL defining miRNA signature is much less evident. Instead other deregulated gene systems like NF-kB also have a role.
Clustering of all DLBCL and BL cases showed that the 38-miRNA signature is able to separate BL from DLBCL with only few exceptions (Figure 1). In addition, a sub-population of cases within the miR-DLBCL cluster exhibits an expression pattern of the 38 miRNAs with overlapping features between the miR-DLBCL and the miRNA-defined BL group, thus challenging their unequivocal definition as miR-DLBCL. Interestingly, four BL cases were grouped into this miR-DLBCL subgroup. Thus, this subgroup of cases is reminiscent of the cases intermediate between BL and DLBCL as detected by mRNA expression profiling.9
In respect to the BL variants, the results derived from the supervised statistical approach argue for only minor molecular differences as (i) only six miRNAs were found to be differentially expressed between eBL and sBL (Table 3) and (ii) their fold changes were much lower as compared with DLBCL and BL. Neither HIV nor EBV infection displayed a significant impact on the miRNA profile in our analysis. Although EBV infection occurs in the BL tumor cells, the restriction to a latency 1 infection pattern might prevent a significant modification of the miRNA profile. However, we cannot completely exclude that latency 1 EBV infection in BL might interfere with some coding mRNAs. Our miRNA-based findings were most recently supported by a parallel study of Piccaluga et al.56 in which the coding mRNA expression profiling also demonstrated a broad similarity between the epidemiologic BL subtypes.
In summary, our analysis reveals that BL differs from DLBCL by a strong and characteristic miRNA signature that is enriched in miRNAs targeted by MYC and in NF-kB pathway-associated miRNAs. In contrast, the small differences in the miRNA transcriptome between sBL and eBL and the absence of differences between the former and HIV-BL underscore the view that the three BL variants are not distinctive subtypes but closely related representatives of the same biological entity.
Supplementary Material
Acknowledgements
We would like to thank the German High Grade Lymphoma Study Group (DSHNL) for supporting this study and the pathologists of the German Reference Centers for Lymph Node Pathology for the panel review of the DLBCL included in the RiCOVER-60 study. We are also grateful to Korinna Jöhrens for evaluating the BL samples for generation of the tissue microarrays and Christoph Loddenkemper for evaluating immunohistochemical stainings. Furthermore, we thank E Berg, H Lammert and S Meier for excellent technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) for the Transregio 54 (TRR54) the Fondazione Monte dei Paschi (MPS), the Kinderkrebsinitiative Buchholz / Holm-Seppensen and the Deutsche Krebshilfe.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author contributions
DL: analyzed and interpreted data, designed research, wrote the paper; LL: initiated and planned the study, designed research, interpreted data, wrote the paper; MH: interpreted data, designed research, wrote the paper; SV: performed statistical analysis; HH, CGL, JP, TA, GDF: performed research; JG, JN, ER, MP: contributed vital new reagents; GO, AR, RS: analyzed and interpreted data; CC: contributed analytical tools; HS: initiated the study, designed research, interpreted data, wrote the paper.
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.WHO classification of tumours of haematopoietic and lymphoid tissues. 4th edn. IARC; Lyon: 2008. [PubMed] [Google Scholar]
- 2.Stein H, Gerdes J, Mason DY. The normal and malignant germinal centre. Clin Haematol. 1982;11:531–559. [PubMed] [Google Scholar]
- 3.Tamaru J, Hummel M, Marafioti T, Kalvelage B, Leoncini L, Minacci C, et al. Burkitt’s lymphomas express VH genes with a moderate number of antigen-selected somatic mutations. Am J Pathol. 1995;147:1398–1407. [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman CJ, Mockridge CI, Rowe M, Rickinson AB, Stevenson FK. Analysis of VH genes used by neoplastic B cells in endemic Burkitt’s lymphoma shows somatic hypermutation and intraclonal heterogeneity. Blood. 1995;85:2176–2181. [PubMed] [Google Scholar]
- 5.Shiramizu B, Barriga F, Neequaye J, Jafri A, Dalla-Favera R, Neri A, et al. Patterns of chromosomal breakpoint locations in Burkitt’s lymphoma: relevance to geography and Epstein-Barr virus association. Blood. 1991;77:1516–1526. [PubMed] [Google Scholar]
- 6.van den Bosch CA. Is endemic Burkitt’s lymphoma an alliance between three infections and a tumour promoter? Lancet Oncol. 2004;5:738–746. doi: 10.1016/S1470-2045(04)01650-X. [DOI] [PubMed] [Google Scholar]
- 7.Pelicci PG, Knowles DM, Magrath I, Dalla-Favera R. Chromosomal breakpoints and structural alterations of the c-myc locus differ in endemic and sporadic forms of Burkitt lymphoma. Proc Natl Acad Sci USA. 1986;83:2984–2988. doi: 10.1073/pnas.83.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellan C, Lazzi S, Hummel M, Palummo N, de Santi M, Amato T, et al. Immunoglobulin gene analysis reveals two distinct cells of origin for EBV positive and EBV negative Burkitt’s lymphomas. Blood. 2005:106–1031. doi: 10.1182/blood-2005-01-0168. [DOI] [PubMed] [Google Scholar]
- 9.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 10.Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 11.Lee RC, Feinbaum RL, Ambros V. The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 12.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 16.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 17.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 19.O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 22.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 25.Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 27.Pfreundschuh M, Schubert J, Ziepert M, Schmits R, Mohren M, Lengfelder E, et al. Six versus eight cycles of bi-weekly CHOP-14 with or without rituximab in elderly patients with aggressive CD20+ B-cell lymphomas: a randomised controlled trial (RICOVER-60) Lancet Oncol. 2008;9:105–116. doi: 10.1016/S1470-2045(08)70002-0. [DOI] [PubMed] [Google Scholar]
- 28.Lazzi S, Ferrari F, Nyongo A, Palummo N, de Milito A, Zazzi M, et al. HIV-associated malignant lymphomas in Kenya (Equatorial Africa) Hum Pathol. 1998;29:1285–1289. doi: 10.1016/s0046-8177(98)90258-1. [DOI] [PubMed] [Google Scholar]
- 29.Noske A, Denkert C, Schober H, Sers C, Zhumabayeva B, Weichert W, et al. Loss of Gelsolin expression in human ovarian carcinomas. Eur J Cancer. 2005;41:461–469. doi: 10.1016/j.ejca.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 30.Ventura RA, Martin-Subero JI, Jones M, McParland J, Gesk S, Mason DY, et al. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn. 2006;8:141–151. doi: 10.2353/jmoldx.2006.050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hummel M, Anagnostopoulos I, Dallenbach F, Korbjuhn P, Dimmler C, Stein H. EBV infection patterns in Hodgkin’s disease and normal lymphoid tissue: expression and cellular localization of EBV gene products. Br J Haematol. 1992;82:689–694. doi: 10.1111/j.1365-2141.1992.tb06945.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3:563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 33.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 35.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M, et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–5546. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 36.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kluiver J, van den BA, de Jong D, Blokzijl T, Harms G, Bouwman E, et al. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 38.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TFE, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 41.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 42.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang CH, Lee DY, Deng Z, Jeyapalan Z, Lee SC, Kahai S, et al. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS One. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 46.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 47.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 48.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 49.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 50.Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ, et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 51.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 52.Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol. 2010;149:484–497. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- 53.Tagawa H, Karube K, Tsuzuki S, Ohshima K, Seto M. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci. 2007;98:1482–1490. doi: 10.1111/j.1349-7006.2007.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robertus JL, Kluiver J, Weggemans C, Harms G, Reijmers RM, Swart Y, et al. MiRNA profiling in B non-Hodgkin lymphoma: a MYC-related miRNA profile characterizes Burkitt lymphoma. Br J Haematol. 2010;149:896–899. doi: 10.1111/j.1365-2141.2010.08111.x. [DOI] [PubMed] [Google Scholar]
- 55.Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piccaluga PP, De Falco G, Kustagi M, Gazzola A, Agostinelli C, Tripodo C, et al. Gene expression analysis uncovers similarity and differences among Burkitt lymphoma subtypes. Blood. 2011;117:3596–3608. doi: 10.1182/blood-2010-08-301556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.