Summary
An often-overlooked aspect of neural plasticity is the plasticity of neuronal composition, in which the numbers of neurons of particular classes are altered in response to environment and experience. The Drosophila brain features several well-characterized lineages in which a single neuroblast gives rise to multiple neuronal classes in a stereotyped sequence during development [1]. We find that in the intrinsic mushroom body neuron lineage, the numbers for each class are highly plastic, depending on the timing of temporal fate transitions and the rate of neuroblast proliferation. For example, mushroom body neuroblast cycling can continue under starvation conditions, uncoupled from temporal fate transitions that depend on extrinsic cues reflecting organismal growth and development. In contrast, the proliferation rates of antennal lobe lineages are closely associated with organismal development, and their temporal fate changes appear to be cell cycle-dependent, such that the same numbers and types of uniglomerular projection neurons innervate the antennal lobe following various perturbations. We propose that this surprising difference in plasticity for these brain lineages is adaptive, given their respective roles as parallel processors vs. discrete carriers of olfactory information.
Results and Discussion
Mushroom body temporal class populations are highly plastic
Developmental plasticity permits individual organisms to alter their phenotypes in response to the particular challenges posed by their environments. Holometabolous insects have several larval instars specialized for feeding and growth and an adult stage specialized for dispersal, mating, and egg deposition. When limiting nutrients slow larval growth and delay development, we might expect the nervous system to adapt in order for precious food sources to be located and recalled more efficiently.
The mushroom body (MB) is an arthropod brain structure shown to be critical for olfactory learning and memory [2]. The adult Drosophila MBs each consist of about 2500 intrinsic neurons that contribute axons to one or more of five lobes (γ, α, α′, β, and β′). Each MB neuroblast (NB) divides repeatedly to generate γ neurons during embryogenesis and early larval development, α′/β′ neurons during late larval development, and α/β neurons (including a pioneer subclass) during metamorphosis [3].
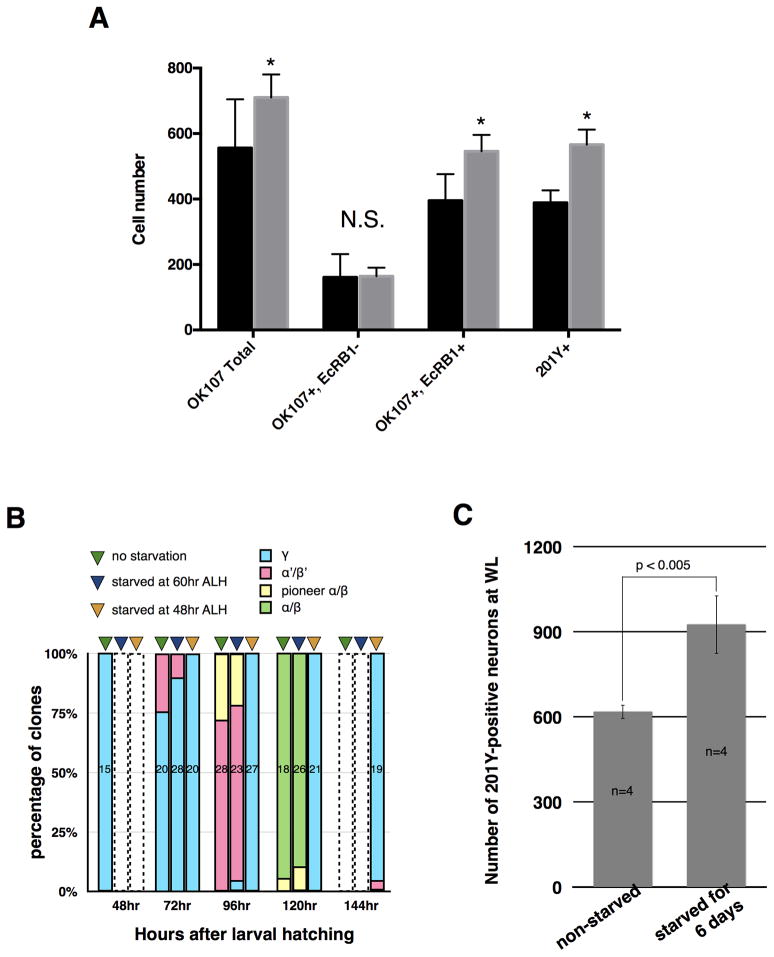
Experience-dependent plasticity has been demonstrated for this structure in many studies, including one in which the numbers of MB fibers increased in female Drosophila cultured at high density as larvae [4]. Moreover, culturing newly hatched larvae in 20% sucrose solution prevents the reactivation of dormant NBs, yet the actively dividing MB NBs continue to proliferate [5–7]. We therefore hypothesized that delaying organismal growth and development via nutrient deprivation might allow the MB NBs to generate greater numbers of neurons. To test this idea, we “protein-starved” newly hatched larvae by transferring them to 20% sucrose solution for 48 hours before placing them on standard fly medium. Protein-starved animals delayed pupariation by ~3 days and produced significantly more MB neurons (710 ± 71, n = 9) by P0 (white puparium stage) than normally fed controls (556 ± 149 n = 12; p < 0.01), based on expression of a UAS-CD8::GFP reporter by OK107-GAL4 [8](Fig. 1A). This increase was due to the expansion of the early born γ class, which is specifically labeled by both monoclonal antibody AD4.4 (anti-EcRB1) [9](546 ± 50, n = 9 vs. 395 ± 81, n = 12; p<1×10−4) and 201Y-GAL4 [10](566 ± 46, n = 13 vs. 388 ± 38, n = 14; p<1×10−10) (Fig. 1A). Thus, the size of the γ neuron population is influenced by nutrient availability during larval life.
Figure 1. Mushroom body γ neuron population size is highly plastic.
(A) Protein starvation of newly hatched larvae results in extra MB neurons (OK107+, p<0.01), which are of the γ class (EcRB1+, p<1×10−4 and 201Y+, p<1×10−10), by P0. No significant difference in other (EcRB1−) classes was observed. Control: Animals of genotype OK107-GAL4 or 201Y-GAL4; UAS-CD8::GFP raised on standard food. Sucrose: Animals of genotype OK107-GAL4 or 201Y-GAL4; UAS-CD8::GFP protein-starved in 20% sucrose solution for 48 hrs ALH then transferred to standard food. Standard error bars are shown, with Student’s t-test used to establish significance.
(B) Percentage of clones induced at different developmental times (48, 72, 96, 120, and 144 hrs ALH) for each type of MB neuron. The three bars represent non-starved larvae (green triangles) and larvae starved from 48 hrs (orange triangles) or 60 hrs (blue triangles) ALH. The colored areas in each bar represent the percentage of clones for γ (blue), α′/β′ (magenta), pioneer α/β (yellow), and α/β (green) neurons, while the number indicates how many clones were analyzed for each treatment. Open bars with dashed outlines: not assayed.
(C) Number of 201Y-GAL4-positive (γ) neurons at wandering third instar for control larvae and larvae protein-starved from 48 hrs ALH for six days before being transferred back to food.
The transition from γ to α′/β′ production normally occurs around mid-third instar [3], as does the attainment of critical weight, the point at which Drosophila larvae will proceed to pupariation even if protein-starved [11]. By using the MARCM technique [12] to label the MB neurons born at particular timepoints, we found that while α′/β′ neurons are normally generated by 72 hrs after larval hatching (ALH), protein starvation starting at 48 hrs ALH delayed this transition, allowing production of γ neurons through 144 hrs ALH (Fig. 1B). These protein-starved animals never pupariated, indicating that they had failed to attain critical weight (Fig. S1). However, if such larvae were replaced on normal medium after six days, they resumed growth and pupariated three days later, at which time they had roughly 50% more γ (201Y+) neurons than normally fed controls (Fig. 1C). In contrast, protein starvation initiated at 60 hrs ALH neither prevented pupariation (Fig. S1) nor delayed the transition to α′/β′ production (Fig. 1B). These results suggest that the plasticity of the γ population ends between 48 and 60 hrs ALH, perhaps due to triggering of the transition to the next temporal class by hormonal signals associated with the attainment of critical weight.
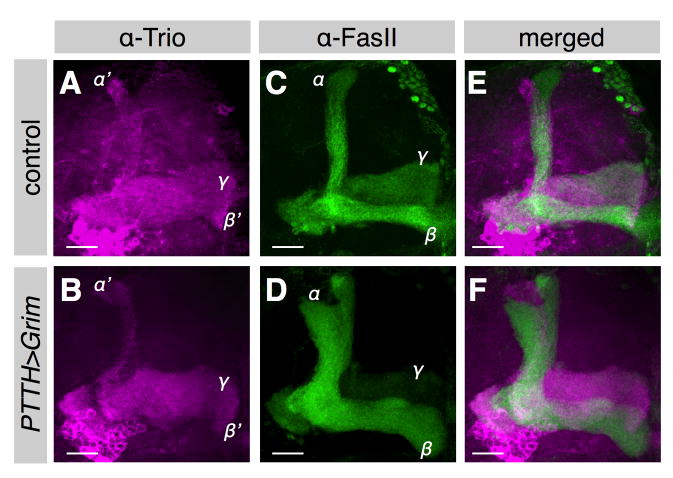
To determine whether the sizes of the later born MB populations were also developmentally plastic, we genetically ablated the neurons that innervate the ring gland and release prothoracicotropic hormone (PTTH), which normally drives the prepupal ecdysone peak that induces puparium formation. PTTH neuron ablation delayed puparium formation by about five days (Fig. S2), as previously reported [13]. We found that the mature MBs of these flies had γ and α′/β′ axonal bundles (labeled by anti-Trio antibody [14]) that were not significantly different in size from those of controls, but their α/β axonal bundles were much thicker (Fig. 2A–F), indicating significant expansion of that population. Moreover, the α/β lobes were already enlarged by 24 hrs after puparium formation (APF) (Fig. S2A,B). Since their NB proliferation rate during early metamorphosis was similar to that of wild type flies (Fig. S2C), we concluded that the excess α/β neurons were produced during the prolonged late larval period. Therefore, the sizes of both γ and α/β neuron populations are highly plastic and influenced by organismal growth and development. In addition, our results suggest that the onset of α/β neuron production is not triggered by puparium formation, although they normally coincide.
Figure 2. Mushroom body α/β neuron population size is highly plastic.
(A–F) The MBs of a wild-type fly (A,C,E) and a fly in which the PTTH neurons had been ablated by overexpressing the death gene grim using PTTH-GAL4 (B,D,F). The MBs were co-labeled with anti-Trio (magenta, γ and α′/β′ neurons) (A,B,E,F) and anti-FasII (green, γ and α/β neurons) (C,D,E,F). Scale bars: 20 μm.
Mushroom body temporal identity transitions are regulated by overall growth, not an internal counting mechanism
What allows the composition of the mushroom body lineage to vary so dramatically? The results of the protein starvation experiments imply that the transition from γ to α′/β′ production depends on an extrinsic cue linked to organismal growth and development rather than on a MB-intrinsic cue that “counts” the number of neuroblast divisions or progeny. Given the increase in γ neurons in the sucrose-cultured animals, we wondered if insulin signaling could be regulating the proliferation rate of MB neuroblasts and/or the temporal transitions of MB neurons.
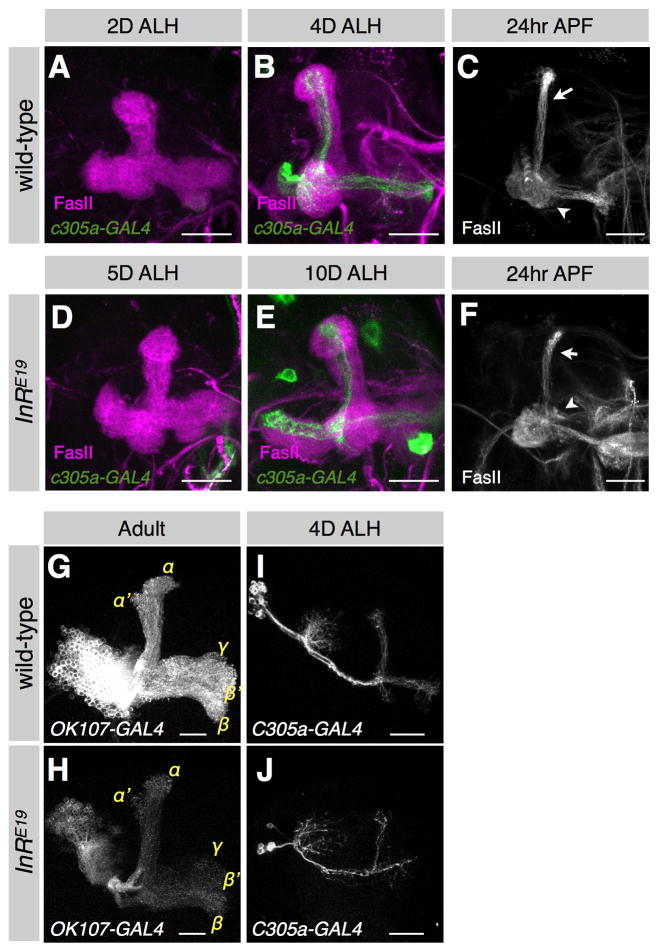
We first followed MB development in flies homozygous for a hypomorphic insulin receptor allele, InRE19, previously shown to control body and organ size [15]. Pupariation was delayed by about 6 days in InRE19 homozygotes, during which time the MB NBs continued to divide, albeit more slowly (Fig. S3A). In wild-type larvae, strong c305a-GAL4 expression identifying α′/β′ neurons [16] was observed at 4 days ALH (Fig. 3A,B). In InRE19 homozygotes, c305a-GAL4 was barely detectable at 5 days ALH (Fig. 3D), and strong expression comparable to that of 4-day-old wild-type larvae was not observed until 10 days ALH (Fig. 3E). Therefore, the γ to α′/β′ transition was significantly delayed in the slow-growing InRE19 animals. However, when we examined the MBs at 24 hrs APF (Fig. 3C,F), the FasII-positive nascent α/β neurons were already visible, suggesting that the transition from α′/β′ to α/β had still occurred around the time of pupariation. Thus, in InRE19 animals, MB NB division rates are reduced but temporal identity transitions are delayed, potentially leaving the final number of γ neurons unchanged. This is supported by the comparable MB sizes in wild-type vs. InRE19 animals just after the γ to α′/β′ transition (Fig. 3B,E).
Figure 3. MB temporal transitions are regulated by overall growth, not an internal counting mechanism.
(A–F) The γ to α′/β′ transition is delayed in insulin receptor mutant animals. The MBs of wild-type (A,B) and InRE19 homozygous (D,E) animals at specific days after larval hatching (ALH). The α′/β′ neurons were labeled by c305a-GAL4 (green), and the γ neurons were immunostained with anti-FasII Ab (magenta). Scale bars: 20 μm. The MBs of wild-type (C) and InRE19 homozygous (F) animals at 24 hrs after puparium formation (APF). The partially pruned γ neurons (arrowheads) and the nascent α/β neurons (arrows) were immunostained with anti-FasII Ab (gray). Scale bars: 20 μm. (G–J) InRE19 homozygous mushroom body clones induced in newly hatched larvae are smaller than wild-type clones but still feature all neuron classes. A wild-type (G) and InRE19 homozygous (H) MB clone labeled by OK107-GAL4 in adult. The α′/β′ neurons labeled by c305a-GAL4 in a wild-type (I) and InRE19 (J) mushroom body clone at 4 days ALH. Scale bars: 20 μm.
Next we sought to determine whether the effects of the insulin receptor on MB neuroblast proliferation and cell type progression were cell-autonomous by creating MB clones homozygous for InRE19 using the MARCM method [12]. Although heterozygous larvae exhibited no obvious growth delay, the InRE19 MB clones contained only 250 ± 69 (n = 4) cells, 35% less than wild-type MB clones (384 ± 38, n = 4) (Fig. 3G,H). These InRE19 MB clones still had all axon lobes in normal proportions, indicating that insulin reception cell-autonomously regulates the proliferation of the MB NBs, but not temporal identity switching. Moreover, we still detected strong expression of the α′/β′ marker c305a-GAL4 in the InRE19 MB clones by 4 days ALH (Fig. 3I,J). Therefore, the delayed γ to α′/β′ transition observed in InRE19 mutant larvae is not due to deficits in MB insulin reception but rather to the delay in organismal growth and development.
We also tested the dependence of the α′/β′ to α/β transition on insulin signaling by knocking down InR specifically in the MB using InR miRNA driven by OK107-GAL4. Consistent with the phenotype of the InRE19 MB clones, the adult OK107>InRi MBs were proportionally smaller than wild-type MBs (Fig. S3B,C). Since we could readily detect the FasII+ nascent α/β bundles at 24 hrs APF (Fig. S3D,E), the α′/β′ to α/β transition was not affected by the cell-autonomous reduction in InR signaling despite the reduction in neuroblast cell cycling. We also overexpressed InR in the MB and found that MB neurons increased by 33% (wild-type: 2598 ± 576, n = 7; OK107>InR MB: 3462 ± 590, n = 5; p<0.05). Interestingly, we found a disproportional enlargement of the γ lobe (Fig. S3F–I), suggesting that InR overexpression caused enhanced cycling in the early larval instars, when proliferation rates are normally low [5]. These results support a tissue-specific role for insulin signaling in the proliferation rate of MB NBs but not in the timing of cell type transitions.
Finally, we examined mutants in slender lobe (sle), which encodes a nuclear protein that positively regulates MB NB proliferation rate [17]. We found that adult sle MBs were smaller than wild-type MBs (as previously reported) and that the population sizes of all neuronal classes had been reduced. The FasII-positive γ and α/β axon lobes [18] were thinner (Fig. S3J,K), while the number of α′/β′ (c305a+) neurons decreased dramatically (Fig. S3L,M), as did the number of pioneer α/β neurons, labeled by c708a-GAL4 [19] (Fig. S3N,O). In addition, the α/β axonal bundles were already clearly visible at 24 hrs APF (Fig. S3P,Q), indicating that the pioneer α/β to α/β transition had not been significantly delayed. These sle data confirm that MB temporal transitions do not depend on the number of NB cell cycle divisions or progeny, but rather on external cues linked to organismal growth and development.
Nutrient deprivation does not alter final neuronal composition of the antennal lobe lineages
Having established that the composition of the MB neuroblast lineage is developmentally plastic for all four identified temporal classes, we next tested whether this finding could be generalized to two other well-characterized neuroblast lineages in the olfactory system. The anterodorsal projection neuron (adPN) and lateral antennal lobe (lAL) lineages both generate uniglomerular projection neurons (PNs) that innervate single glomeruli in the antennal lobe (AL) and relay sensory neuron activity patterns to higher brain centers. The adPN lineage is comprised of 40 classes of Acj6-expressing PNs, 35 of which are uniglomerular PNs that are generated in a stereotyped sequence during development [20–24]. The lAL NB is mitotically active at hatching and produces 12 classes of Acj6- uniglomerular PNs, paired with Acj6- multiglomerular local interneuron (LN) siblings and temporally interspersed with Acj6+ neurons that do not innervate the AL [24,25]. All uniglomerular lAL PNs and the majority of uniglomerular adPNs can also be labeled using the driver GH146-GAL4 [26].
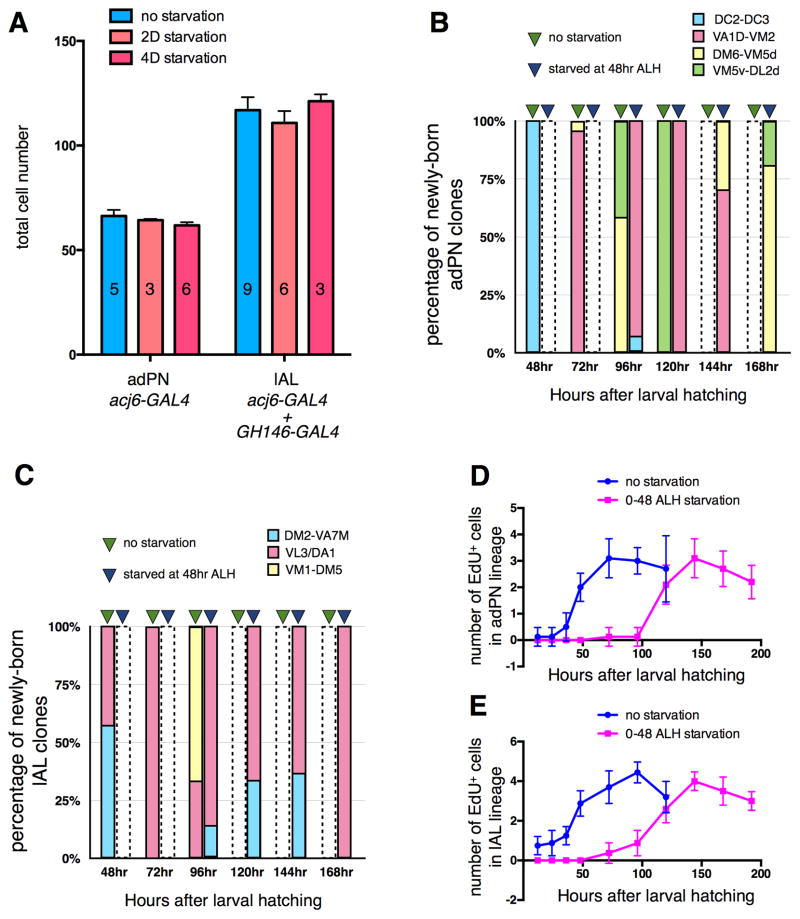
We examined the plasticity of these two AL lineages using many of the same manipulations shown to alter the neuronal composition of the MB. We first used the MARCM method with either acj6-GAL4 or GH146-GAL4 to label the postembryonic antennal lobe lineages by heat-shocking newly hatched larvae. Protein-starving these larvae for 48 hours and then replacing them on normal food changed neither the final numbers of Acj6+ or GH146+ cells in neuroblast clones (Fig. 4A) nor the patterns of glomerular innervation (Fig. S4A–D), implying that extra PNs were not generated by delaying early larval development.
Figure 4. Protein starvation delays temporal identity transitions without altering final neuronal composition of the antennal lobe lineages.
(A) Comparison of adult antennal lobes from fed control larvae to those of protein-starved larvae. 2D starvation: larvae were protein-starved from 0–48 hrs ALH then replaced on normal food. 4D starvation: larvae were starved from 48 hrs ALH for 4 days then replaced on normal food. adPN NB clones were labeled by acj6-GAL4, and lAL NB clones were labeled by acj6-GAL4 and GH146-GAL4.
(B,C) The uniglomerular PN classes labeled by heat shock at different developmental timepoints (48, 72, 96, 120, 144 and 168 hrs ALH). The two bars at each timepoint represent fed control larvae (green triangles) and larvae starved from 48 hrs ALH (blue triangles). The colored areas in each bar represent the percentage of clones that belonged to particular temporal classes: DC2-DC3 (blue), VA1d-VM2 (magenta), DM6-VM5d (yellow) and VM5v-DL2d (green) neurons in adPN (B), and DM2-VA7m (blue), VL3/DA1 (magenta) and VM1-DM5 (yellow) neurons in lAL (C). Open bars with dashed outlines: not assayed.
(D,E) Proliferation speeds of adPN (D) and lAL (E) NBs for fed controls (blue line) and animals protein-starved from 0–48 hrs ALH (magenta line). Brains were dissected at different developmental timepoints and incubated in S2 medium containing EdU (100 μg/ml) for 2 hrs at 25°C before fixation.
Similarly, when these larvae were transferred to sucrose solution at 48 hrs ALH for four days, then replaced on normal medium, there was no difference in total neuron number in adulthood for either lineage as compared to normally fed controls (Fig. 4A). Moreover, all appropriate glomeruli were innervated in the adult AL (Fig. S4E,F), implying production of even the latest born uniglomerular PN classes. Thus, unlike in the MB, the AL lineages did not produce additional neurons of early born classes when development was delayed due to nutrient deprivation. Final AL lineage composition was not affected in developmentally delayed InRE19 or PTTH neuron-ablated animals, or by lineage-autonomous loss or gain of insulin receptor function (data not shown).
We propose that these differences in plasticity make sense, given the roles of MB neurons vs. AL uniglomerular projection neurons. Because the MB uses parallel processing by many neurons of each class, an increase or decrease in number would not be expected to eliminate function. It is even possible that this plasticity is adaptive: low nutrient conditions result in a bigger MB, conferring better olfactory memory when food sources are scarce, while investment in fewer neurons will suffice when food sources are plentiful. In contrast, olfactory discrimination depends on a large diversity of uniglomerular PN types in the AL to relay signals from the reception of many different possible odorants, so plasticity - particularly the loss of specific neuron classes - would be expected to have a detrimental effect on function. An analogous preservation of neuronal diversity during nutrient restriction was recently reported for the Drosophila visual system [27].
Nutrient deprivation delays both neuroblast proliferation and temporal fate transitions in the antennal lobe lineages
The lack of expansion of the AL lineages under our protein starvation protocol made us wonder whether temporal transitions were still occurring normally in the developmentally delayed larvae. We used the MARCM method with acj6-GAL4 or GH146-GAL4 to label newly born adPN and lAL neurons. Control animals generated single cell adPN clones of the expected uniglomerular classes when heat-shocked during specific windows. To test the effects of protein starvation, at 48 hrs ALH we transferred larvae to sucrose solution and cultured them for various lengths of time before returning them briefly to normal food and heat-shocking them to induce clones. The PN classes labeled in the adult AL indicated that temporal fate transitions in these protein-starved larvae had been delayed by roughly three days in the adPN lineage (Fig. 4B) and by even longer in the lAL lineage (Fig. 4C).
To explain these apparently contradictory results, we hypothesized that protein starvation reduced neuroblast proliferation rates in the AL lineages as well as delaying temporal transitions. EdU labeling showed that transient protein starvation of newly hatched larvae delayed the onset of adPN neuroblast proliferation by about three days (Fig. 4D) and halted division of the lAL neuroblast until 24 hrs after the larvae were given normal food again (Fig. 4E). Interestingly, the lAL and MB NBs are the only NBs that are actively cycling at hatching [5,7], but unlike the lAL NB, the MB NBs continue to divide during early protein starvation [6]. This differential response to protein starvation suggests that the respective degrees of plasticity in the MB vs. AL lineages may be adaptive responses to the larval environment, rather than resulting merely from an accidental difference in cell division dynamics.
Taken together, these results suggest that both cell cycle and temporal identity transitions in the AL lineages are dynamically regulated in accordance with organismal development. They are comparably slowed down and delayed upon prolongation of larval development by nutrient deprivation (Fig. 4B–E) or other manipulations including PTTH neuron ablation (data not shown), ultimately generating lineages of unaltered neuronal composition. This further implies that the temporal identity changes in the AL NB lineages are likely to be cell cycle-dependent.
Conclusions
We have found that environmental factors, including nutrition, sculpt different lineages of the Drosophila olfactory system to widely varying degrees. The intrinsic MB lineage is highly plastic, with numbers of each class changing in response to growth conditions and neuroblast proliferation rates. In contrast, the adPN and lAL lineages consist of an invariant number of uniglomerular neurons following similar manipulations, suggesting that both neuroblast proliferation rates and temporal transitions are altered in accordance with organismal development. We propose these differences in plasticity of composition to be adaptive, ensuring a functional olfactory system under a wide variety of conditions, yet enhancing olfactory learning and memory in a resource-scarce environment.
Supplementary Material
Highlights.
Mushroom body sublineages are plastic and expand with developmental delay
Mushroom body lineage cell cycle and temporal fate progression can be uncoupled
Antennal lobe lineages are resistant to alteration by developmental delay
Antennal lobe lineage and organismal developmental dynamics are tightly coupled
Acknowledgments
We thank Chihiro Hama at Kyoto Sangyo University for sle[057], Ulrike Heberlein at HHMI Janelia Farm Research Campus for the UAS-InR flies, Julie Simpson at HHMI Janelia Farm Research Campus for the nSyb-GAL4 flies, Carl Thummel at the University of Utah for the AD4.4 antibody, and members of the Marin and Lee groups for thoughtful comments on the manuscript. JWT was funded by the Howard Hughes Medical Institute and a grant from the National Institutes of Health [NS13079]. TL was funded by the Howard Hughes Medical Institute and a grant from the National Institutes of Health [MH080739]. The authors declare that they have no competing interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kao CF, Lee T. Birth time/order-dependent neuron type specification. Curr Opin Neurobiol. 2010;1:14–21. doi: 10.1016/j.conb.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 3.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;18:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 4.Heisenberg M, Heusipp M, Wanke C. Structural plasticity in the Drosophila brain. J Neurosci. 1995;3(Pt 1):1951–1960. doi: 10.1523/JNEUROSCI.15-03-01951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;1:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 6.Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;11:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- 7.Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;1:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- 8.Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O’Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;5295:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- 9.Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;3:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- 10.Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;1:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- 11.Mirth CK, Riddiford LM. Size assessment and growth control: how adult size is determined in insects. Bioessays. 2007;4:344–355. doi: 10.1002/bies.20552. [DOI] [PubMed] [Google Scholar]
- 12.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;3:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 13.McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, Thummel CS, Dauphin-Villemant C, Gilbert LI, O’Connor MB. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell. 2007;6:857–871. doi: 10.1016/j.devcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, Hama C. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron. 2000;1:119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- 15.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;4:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 16.Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron. 2007;1:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orihara-Ono M, Suzuki E, Saito M, Yoda Y, Aigaki T, Hama C. The slender lobes gene, identified by retarded mushroom body development, is required for proper nucleolar organization in Drosophila. Dev Biol. 2005;1:121–133. doi: 10.1016/j.ydbio.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn Mem. 1998;1–2:38–51. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;2:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 20.Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;6860:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- 21.Yu HH, Kao CF, He Y, Ding P, Kao JC, Lee T. A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM. PLoS Biol. 2010;8:e1000461. doi: 10.1371/journal.pbio.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komiyama T, Johnson WA, Luo L, Jefferis GS. From lineage to wiring specificity. POU domain transcription factors control precise connections of Drosophila olfactory projection neurons. Cell. 2003;2:157–167. doi: 10.1016/s0092-8674(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 23.Marin EC, Watts RJ, Tanaka NK, Ito K, Luo L. Developmentally programmed remodeling of the Drosophila olfactory circuit. Development. 2005;4:725–737. doi: 10.1242/dev.01614. [DOI] [PubMed] [Google Scholar]
- 24.Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;17:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- 25.Lin S, Kao CF, Yu HH, Huang Y, Lee T. Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions. PLoS Biol. 2012;11:e1001425. doi: 10.1371/journal.pbio.1001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;5:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 27.Lanet E, Gould AP, Maurange C. Protection of Neuronal Diversity at the Expense of Neuronal Numbers during Nutrient Restriction in the Drosophila Visual System. Cell Rep. 2013;3:587–594. doi: 10.1016/j.celrep.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.