Abstract
The DNA-binding protein recombination signal binding protein-Jκ (RBP-J) mediates transcriptional activation of the Notch intracellular domain (NIC). In the absence of transcriptional activators, RBP-J suppresses transcription by recruiting co-suppressors. KyoT2 is a LIM domain protein that inhibits the RBP-J-mediated transcriptional activation. Here we provide evidence that the polycomb group protein RING1 interacts with the LIM domains of KyoT2 in yeast and mammalian cells. The interaction between KyoT2 and RING1 was detected both in vitro and in vivo. By using a co-immunoprecipitation assay, we also showed that, though RING1 and RBP-J did not associate directly, the two molecules could be co-precipitated simultaneously by KyoT2, probably through the LIM domains and the RBP-J-binding motif of KyoT2, respectively. These results suggested the formation of a three-molecule complex consisting of RBP-J, KyoT2 and RING1 in cells. Moreover, we found that overexpression of RING1 together with KyoT2 in cells inhibited transactivation of RBP-J by NIC. Suppression of the NIC- mediated transactivation of RBP-J by RING1 was abrogated by overexpression of KBP1, a molecule that competed with RING1 for binding to LIM domains of KyoT2, suggesting that suppression of RBP-J by RING1 was dependent on its associating with KyoT2. Taken together, our data suggested that there might be at least two ways of the KyoT2-mediated suppression of RBP-J, namely competition for binding sites with transactivators, and recruitment of suppressors such as RING1.
INTRODUCTION
The Notch signaling pathway plays a pivotal role in cell fate specification in development and is conserved during evolution from worm through to man (1,2). In Drosophila, the loss of function mutation of the Notch receptor or other essential molecules of the Notch signaling pathway during early embryogenesis leads to abnormal cell fate determination in numerous differentiation steps (3). In mammals, the Notch signaling pathway is also critical for embryonic development, as targeted disruption of Notch1 or its downstream transcription factor RBP-J (recombination signal binding protein-Jκ) in mice results in disorganization of somites and delayed closure of the neurotube, which finally leads to early embryonic lethality (4–6). More recently, conditional gene knock-out mice of Notch1 and RBP-J have been constructed by Radtke et al. (7) and Han et al. (8), respectively. Phenotype analyses of these mice have demonstrated that disruption of Notch1 or RBP-J in adult mice caused abnormal differentiation of common lymphoid progenitors, resulting in blockade of T cell fate determination and induction of B cell differentiation in the thymus where normally most T lymphocytes but few B lymphocytes are generated (7,8). Therefore, the Notch signaling pathway also plays a critical role in the regulation of cell differentiation in adults.
Notch is a family of type I transmembrane protein that includes four members in mouse and man (Notch1–4) (1). DNA binding protein RBP-J is the main signaling adaptor in nucleus for all the four kinds of Notch receptors (9–11). When Notch is triggered by direct interaction with its ligands, the Delta family proteins expressed on neighboring cells, the intracellular domain of the Notch receptor (NIC) is released from the membrane after receptor cleavage executed by a γ-secretase-like protease (12–14). NIC translocates to nucleus and associates with RBP-J through its N-terminal RAM (RBP-J association molecule) (15,16) domain, and transactivates promoters harboring RBP-J-binding sites through recruiting co-activators such as GCN5 and p300/CBP (17,18). In addition to NIC, RBP-J also mediates transactivation of Epstein–Barr (EB) virus nuclear antigen (EBNA) 2 and therefore may play a role in the immortalization of cells infected by EB virus (19,20).
On the other hand, in the absence of transactivators such as NIC or EBNA2, RBP-J suppresses transcription of promoters recognized by RBP-J (21). This is mostly attributable to multiple co-suppressors and/or adaptor molecules recruited by RBP-J. Kao et al. (22) and Zhou and Hayward (23) reported that SMRT/N-CoR interacts with RBP-J and suppresses the RBP-J-mediated transcription by competing with NIC and by recruiting histone deacetylases (HDACs), which renders the chromatin into architecture that is inaccessible to the general transcriptional machinery. In addition, CIR (CBF1 interacting co-repressor), another RBP-J-interacting protein, also suppresses the RBP-J-mediated transactivation by associating with HDAC2 and SAP30 (24). More recently, Oswald et al. (25) and Kuroda et al. (26) demonstrated that MINT (MSX2-interacting nuclear target protein) (27), a nuclear matrix protein interacting with SMRT/N-CoR (28), also interacts with RBP-J and suppresses the RBP-J-mediated transactivation by competing for binding sites and by recruiting co-repressors, and may thus regulate the Notch signaling pathway.
KyoT is a LIM domain protein that was cloned through its interaction with RBP-J using the yeast two hybrid system (29). While two isoforms of KyoT (KyoT1 and KyoT2) were identified, only KyoT2 possesses an RBP-J-binding motif probably generated by alternative mRNA splicing. Biochemical and molecular biological analyses performed by Taniguchi et al. (29) have shown that, through competing stochastically for a binding site on RBP-J with NIC and EBNA2, KyoT2 inhibits transactivation of promoters containing RBP-J recognition sites by NIC and EBNA2. However, a large body of evidence has suggested that LIM domains function as an interface of protein– protein interaction (30–32). We therefore assumed that, in addition to competing for binding sites, KyoT2 might also regulate the RBP-J-mediated transactivation by recruiting other co-suppressor molecule(s) to RBP-J through its LIM domains.
We assessed this assumption by screening a cDNA library to isolate KyoT-interacting molecules using the yeast two hybrid system (33). The results showed that the cDNA fragments of RBP-J with different length were isolated three times among the positive clones, indicating that the screening was reliable. Sequence analysis of other positive clones showed that one of them was RING1, a member of the polycomb group (PcG) proteins that function as transcriptional suppressors (34–36). In the current study, we showed that RING1 and KyoT2 interacted through the C-terminal fragment of RING1 and the LIM domains of KyoT2 in yeast and in mammalian cells, both in vitro and in vivo. Because KyoT2 associates with RING1 by its LIM domains (this study) and with RBP-J by the C-terminal motif (29), RING1, KyoT2, and RBP-J may form a three-molecule complex in cells. Moreover, we found that overexpression of RING1 and KyoT2 suppressed transactivation of RBP-J by NIC. The RING1-mediated suppression of RBP-J was abrogated by overexpression of KBP1 (for KyoT-binding protein 1) (33), a protein competing with RING1 for binding to the LIM domains of KyoT2, suggesting that suppression of RBP-J by RING1 was dependent on its association with KyoT2. We suggested that there might be at least two approaches for the KyoT2-mediated suppression of RBP-J, namely competition for binding sites with transactivators and recruitment of suppressors such as RING1 through the LIM domains of KyoT2.
MATERIALS AND METHODS
RT–PCR
Total cellular RNA from cultured Hela cells was prepared using the Trizol reagent (Gibco BRL, Gaithersberg, MD) according to the manufacturer’s instructions. cDNA was synthesized using a kit from Gibco BRL with total RNA as a template and oligo-dT as a primer. The coding region of human RING1 cDNA was amplified by PCR using: 5′ primer, 5′-GAATTCATGGATGGCACAGAGATTGC; and 3′ primer, 5′-TCACTTTGGATCCTTGGTGGGAGC. The amplified fragments were cloned into a T-vector (Promega, Germany) and confirmed by DNA sequencing.
Yeast two hybrid assay
Coding regions of KyoT1 and KyoT2 cDNA were amplified from the full-length KyoT cDNA clones (kindly provided by T. Honjo) by PCR using primers K1 (5′-CATATGTCGGAGAAGTTCGAC) and K2 (5′-GAATTCTTACAGCTTTTTGGACAAG), and were inserted in-frame into the multiple cloning sites of pGBKT7 of the yeast two hybrid system 3 (Clontech, Palo Alto, CA), to generate bait plasmids pGBK-KyoT1 and pGBK-KyoT2, respectively. The two LIM domains (named LIM1 and LIM2 from the N-terminal of KyoT2) were also cloned by PCR using primers K1 plus K3 (5′-CTAGTCCTCCCGAGTAGCGCAC) for LIM1, and K2 plus K4 (5′-GAATTCGCTACTCGGGAGGACTCC) for LIM2, and were inserted in-frame into pGBKT7 (pGBK-LIM1 and pGBK-LIM2). The coding region of the full-length RING1 cDNA was inserted in-frame into pGADT7 to construct a prey plasmid (pGAD-RING1). Prey plasmids with truncated RING1 (pGAD-RING1-N with amino acids 1–242, and pGAD-RING1-C with amino acids 243–377) were generated by restriction digestion and ligation. The bait and prey plasmids were all confirmed by DNA sequencing and were used to transform the yeast strain AH109 by the LiAc method. The resulting yeast clones were plated on SD/-trp-leu-his-ade plates and single colonies were amplified in medium. Yeast cells were collected as the cultures reached exponential stage and were lysed by repeated freeze and thaw in liquid nitrogen, followed by liquid β-galactosidase assay for β-galactosidase activity.
Cell culture and transfection
HEK-293, 293T and COS7 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 2 mM glutamine (Gibco BRL). Cells were plated in six-well plates and were transfected the next day using Lipofectamine™ 2000 (Invitrogen Life Technologies, Carlsbad, CA) at a confluency of about 80%. Briefly, plasmid DNA (with amounts shown in each figure legend, brought up to 2 µg in total with empty vectors) was mixed with Lipofectamine™ 2000 in serum-free DMEM and incubated at room temperature for 20 min. The DNA–Lipofectamine™ 2000 complexes formed were added to the cells, and the transfection was allowed to proceed for 6 h. After DMEM containing 20% FCS was added to each well, cells were cultured further in medium for indicated periods of time, and then collected for further experiments.
GST-pull down assay
The N- and C-terminal fragments of RING1 were recovered from pGAD-RING1-N and pGAD-RING1-C, respectively, and were inserted in-frame into the multiple cloning sites of pGEX-4T-1 (Pharmacia, Uppsala, Sweden) and confirmed by sequencing (pGEX-RING1-N and pGEX-RING1-C). Fusion proteins composed of GST and the RING1 N-terminal fragment (GST-RING1-N) as well as of GST and the RING1 C-terminal fragment (GST-RING1-C) were produced in Escherichia coli, and were purified from the bacterial lysates using the glutathione-Sepharose 4B beads (Pharmacia). The coding region of full-length KyoT2 cDNA was inserted into p CMV2-Flag, and the generated plasmid (pCMV2-KyoT2-Flag) was used to transfect 293T cells. Cell lysates were prepared 60 h after transfection and were mixed with the purified GST-RING1-N or GST-RING1-C fusion protein bound to glutathione-Sepharose 4B beads. The beads were incubated at 4°C for 2 h, and were washed with a buffer (142.5 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.6, 1 mM EDTA, 0.2% NP-40) five times. The associated proteins were then separated by SDS–PAGE with a polyacrylamide concentration of 12%, followed by western blotting with an anti-Flag antibody (M2, Sigma, 1:1000). GST was used as a negative control for the experiment.
Competitive GST-pull down assay among RING1, KBP1 and KyoT2 was carried out in a similar way, using GST-RING1-C and His-tagged KBP1 and LIM1 and LIM2 fragments of KyoT2 expressed in E.coli.
Co-immunoprecipitation
Myc-tagged KyoT2 (pCMV-KyoT2-myc) and RBP-J (pCMV-RBP-J-myc) were constructed by inserting full-length KyoT2 and RBP-J cDNA into pCMV-myc (Clontech). Flag-tagged RING1 (pCMV-RING1-Flag), KBP1 (pCMV-KBP1-Flag), and RBP-J (pCMV-RBP-J-Flag) were constructed by inserting full-length RING1, KBP1 and RBP-J cDNA into pCMV2-Flag. A luciferase gene fragment was recovered from pGL3-promoter (Promega) and inserted into pCMV-myc to construct Myc-tagged luciferase (pCMV-luc-myc) used as a negative control. All of the constructs were confirmed by DNA sequencing. Cells were transfected using Lipofectamine™ 2000 with plasmids in combinations as described in the results. Cell lysates were prepared 60 h after transfection by lysing cells with the phospho-lysis buffer (50 mM Tris–Cl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40, 1 mg/ml BSA, 0.1 mM PMSF). The cell lysates with equal amounts of total proteins were incubated with an anti-Myc (9E10, Santa Cruz) or the anti-Flag antibody, and were precipitated with protein A-Sepharose 4B beads (Pharmacia). After washing with the phospho-lysis buffer, the co-precipitated proteins were analyzed by SDS–PAGE followed by western blotting using the anti-Flag or the anti-Myc antibody, respectively.
Reporter assay
For the mammalian two hybrid experiments, coding regions of the full-length cDNA of KyoT2 and RING1 were inserted in-frame into the multiple cloning sites of pCMX-GAL4-DBD and pCMX-VP16 (NLS) (generously provided by T. Honjo), to generate plasmids pCMX-GAL4DBD-KyoT2 and pCMX-VP16-RING1, respectively. The N- and C-terminal fragments of RING1 were recovered from pGAD-RING1-N and pGAD-RING1-C, and were also inserted in-frame into pCMX-VP16 (NLS) to construct pCMX-VP16-RING1-N and pCMX-VP16-RING1-C. The pCMX-VP16-KBP1 was constructed by inserting KBP1 cDNA into pCMX-VP16 (NLS). All constructs were confirmed by DNA sequencing. Cells were transfected with pCMX-GAL4DBD-KyoT2 in combination with one of the plasmids expressing VP16 fusion proteins. The reporter construct (TK MH100 × 4 Luc) was also included in the transfection. Cells were collected 48 h after transfection and were lysed in a hypotonic buffer (91 mM K2HPO4, 9 mM KH2PO4, 10% glycerol, 1 mM DTT, 10% Triton X-100). Supernatants were collected after centrifugation and were used to react with Pikagene luciferase reagent (Toyo Ink, Tokyo, Japan). The luciferase activity was read out using a chemo-luminator. Transfection efficiency was calibrated by including pSV-β-gal in transfections, followed by examining the β-galactosidase activity in cell lysates. Each experiment was repeated three times and the data were analyzed with the Student’s t-test.
Transactivation of RBP-J-responsive promoters was detected as described (26). Briefly, expression vectors for KyoT2 (pEFBOS-KyoT2), RING1 (pEFBOS-RING1) and NIC (pEFBOS-NIC) were constructed by insertion of full-length cDNA of KyoT2, RING1, as well as NIC into pEFBOS-neo. pCMX-VP16-RBP was obtained from T. Honjo. Cells were cultured and transfected with the expression vectors together with the reporter construct pGa981–6, which contains a hexamerized 50 bp EBNA2 response element of the TP-1 promoter and is strictly dependent on RBP-J (37). Cells were collected 48 h after transfection and the luciferase activity was examined as above. pSV-β-gal was included in each transfection as an internal control of the transfection efficiency.
RESULTS
KyoT2 suppresses constitutively active RBP-J
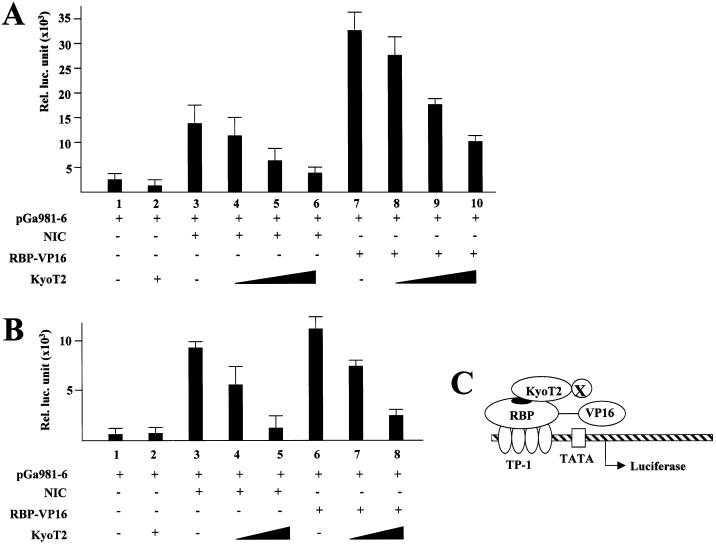
KyoT2 is a LIM domain protein suppressing transactivation of RBP-J by NIC and EBNA2 through a competition mechanism, as suggested by Taniguchi et al. (29). We tested whether KyoT2 could actively suppress transcription by employing a constitutively active form of RBP-J, the RBP-J-VP16, which strongly activates RBP-J-dependent promoters (37). When co-transfected with the reporter construct, NIC showed strong transactivation of the reporter gene, which was suppressed by KyoT2 in a dose-dependent manner (Fig. 1A). This is consistent with the competition model, which predicted that KyoT2 suppressed transcription by excluding transactivators such as NIC from RBP-J. This model also predicted that KyoT2 might not suppress transcription mediated by the constitutively active RBP-J-VP16, in which the transactivation domain of VP16 was fused to the C-terminal of RBP-J. However, in this study, as shown in Figure 1A, KyoT2 also suppressed the RBP-J-VP16 moderately but significantly. Similar results were seen when COS7 cells were transfected with the same groups of plasmids (Fig. 1B). These results suggested that KyoT2 might recruit co-suppressor(s) to actively suppress the RBP-J-mediated transactivation, most probably through its LIM domains that have been regarded as protein-interacting interfaces (30–32) (Fig. 1C).
Figure 1.
KyoT2 suppresses the constitutively active RBP-J, RBP-J-VP16. HEK-293 (A) and COS7 (B) cells were co-transfected with expression vectors of NIC (0.2 µg) or RBP-J-VP16 (0.2 µg), together with increasing amounts (0.2, 0.5 and 1.0 µg) of plasmid expressing KyoT2, and transactivation of RBP-J was detected using a reporter construct (0.2 µg) with luciferase gene under control of the TP-1 promoter (37). (C) A schematic representative showing that KyoT2 may recruit co-suppressor(s) (X) to suppress the RBP-J-mediated transactivation.
KyoT2 interacts with RING1 through LIM domains
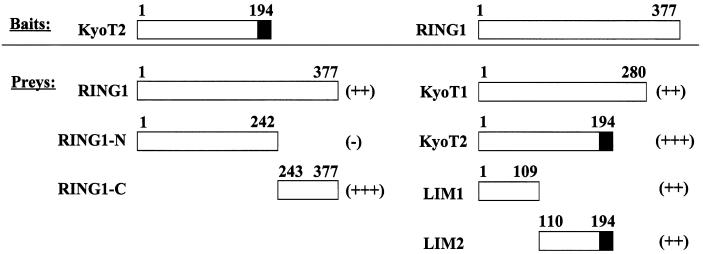
We screened KyoT2-interacting molecules using the yeast two hybrid system with KyoT2 as bait. In total, 5 × 106 clones from a human lymph node cDNA library were screened, and the cDNA fragments of RBP-J with different lengths were picked up three times, indicating that the screening was reliable (33). Among other known and unknown proteins, RING1 was isolated as a candidate of KyoT2-binding proteins. To confirm the interaction between KyoT2 and RING1, and to identify the potential interacting domains between KyoT2 and RING1, we performed a yeast two hybrid assay with full-length KyoT1 or KyoT2 and RING1 molecules, as well as their truncated derivatives (Fig. 2). When co-expressed in host yeast cells, significant interaction was detected between KyoT2 and RING1 as well as KyoT1 and RING1, suggesting that KyoT interacted with RING1 through its LIM domains, as KyoT1 is a LIM-only protein and does not possess an RBP-J-binding motif (29). This was confirmed by yeast two hybrid assay using single LIM domain (LIM1 or LIM2 from the N-terminal) of KyoT2 as bait, which also showed positive interaction with RING1. On the other hand, while the C-terminal fragment of RING1 showed positive interaction with KyoT2 in yeast, its N-terminal fragment did not interact with KyoT2 in yeast. These results indicated that the LIM domains of KyoT2 and the C-terminal fragment of RING1 might be responsible for the interaction between KyoT2 and RING1 in yeast.
Figure 2.
Identification of interaction domains between KyoT2 and RING1 in yeast. Full-length or truncated fragments of KyoT2, KyoT1 and RING1 cloned in the yeast two hybrid (system 3) vectors were used to transform yeast cells. The β-galactosidase activity in cell lysates was examined and is shown: (–), ≤0.25; (+), 0.25–0.50; (++), 0.50–1.00; (+++), ≥1.00.
Physical interaction between KyoT2 and RING1
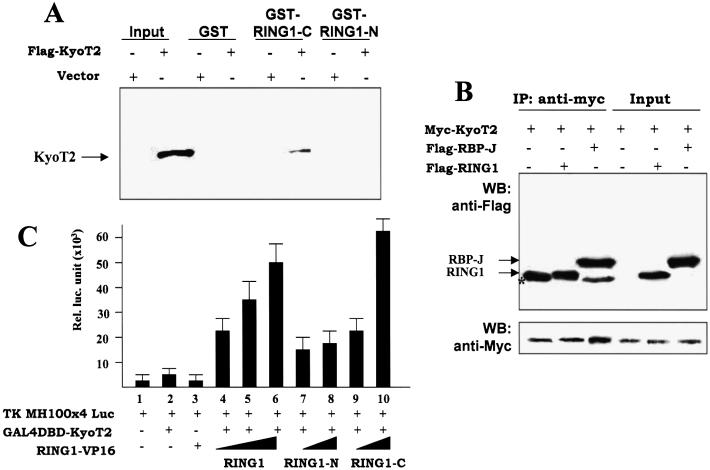
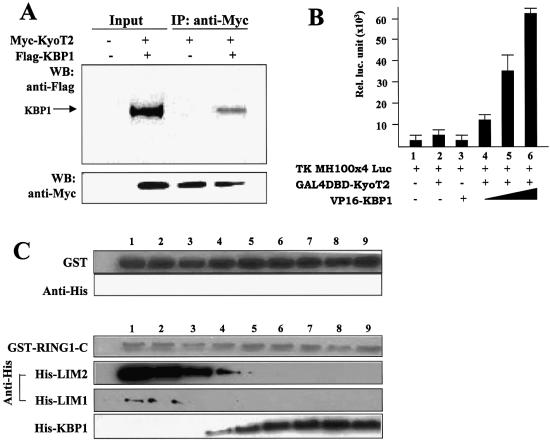
We examined the direct physical interaction between KyoT2 and RING1 using GST-pull down and co-immunoprecipitation assays. Because full-length RING1 was hardly expressed as GST fusion protein in our hands, we employed GST fusion proteins of the N- and C-terminal fragments of RING1 for the GST-pull down experiment. The GST-RING1-N and GST-RING1-C fusion proteins were generated in E.coli and purified, and were used to interact with the Flag-tagged KyoT2 protein expressed by transfection of cultured 293T cells. The GST protein was also prepared and was used as a negative control. The Flag-KyoT2 interacting with RING1 fragments was pulled down and detected by an anti-Flag antibody after western blotting. As shown in Figure 3A, consistent with the yeast two hybrid assay, Flag-KyoT2 interacted with GST-RING1-C instead of GST-RING1-N or GST, suggesting that KyoT2 directly interacted with RING1.
Figure 3.
Interaction between KyoT2 and RING1 in vitro and in vivo. (A) GST-pull down assay. GST, GST-RING1-C and GST-RING1-N were generated in E.coli and purified. Cells (293T) were transfected with the Flag-KyoT2-expressing vector. Cell lysates were prepared 60 h after transfection, and incubated with purified GST, GST-RING1-C and GST-RING1-N, followed by precipitation with glutathione-Sepharose 4B beads. Bound proteins were analyzed by western blotting using the anti-Flag antibody. (B) Immunoprecipitation assay. Vectors for expression of the Myc-tagged KyoT2 and the Flag-tagged RING1 or the Flag-tagged RBP-J were used to transfect 293T cells. Cell lysates were prepared 60 h after transfection and were immunoprecipitated with the anti-Myc antibody. Co-precipitated proteins were detected with the anti-Flag antibody after western blotting. The asterisk indicates the heavy chain of the anti-Myc antibody. (C) Mammalian two hybrid assay. HEK-293 cells were co-transfected with expression vectors of GAL4-DBD-KyoT2 (0.2 µg) and with increasing amounts (0.2, 0.4 and 0.8 µg) of expressing vectors of VP16-RING1, VP16-RING1-N or VP16-RING1-C, and a luciferase reporter plasmid (0.4 µg). Luciferase activity was detected in cell lysates 48 h after transfection.
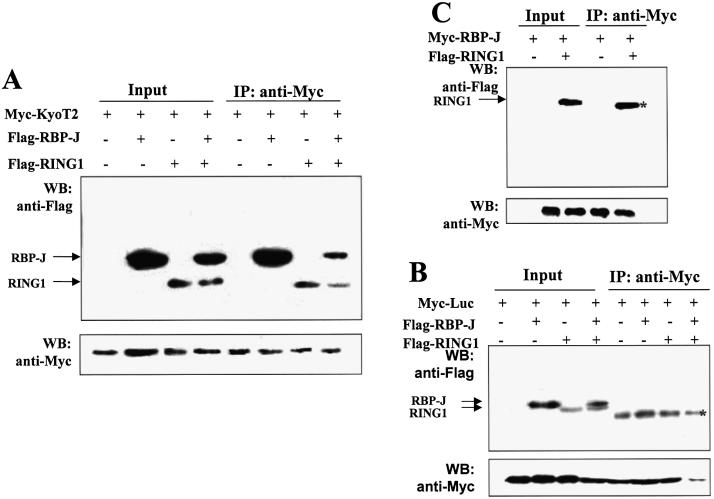
A co-immunoprecipitation assay was performed to detect interaction between KyoT2 and RING1 in cells. Cells (293T) were transfected with expression vectors of the Myc-tagged KyoT2 and the Flag-tagged RING1, or the Flag-tagged RBP-J as a positive control. Cell lysates were prepared 60 h after transfection and were incubated with the anti-Myc antibody and protein-A beads, followed by western blotting with the anti-Flag antibody. The results showed that Flag-RING1 and Flag-RBP-J were co-precipitated by Myc-KyoT2 (Fig. 3B). On the other hand, as a negative control, Myc-luciferase was co-transfected with Flag-RING1 or Flag-RBP-J, but none of these Flag-tagged proteins was detected by anti-Flag after co-immunoprecipitation using the anti-Myc antibody (data not shown and Fig. 4B). These results indicated that KyoT2 and RING1 interacted physically both in vitro and in vivo.
Figure 4.
RING1 binds to RBP-J through KyoT2. (A) Expression vectors of the Myc-tagged KyoT2 and the Flag-tagged RBP-J and RING1 were used to transfect 293T cells. Cell lysates were prepared 60 h after transfection and were immunoprecipitated with the anti-Myc antibody. Co-precipitated proteins were detected with the anti-Flag antibody after western blotting. (B) A similar experiment was performed as in (A) except that the bait for immunoprecipitation was the Myc-tagged luciferase instead of KyoT2. (C) Immunoprecipitation was carried out using the Myc-tagged RBP-J and the Flag-tagged RING1. The asterisks indicate the heavy chain of the anti-Myc antibody.
To examine further whether KyoT2 and RING1 interact in mammalian cells, we employed the mammalian two hybrid assay. We constructed vectors expressing fusion proteins GAL4DBD-KyoT2 and RING1-VP16 as well as RING1-N-VP16 and RING1-C-VP16, and co-transfected these expression vectors with a reporting construct, in which the luciferase gene is under control of a promoter containing GAL4 recognition sites. Luciferase activity in cell lysates was examined 60 h after transfection. The result (Fig. 3C) showed that, while transfection with only GAL4DBD-KyoT2 or only RING1-VP16 expression vector did not activate expression of luciferase in cells, co-transfection of the two vectors stimulated a high luciferase activity in the cell lysates. This suggested that KyoT2 and RING1 could interact in mammalian cells. Moreover, while RING1-N-VP16 showed mild transactivation activity on the reporter gene, RING1-C-VP16 strongly transactivated the expression of the luciferase gene, suggesting that the interaction between KyoT2 and RING1 was mostly mediated through the C-terminal fragment of RING1.
RING1 interacts with RBP-J through KyoT2 in cells
The KyoT2 molecule possesses two LIM domains on its N-terminal and an RBP-J-binding motif on the C-terminal (29), suggesting that KyoT2 may interact simultaneously with RING1 through the LIM domains, and with RBP-J through the RBP-J-binding motif. We tested the formation of a three-molecule complex of KyoT2, RING1 and RBP-J in cells using the co-immunoprecipitation assay. We co-expressed the Flag-tagged RING1 and the Flag-tagged RBP-J in 293T cell with the Myc-tagged KyoT2. Immunoprecipitation was carried out using the anti-Myc antibody 48 h after transfection, and the co-precipitated proteins were detected with the anti-Flag antibody after western blotting. As shown in Figure 4A, Flag-RBP-J and Flag-RING1 were simultaneously co-precipitated with KyoT2, suggesting that KyoT2, RING1 and RBP-J can form a three-molecule complex in cells. The Myc-tagged luciferase, which was used as a negative control, did not co-precipitate with either RING1 or RBP-J (Fig. 4B). Because RING1 did not directly associate with RBP-J in the yeast two hybrid assay (data not shown) or the co-immunoprecipitation assay (Fig. 4C), RING1 might be recruited to RBP-J through KyoT2.
RING1 suppresses constitutively active RBP-J as well as transactivation of RBP-J by NIC
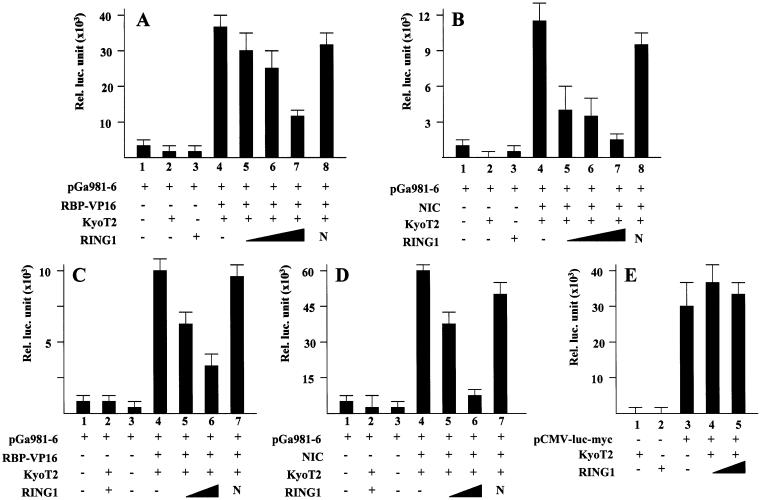
KyoT2 is a LIM domain protein that interacts with and suppresses transcription factor RBP-J by competing with transactivators such as NIC and EBNA2 for binding with RBP-J (29). Our data presented here have shown that KyoT2 may recruit RING1 to RBP-J through its LIM domains. RING1 is a member of the PcG proteins and has transcriptional suppression activity (34). We therefore tested whether RING1 inhibited the RBP-J-mediated transactivation through interaction with KyoT2 using reporter assays. RBP-J and KyoT2 were expressed in HEK293 cells, while RING1 mRNA could not be detected in the same cells after extensive RT–PCR amplification (45 cycles) (data not shown). To test whether RING1 suppresses transcription mediated by RBP-J-VP16, we transfected HEK293 and COS7 cells using the vectors expressing KyoT2 and RING1, together with the expression vector of RBP-J-VP16 and the RBP-J-specific reporter construct, pGa981-6 (37). The results showed that in HEK293 as well as COS7 cells, in the presence of KyoT2, co-transfection of RING1 suppressed transcriptional activation by the constitutively active RBP-J, the RBP-J-VP16 fusion protein (Fig. 5A and C).
Figure 5.
RING1 inhibits the RBP-J-mediated transcription. (A and C) RING1 suppresses transactivation by the constitutively active RBP-J. HEK-293 (A) and COS7 (C) cells were co-transfected with plasmids expressing RBP-VP16 (0.2 µg), KyoT2 (0.1 µg) and increasing amounts (0.2, 0.5 and 1.0 µg) of plasmid expressing RING1, together with the TP-1 reporter constructs (0.2 µg). Transactivation of the reporter construct was detected using luciferase assay. (B and D) RING1 suppresses transactivation of the RBP-J-dependent promoter by NIC. HEK-293 (B) and COS7 (D) cells were transfected and examined as in (A) and (C) except that plasmid expressing NIC instead of RBP-J-VP16 was used. (E) RING1 did not suppress CMV promoter. The reporter construct with CMV promoter (0.4 µg) was co-transfected with expression vectors of KyoT2 (0.1 µg) and increasing amounts (0.25, 0.5 and 1.0 µg) of plasmid expressing RING1. Relative luciferase activity was examined.
We also tested whether overexpression of RING1 in cultured cells could suppress transactivation of the RBP-J-mediated transcription by one of its physiological transactivators, NIC. As shown in Figure 5B and D, in both HEK293 and COS7 cells, NIC showed a strong transactivation of the RBP-J-dependent reporter construct. This transactivation was suppressed by co-transfection of the full-length RING1 cDNA in a dose-dependent manner in the presence of KyoT2. The N-terminal fragment of RING1 had no suppressive activity to either RBP-J-VP16 or NIC (Fig. 5A–D). Because RING1 interacts with KyoT2 and KyoT2 associates with RBP-J, we proposed that KyoT2 recruited RING1 to suppress the RBP-J-mediated transcription in cells. Therefore, KyoT2 may suppress the RBP-J-mediated transcription through two ways: one is competition with transactivators for binding sites (29), and the other is recruitment of co-repressors such as RING1.
We excluded a pan-suppressive activity of RING1 on general promoters using a reporter construct in which luciferase gene was driven by a CMV IE promoter (pCMV-luc-myc). No significant suppression of luciferase expression was detected with increasing amounts of RING1, as shown in Figure 5E.
Suppression of RBP-J by RING1 is abrogated by KBP1, which competes with RING1 for binding to LIM domains of KyoT2
To assess the mechanism of the RING1-mediated suppression of RBP-J, we employed KBP1, which encodes 210 amino acids and was identified as another KyoT2-binding protein (33). In yeast, KBP1 associated with both LIM domains KyoT2 (H.Han, unpublished data). To look at the interaction between KBP1 and KyoT2, we carried out co-immunoprecipitation and mammalian two hybrid assays. As in Figure 6A and B, respectively, both assays showed that KBP1 and KyoT2 interacted with each other in mammalian cells. Moreover, we tested whether KBP1 and RING1 competed for binding to LIM domains of KyoT2 using the in vitro GST-pull down assay. As shown in Figure 6C, while neither of the LIM domains interacted with GST, both showed interaction with the GST-RING1-C fusion protein, which was consistent with the previous result (Fig. 3A). However, when the recombinant KBP1 protein was included in this assay system, with the amount of KBP1 increasing, the LIM protein associated with GST-RING1-C decreased (Fig. 6C). This result suggested that KBP1 blocked the interaction between RING1 and LIM domains of KyoT2 probably by binding to KyoT2 at the same sites as RING1.
Figure 6.
KBP1 competes with RING1 for binding with the LIM domains of KyoT2. (A and B) Interaction between KBP1 and KyoT2 in cells was detected by co-immunoprecipitation assay (A) and mammalian two hybrid assay (B). (C) GST-pull down assay. GST, GST-RING1-C, His-KBP1, His-LIM1 and His-LIM2 were produced in E.coli and purified. A constant amount of GST-RING1-C was incubated with His-LIM1 or His-LIM2, and an increasing amount of His-KBP1. Associated proteins were pulled down using glutathione-Sepharose beads, and were detected with the anti-His antibody. GST was used as a negative control.
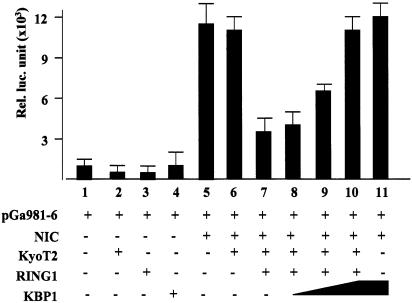
We then tried to block the RING1-mediated suppression of RBP-J by overexpression of KBP1 in cells. When transfected with the reporter construct, KBP1 did not activate or suppress transcription (Fig. 7, lanes 4 and 11). However, when transfected together with RING1, we found that overexpression of KBP1 abrogated the suppressive effect of RING1 on RBP-J (Fig. 7). As KBP1 might compete for binding sites on KyoT2 with RING1 (Fig. 6C), this result suggested that the suppressive activity of RING1 might be dependent on its association with KyoT2.
Figure 7.
Overexpression of KBP1 abrogates suppressive effect of RING1 on RBP-J. HEK293 cells were co-transfected with expressing vectors of NIC (0.5 µg), KyoT2 (0.1 µg), RING1 (0.2 µg) and increasing amounts (0.2, 0.5 and 1.0 µg) of plasmid expressing KBP1 as well as pGa981-6 (0.2 µg). Relative luciferase activity was examined 48 h after transfection.
DISCUSSION
RBP-J is the main intra-nuclear target of the Notch signaling pathway and mediates transactivation of all the four Notch receptors as well as the EB virus transactivator EBNA2 (2,11). These transactivators activate transcription of the promoters recognized by RBP-J through binding to a motif of RBP-J and recruiting co-activators such as p300/CBP and GCN5 to the promoter (17,18). In the absence of transactivators, RBP-J suppresses transcription (21,38). Several transcription co-suppressors such as MINT have been shown to compete with transactivators for the binding motif of RBP-J. More importantly, these suppressors also recruit other co-suppressors including HDACs to RBP-J to suppress the RBP-J-mediated transcription by reducing the accessibility of the chromatin. KyoT2 also suppresses the RBP-J-mediated transcription through competing with transactivators for the binding motif of RBP-J (29). In the current study, we showed that KyoT2 might also inhibit the RBP-J-mediated transcriptional activation by recruiting the PcG protein RING1 to RBP-J through its LIM domains, in addition to competing for binding sites on RBP-J with transcription activators. Our results indicated that KyoT2 interacts with RING1 through its LIM domains in yeast, and the interaction was also detected in mammalian cells. When recruited to RBP-J through KyoT2, RING1 suppressed not only the transactivation of RBP-J by NIC, but also the activity of the RBP-VP16 fusion protein, a constitutively active form of RBP-J, indicating that competition for binding sites may not be the only way for the KyoT2-mediated transcription suppression of RBP-J. There fore, there might be at least two ways for the KyoT2-mediated suppression of RBP-J. One is competition for binding sites with transactivators, and the other is recruitment of suppressors such as RING1.
LIM domain is a zinc-containing protein domain implicated in mediating protein–protein interactions (30–32). Proteins with LIM domains are divided into two classes, namely the LIM-only proteins composed of only LIM domains, and LIM-homeodomain proteins that contain homeodomains in addition to LIM domains. While the function of LIM-only proteins is largely unknown, the LIM-homeodomain proteins play critical roles in cellular differentiation through binding to DNA by homeodomains and recruiting regulators of transcription through LIM domains (30–32). KyoT1 is a LIM-only protein. However KyoT2 generated by alternative splicing may be recruited to chromatin through interaction with RBP-J, and thus may function in a similar way to LIM-homeodomain proteins. Therefore, KyoT2 is able to recruit transcription regulators by its LIM domain to promoters through associating with DNA-binding protein RBP-J. In this regard, KyoT2 is a LIM domain protein with unique properties.
Our results showed that RING1 associated with both LIM domains of KyoT2, suggesting that multiple RING1 molecules were recruited to RBP-J by KyoT2. RING1 may suppress the RBP-J-mediated transcriptional activation through two mechanisms. First, RING1 itself has a transcriptional repressor activity. As shown by Satijn et al. (34), when tethered to a promoter of a reporter construct, RING1 actively suppressed transcription from the promoter. Second, RING1 associates with the PcG protein complex (35). The PcG proteins form a large protein complex in the nucleus and suppress transcription by reducing accessibility of the targeted chromatin regions (35). Recently, it has been reported that the PcG complex could also be recruited to promoters through interaction with specific transcription factors such as the retinoblastoma (Rb) protein, and suppressed transcriptional activation of the promoter (36).
Transcription memory is supposed to play an essential role in the regulation of cell differentiation (39,40). One of the main participants of transcriptional memory is the PcG protein complex. During embryonic development of Drosophila, the PcG proteins are responsible for ‘memorizing’ the transcriptional silencing status of the homeotic genes established by the Gap proteins, while the Trithorax group (TrG) proteins are responsible for ‘memorizing’ the active status of the homeotic genes created by the Pair-rule proteins. Multiple members of the PcG proteins have been identified in mammals. In this study, we reported that the transcription factor RBP-J might be a target of the PcG-mediated transcriptional silencing. As RBP-J is the main nuclear target of Notch signaling, and the Notch signaling pathway plays a pivotal role in the control of cell differentiation, PcG proteins might play a role in regulation of Notch-mediated cell fate specification through memorizing of the transcription status established by the Notch/RBP-J signaling pathway.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr T. Honjo for plasmids. We thank Ms Yan-Xia Zhang and Ms Ling Zhang for secretarial assistance and technical support. We also thank Prof. Kai-Chun Wu and Prof. Wen-Li Yan for critical reading of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (39970376, 30200148) and from the Ministry of Science and Technology of China (001CB509906).
REFERENCES
- 1.Artavanis S., Rand,M. and Lake,R.J. (1999) Notch signaling: cell fate control and signal integration in development. Science, 284, 770–776. [DOI] [PubMed] [Google Scholar]
- 2.Honjo T. (1996) The shortest path from the surface to the nucleus: RBP-Jk/Su(H) transcription factor. Genes Cells, 1, 1–9. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas S., Matsuno,K. and Fortini,M.E. (1995) Notch signaling. Science, 268, 225–232. [DOI] [PubMed] [Google Scholar]
- 4.Swiatek P.J., Lindsell,C.E., del Amo,F.F., Weinmaster,G. and Gridley,T. (1994) Notch1 is essential for postimplantation development in mice. Genes Dev., 8, 707–719. [DOI] [PubMed] [Google Scholar]
- 5.Conlon R.A., Reaume,A.G. and Rossant,J. (1995) Notch1 is required for the coordinate segmentation of somites. Development, 121, 1533–1545. [DOI] [PubMed] [Google Scholar]
- 6.Oka C., Nakano,T., Wakeham,A., de la Pompa,J.L., Mori,C., Sakai,T., Kawaichi,M., Shiota,K., Mak,T.M. and Honjo,T. (1995) Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development, 121, 3291–3301. [DOI] [PubMed] [Google Scholar]
- 7.Radtke F., Wilson,A., Stark,G., Bauer,M., van Meerwijk,J., MacDonald,H.R. and Aguet,M. (1999) Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity, 10, 547–558. [DOI] [PubMed] [Google Scholar]
- 8.Han H., Tanigaki,K., Yamamoto,N., Kuroda,K., Yoshimoto,M., Nakahata,T., Ikuta,K. and Honjo,T. (2002) RBP-J deficient mouse reveals essential role of Notch in T versus B lineage determination but not in later stages of thymocyte development. Int. Immunol., 14, 637–645. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa T., Maruyama,S., Kawaichi,M. and Honjo,T. (1992) The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell, 69, 1191–1197. [DOI] [PubMed] [Google Scholar]
- 10.Schweisguth F. and Posakony,J.W. (1992) Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell, 69, 1199–1212. [DOI] [PubMed] [Google Scholar]
- 11.Kato H., Sakai,T., Tamura,K., Minoguchi,S., Shirayoshi,Y., Hamada,Y., Tsujimoto,Y. and Honjo,T. (1996) Functional conservation of mouse Notch receptor family members. FEBS Lett., 395, 221–224. [DOI] [PubMed] [Google Scholar]
- 12.Struhl G. and Adachi,A. (1998) Nuclear access and action of Notch in vivo. Cell, 93, 649–660. [DOI] [PubMed] [Google Scholar]
- 13.Schroeter E.H., Kisslinger,J.A. and Kopan,R. (1998) Notch-1 signaling requires ligand-induced proteolytic release of intracellular domain. Nature, 393, 382–386. [DOI] [PubMed] [Google Scholar]
- 14.DeStrooper B., Annaert,W., Cuppers,P., Saftig,P., Craessaerts,K., Mumm,J., Schroeter,E.H., Schrijvers,V., Wolfe,M.S., Ray,W.J., Goate,A. and Kopan,R. (1999) A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature, 398, 518–522. [DOI] [PubMed] [Google Scholar]
- 15.Tamura K., Taniguchi,Y., Minoguchi,S., Sakai,T., Tun,T., Furukawa,T. and Honjo,T. (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr. Biol., 5, 1416–1423. [DOI] [PubMed] [Google Scholar]
- 16.Jarriault S., Brou,C., Logeat,F., Schroeter,E.H., Kopan,R. and Isreal,A. (1995) Signaling downstream of activated mammalian Notch. Nature, 377, 355–358. [DOI] [PubMed] [Google Scholar]
- 17.Kurooka H., Kuroda,K. and Honjo,T. (1998) Roles of the ankyrin repeats and C-terminal region of the mouse Notch1 intracellular region. Nucleic Acids Res., 26, 5448–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurooka H. and Honjo,T. (2000) Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferase PCAF and GCN5. J. Biol. Chem., 275, 17211–17220. [DOI] [PubMed] [Google Scholar]
- 19.Waltzer L., Logeat,F., Brou,C., Isreal,A., Sergeant,A. and Manet,E. (1994) The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein–Barr virus EBNA2 protein to its DNA responsive elements. EMBO J., 13, 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henkel T., Ling,P.D., Hayward,S.D. and Peterson,M.G. (1994) Mediation of Epstein–Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science, 265, 92–95. [DOI] [PubMed] [Google Scholar]
- 21.Dou S., Zeng,X., Cortes,P., Erdjument Bromage,H., Tempst,P. and Honjo,T. (1994) The recombination signal sequence-binding protein RBP-2N functions as a transcription repressor. Mol. Cell. Biol., 14, 3310–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao H.Y., Ordentlich,P., Koyano-Nakagawa,N., Tang,Z., Downes,M., Kintner,C.R., Evans,R.M. and Kadesch,T. (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev., 12, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou S. and Hayward,S.D. (2001) Nuclear localization of cbf1 is regulated by interactions with smrt coreppressor complex. Mol. Cell. Biol., 21, 6222–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh J.J., Zhou,S., Chen,L., Young,D.B. and Hayward,S.D. (1999) CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl Acad. Sci. USA, 96, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oswald F., Kostezka,U., Astrahantseff,K., Bourteele,S., Dillinger,K., Wilda,M., Hameister,H., Knochel,W., Liptay,S. and Schmid,R. (2002) SHARP is a novel component of the Notch/RBP-Jk signaling pathway. EMBO J., 21, 5417–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda K., Han,H., Tani,S., Tanigaki,K., Tun,T., Furukawa,T., Taniguchi,Y., Kurooka,H., Hamada,Y., Toyokuni,S. and Honjo,T. (2003) Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity, 18, 301–312. [DOI] [PubMed] [Google Scholar]
- 27.Newberry E.P., Latifi,T. and Towler,D. (1999) The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry, 38, 10678–10690. [DOI] [PubMed] [Google Scholar]
- 28.Horlein A.J., Naar,A.M., Heinzel,T., Torchia,J., Gloss,B., Kurokawa,R., Ryan,A., Kamei,Y., Soderstrom,M., Glass,C.K. and Rosenfeld,M.G. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature, 377, 397–404. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi Y., Furukawa,T., Tun,T., Han,H. and Honjo,T. (1998) LIM protein KyoT2 negatively regulates transcription by association with RBP-J DNA-binding protein. Mol. Cell. Biol., 18, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Garcia I. and Rabbits,T.H. (1994) The LIM domain, a new structural motif found in zinc-finger-like protein. Trends Genet., 10, 315–321. [DOI] [PubMed] [Google Scholar]
- 31.Feuerstein R., Wang,X., Song,D., Cooke,N.E. and Liebhaber,S.A. (1994) The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc. Natl Acad. Sci. USA, 91, 10655–10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmeichel K.L. and Beckerle,M.C. (1994) The LIM domain is a molecular protein-binding interface. Cell, 79, 211–219. [DOI] [PubMed] [Google Scholar]
- 33.Li R., Wang,J., Sun,Q., Wang,J., Yang,X., Huang,H., Zhou,P. and Han,H. (2002) Screening of molecules interacting with LIM domain protein KyoT. Yi Chuan Xue Bao, 29, 175–180. [PubMed] [Google Scholar]
- 34.Satijn D., Gunster,M.J., van der Vlag,J., Hamer,K.M., Schul,W., Alkema,M.J., Saurin,A.J. and Freemont,P.S. (1997) RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell. Biol., 17, 4105–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francis N.J. and Kingston,R.E. (2001) Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol., 2, 409–421. [DOI] [PubMed] [Google Scholar]
- 36.Dahiya A., Wong,S., Gonzalo,S., Gavin,M. and Dean,D.C. (2001) Linking the Rb and polycomb pathways. Mol. Cell, 8, 557–568. [DOI] [PubMed] [Google Scholar]
- 37.Kato H., Taniguchi,Y., Kurooka,H., Minoguchi,S., Sakai,T., Nomura-Okazaki,S., Tamura,K. and Honjo,T. (1997) Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development, 124, 4133–4141. [DOI] [PubMed] [Google Scholar]
- 38.Lai E.C. (2002) Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep., 3, 840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoudi T. and Verrijzer,C.P. (2001) Chromatin silencing and activation by polycomb and trithorax group proteins. Oncogene, 20, 3055–3066. [DOI] [PubMed] [Google Scholar]
- 40.van Lohuizen M. (1999) The trithorax-group and polycomb-group chromatin modifiers: implications for disease. Curr. Opin. Genet. Dev., 9, 355–361. [DOI] [PubMed] [Google Scholar]