Abstract
The interaction of proteins with DNA recognition motifs regulates a number of fundamental biological processes, including transcription. To understand these processes, we need to know which motifs are present in a sequence and which factors bind to them. We describe a method to screen a set of DNA sequences against a precompiled library of motifs, and assess which, if any, of the motifs are statistically over- or under-represented in the sequences. Over-represented motifs are good candidates for playing a functional role in the sequences, while under-representation hints that if the motif were present, it would have a harmful dysregulatory effect. We apply our method (implemented as a computer program called Clover) to dopamine-responsive promoters, sequences flanking binding sites for the transcription factor LSF, sequences that direct transcription in muscle and liver, and Drosophila segmentation enhancers. In each case Clover successfully detects motifs known to function in the sequences, and intriguing and testable hypotheses are made concerning additional motifs. Clover compares favorably with an ab initio motif discovery algorithm based on sequence alignment, when the motif library includes only a homolog of the factor that actually regulates the sequences. It also demonstrates superior performance over two contingency table based over-representation methods. In conclusion, Clover has the potential to greatly accelerate characterization of signals that regulate transcription.
INTRODUCTION
A transcription factor typically interacts with DNA sequences that reflect a common pattern, or motif, characteristic of the factor. Such a motif can be represented by a consensus sequence or, less crudely, by a W × 4 matrix q, where W is the motif’s size in base pairs, and each matrix element q(k,X) is the probability of observing nucleotide X (A, C, G or T) at position k in the motif. It is then possible to scan this matrix along a DNA sequence, assigning a similarity score to each W-long subsequence using a standard log likelihood ratio formula (1). Typically, any subsequence with a similarity score above some threshold is counted as a ‘match’. Unfortunately, these matrices do not contain sufficient information to locate functional in vivo binding sites accurately; at thresholds low enough to recover genuine binding sites, spurious matches occur at a high rate (2). It seems that transcription factors must be guided to their in vivo binding sites by contextual factors such as chromatin structure and interactions with other transcription factors, in addition to their innate DNA binding preferences. It is widely accepted that knowledge of transcription factor binding motifs is not in itself adequate to elucidate transcriptional control mechanisms. In addition to directly investigating contextual factors, another powerful approach to elucidating regulatory mechanisms is to gather DNA sequences that share a common regulatory property, and search for motifs shared by these sequences.
Two general ways of finding shared motifs can be envisaged. The first is to apply ab initio motif discovery algorithms which search for recurring patterns of any kind. The second is to compile a library of all previously characterized motifs and assess whether any of these motifs are statistically over-represented in the sequences. Even though we expect to observe many spurious matches for each motif, it is plausible that if a motif is functionally present in many of the sequences, then the number of matches will be greater than would be expected by chance. The greater generality of ab initio methods is a double-edged sword: they can find completely novel motifs not in any precompiled library, but the motifs must be stronger in order to be statistically significant and detectable, as compared with library-based methods. In addition, ab initio methods tell us nothing about which factor might bind to a predicted motif, whereas precompiled libraries generally include annotations of which motifs are bound by which factors, or families of factors. Much research effort has been devoted to ab initio motif discovery algorithms [see Frith et al. (3) for references], but until recently library-based methods have been neglected, despite the promising aspects of this approach.
Several techniques for testing whether a motif is over-represented in a target set of DNA sequences have recently been published (4–9), and it is instructive to draw connections among these methods, as most of them ultimately reduce to the statistics of contingency tables. All of these methods scan the motif matrix across the target sequences and a set of control sequences, recording matches with similarity score greater than some threshold. Liu et al. (4) proposed counting the number of target and control sequences with and without a match, and deemed the motif over-represented if matching sequences were at least twice as frequent in the target set as the control set. While this 2-fold excess criterion is intuitive, a more rigorous test using the hypergeometric distribution is available (8,10). More explicitly, the data can be cast as a 2 × 2 contingency table (Fig. 1), where A is the number of target sequences with a match, B is the number of control sequences with a match, C is the number of targets without a match and D is the number of controls without a match. A chi-square test or Fisher’s exact test (the hypergeometric distribution) can be used to test the null hypothesis that the sequences with motif matches are evenly distributed among the target and control sets. Elkon et al. (7) use a more intricate procedure, counting the number of sequences with zero matches, one match, two matches or three or more matches in the target and control sets. These data can be cast as a 4 × 2 contingency table and tested using a multivariate hypergeometric distribution.
Figure 1.
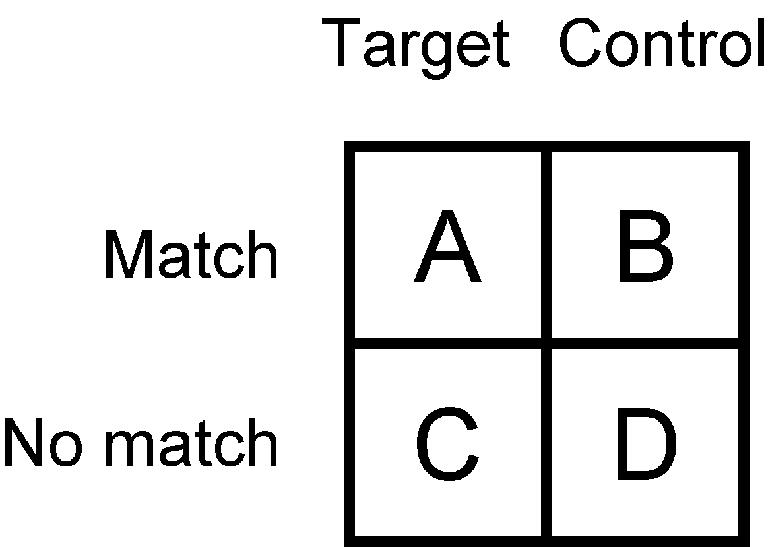
A 2×2 contingency table.
The methods described above can only be applied sensibly if all the target and control sequences have the same length, which is not always easy to arrange. In addition, they may lose statistical power by not counting all matches in each sequence. Several publications have suggested counting all matches in the target and control sequences, and two different binomial formulas have been proposed to test for over-representation (5,6,8,9). In fact, these data can also be cast as a 2 × 2 contingency table (Fig. 1), where A is the number of matches in target sequences, B is the number of matches in control sequences, C is the number of W-long segments in target sequences that do not match and D is the number of W-long segments in control sequences that do not match. To test the null hypothesis, that matches are evenly distributed among the target and control sets, we can imagine randomly drawing A + B matching segments from a pool of A + B + C + D segments of target and control sequences. Equivalently, we can imagine drawing A + C target segments from a pool of A + B + C + D matching and non-matching segments. These two viewpoints lead to the same hypergeometric formula, but to two different binomial approximations of it, which are precisely those described by Sharan et al. (8) versus Aerts et al. (5), Zheng et al. (6) and Haverty et al. (9) These methods assume that occurrence of a match at each W-long segment is independent, which is not quite true because the segments overlap one another, and correlations are also introduced by the presence of repetitive elements in DNA. For these reasons, Zheng et al. (6) needed to treat palindromic motifs specially, and some of their results were greatly influenced by the presence of repeats.
All the previous methods discard potentially useful information by collapsing matrix scores at each location to a binary quantity: above or below the threshold. They also reveal uncertainty regarding whether to count one match per sequence, a few matches per sequence or all matches in each sequence. Regulatory regions of higher eukaryotes often contain multiple binding sites for the same transcription factor, with weaker ‘shadow’ copies of the motif also being observed (11). So consideration of multiple matches per sequence seems likely to help in discovering functional motifs by statistical over-representation. The reason for this site multiplicity is unclear: it might indicate cooperative binding by several factor molecules, it could constitute a mechanism for lateral diffusion of the factor along the DNA and/or the shadow sites might be fossils from the process of binding site turnover (12). Here we report a novel method of combining multiple matches per sequence, which is motivated by a simple thermodynamic model. The matrix score ideally reflects the factor’s binding energy at each location; therefore the score’s exponential should be proportional to the factor’s equilibrium occupancy of that site (1). We suppose that multiple sites simply serve to increase the total occupancy for the sequence, which we estimate by summing the exponentiated matrix score of each site. Finally, we assess whether the estimated total occupancies of the target sequences are greater than would be expected by chance. Thus our method incorporates information from the matrix scores, and combines information from all possible sites per sequence in a biophysically motivated way.
MATERIALS AND METHODS
Our aim is to compare a motif matrix against a set of DNA sequences and assess whether the motif is statistically over-represented (or under-represented) in those sequences. The method proceeds in two steps. We first calculate a single number, which we call the raw score, to quantify the degree of the motif’s presence in the test sequences. The second step is to estimate a P-value for this raw score: the probability of obtaining a raw score of this size or greater merely by chance, computed using background sequence sets. If the P-value is very low (e.g. <0.01), the motif is significantly over-represented in the test sequences, suggesting that it is present for a reason, such as to perform a biological function. If the P-value is very high (e.g. >0.99), the motif is significantly over-represented in the background sequence set. In comparison, it is under-represented in the test sequences, suggesting that it is absent for some good reason, perhaps because its presence in these sequences would be harmful to the organism.
Calculation of the raw score
The raw score calculation is simply a repeated averaging of likelihood ratios (LRs). We begin by calculating the likelihood ratio for a motif’s being present at one particular location in one sequence:
![]()
where W is the width of the motif, L denotes the location being considered, Lk is the nucleotide at position k within this location and p(X) is the background probability of observing nucleotide X, estimated from the frequency of X in that sequence. LR1 is the exponent of the standard motif matrix score and is proportional to the factor’s equilibrium occupancy of this site in a simple thermodynamic model (1,13,14). The likelihood ratio for a motif being present at any location in a sequence S is the average of LR1 taken over all locations in S (on both strands):
![]()
where MS is the number of locations in the sequence. LR2(S) is proportional to the factor’s total equilibrium occupancy of the sequence. Note that LR2(S) is a function of the length of promoter sequence S. If S is extended to include nucleotides that do not include the motif, LR2(S) would decrease. Thus the user is advised to keep promoter sequences short, provided that regions that are most likely to contain motifs have been included. Given sets of equal-length target and control sequences, it is possible to test for motif over-representation by ranking the LR2 scores from both sets and applying the Wilcoxon rank-sum test. Since the sequences are generally not of equal length, we take a different approach.
We would now like to combine the LR2 values for each sequence into one overall number reflecting the motif’s presence in the sequence set as a whole. One possibility is to take the product of the LR2 values, which corresponds to the hypothesis that the motif is present in every sequence. However, this hypothesis is too strict; in realistic applications, the motif is likely to be absent from some fraction of the sequences owing to experimental error in gathering the sequences or to heterogeneity of biological regulatory mechanisms. Instead, we suppose that the motif is present in some number i out of N sequences, where we attach equal prior probability to i taking any value between 1 and N. There are NCi ways of selecting i out of N sequences, and the likelihood ratio for the motif being present in any i sequences is the average over all of these ways:
![]()
In this equation, A runs over all sets of i out of N sequences. The final likelihood ratio is the average over all values of i:
![]()
The raw score is defined to be ln(LR4). The raw score increases when more of the sequences contain good motif matches, and also when there are more good matches in a sequence. It favors cases where motifs are distributed across many of the sequences rather than concentrated in a few of them. LR4 can be interpreted as the factor’s average equilibrium occupancy in a set of sequences.
A fast algorithm for the average of all products
On the face of it, equation 3 requires the enumeration of all ways of selecting i out of N objects, which rapidly becomes infeasible even for moderate values of N and i. Fortunately, there is a recurrence relation that allows us to calculate LR3(i) for all values of i in time proportional to N2. Let Ti j denote the sum of all products of i terms from among the first j elements of the vector LR2. Then
![]()
The boundary conditions are T0 j = 1 and Ti i–1 = 0. LR3(i) is equal to Ti N/NCi. In order to avoid overflow errors, the division by NCi can be folded into the recurrence formula
![]()
The same boundary conditions apply to Aij as to Tij, and LR3(i) is identical to AiN.
Estimation of P-values
We would like to know the probability of observing a given raw score or greater by chance, but there are multiple meanings of the phrase ‘by chance’. Some examples, in order of increasing conservatism, are the probability of obtaining this score for randomly shuffled DNA sequences, for randomly chosen fragments of the organism’s genome or for random promoter sequences from the organism. It would not be surprising if a motif such as the TATA box were over-represented by one of these standards but not by another, and the ‘right answer’ is context dependent.
Our method provides several different ways of estimating P-values. The first is to shuffle the nucleotides randomly within each sequence and calculate the motif’s raw score for the shuffled sequences. This shuffling and raw score recalculation is repeated many times (e.g. 1000), and the fraction of times that the randomized raw score exceeds the real raw score becomes the P-value. The second approach is to count the frequencies of the 16 dinucleotides in each sequence, generate random sequences of the same lengths as the originals based on these dinucleotide abundances and recalculate raw scores as above. This technique takes into account the reduced abundance of the CpG dinucleotide in mammalian sequences, which is important for assessing motifs such as E2F that contain this dinucleotide in conserved positions (15). The third approach is to shuffle the columns of the motif matrix, i.e. each vector of four numbers for A, C, G and T is kept intact internally but the order of these vectors is shuffled. The raw scores of these shuffled matrices against the real sequences are used to obtain a P-value. Finally, a set of background DNA sequences may be supplied to the algorithm. This background set should be much larger than the sequence set being studied; we typically use a whole chromosome, or a large set of promoters from the organism. The algorithm repeatedly extracts random fragments of the background sequences, matched by length to the target sequences, calculates a raw score for each set of fragments and uses these to derive a P-value. For the results presented in this study, P-values were obtained using either the nucleotide shuffling or background sequence approach, with 1000 randomizations.
Multiple testing
We typically test whether each motif from a library of ≥100 is significantly over- or under-represented in a given sequence set, which means that by chance alone it is likely that a few motifs will have P-values more significant than 0.01. Nevertheless, all the P-values in this paper were obtained by performing 1000 randomizations, since it becomes computationally tedious to do more, and motifs with P-values ≤0.01 or ≥0.99 are listed. In practice, we find many motifs with P-values of dead zero, i.e. the raw scores were never equaled in 1000 randomizations, which is highly unlikely to occur by chance (Tables 1A–5). We also find more motifs with P-values ≤0.01 than the handful expected by chance, and in most cases P-values were obtained relative to multiple different backgrounds, and only motifs with significant P-values relative to all backgrounds are listed. Therefore we are confident that the vast majority of motif predictions made here are not merely due to chance.
Table 1A. Significant Jaspar motifs in sequences upstream of human dopamine responsive genes.
| Motif | Raw score | P-value relative to | ||
|---|---|---|---|---|
| Human chrom. 20 | Human promoters | Human CpG islands | ||
| *HFH-2 Forkhead | 40.1 | 0 | 0.002 | 0.004 |
| *HFH-3 Forkhead | 29.2 | 0 | 0.002 | 0.005 |
| *MEF2 MADS | 13.1 | 0 | 0 | 0.007 |
| *AGL3 MADS | 11.8 | 0 | 0 | 0 |
| *SRF MADS | 7.55 | 0 | 0 | 0 |
| *Agamous MADS | 6.59 | 0 | 0 | 0.004 |
| *CREB bZIP | 0.0429 | 0.003 | 0.002 | 0.009 |
| *bZIP910 bZIP | –0.00662 | 0 | 0 | 0.001 |
Table 5. Significant Jaspar motifs in Drosophila embryonic regulatory regions.
| Motif | Raw score | P-value relative to Drosophila chrom. 2R |
|---|---|---|
| Hunchback Zn-finger | 33 | 0 |
| *HMG-IY HMG | 22.7 | 0 |
| *Gklf Zn-finger | 9.01 | 0.001 |
| *Dorsal_1 REL | 8.69 | 0 |
| *Dof3 Zn-finger | 7.06 | 0.005 |
| *c-REL REL | 5.48 | 0.007 |
| cEBP bZIP | 5.46 | 0.009 |
| *c-ETS ETS | 3.84 | 0 |
| *E74A ETS | 3.65 | 0.009 |
| *SPI-1 ETS | 2.15 | 0.008 |
| *RORalpha-1 nuclear receptor | 0.555 | 0.005 |
| *EN-1 homeo | –1.38 | 0.992 |
| *FREAC-7 Forkhead | –1.68 | 1 |
| *Snail Zn-finger | –2.87 | 0.993 |
Motifs indicated with an asterisk are derived from in vitro site selection experiments.
Treatment of masked nucleotides
We learned from experience that it is necessary to treat masked nucleotides carefully in order to avoid artefactual results. Although not performed for the results shown here, it is possible to mask, i.e. replace with ‘n’, nucleotides that occur in repetitive elements, prior to searching for over-represented motifs. Locations that overlap masks are not counted in equation 2. When shuffling sequences, masks are left in place and only non-masked bases are shuffled. When generating dinucleotide-based sequences, unmasked sequences are generated initially, and then masks are copied from the original sequences to the corresponding locations in the generated sequences. When comparing with background sets, fragments are chosen from entirely unmasked portions of the background sequences, and masks are copied from the target sequences as above. These measures ensure that the control sequences resemble the targets regarding masks.
Motif libraries
Our method requires an extensive precompiled library of motif matrices. Two such libraries are used here: Jaspar (16), with 108 motifs, and the 428 vertebrate motifs from the Transfac Professional database version 6.3 (17). Jaspar is in some ways more convenient since it lacks commercial restrictions and it attempts to be non-redundant, but the Transfac motif collection is more extensive.
RESULTS AND DISCUSSION
We wrote a C++ program called Clover (Cis-eLement OVer-representation), which determines which motifs from a precompiled motif library are over- or under-represented in a set of DNA sequences. This program is available for downloading at http://zlab.bu.edu/clover/. Further details of the sequence sets studied in this paper are available at http://zlab.bu.edu/clover/sup/.
Comparison with ab initio motif finding
It is instructive to compare our library-based motif finder with an ab initio method that performs multiple local sequence alignment. While alignment methods can find novel motifs not in any precompiled library, library-based techniques may have greater power to detect weak motifs in long sequences, since they restrict the types of motif to be searched for. We tested Clover’s ability to find mammalian estrogen response elements (EREs) embedded in randomly generated DNA sequences of varying length, using the Jaspar collection of 108 motifs. This test is particularly apt because Jaspar does not contain an ERE; however, it contains a PPARγ motif that closely resembles an ERE (Fig. 2), in addition to six nuclear receptor motifs that contain ERE half-sites. In real applications it is quite likely that the sequences to be analyzed will contain motifs that are absent from the library, although the library may contain similar motifs for related binding factors. Jaspar’s PPARγ motif differs from the ERE mainly in having strong base preferences outside the conserved region of the ERE (e.g. the T at position 2); it also exhibits slightly stricter preferences within the GGTCA half-sites.
Figure 2.

Pictogram representations of the ERE (3) and the Jaspar PPARγ motif (C Burge and F White, http://genes.mit.edu/pictogram.html).
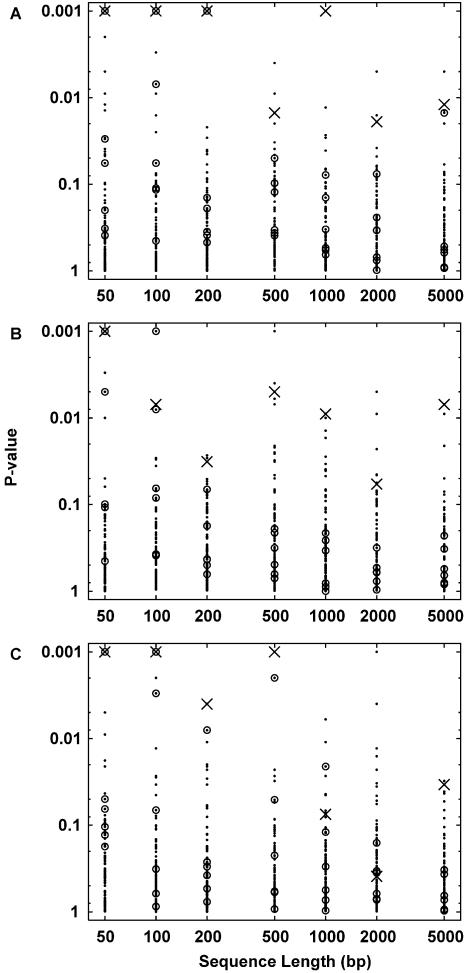
Each of 15 EREs was embedded into a random DNA sequence, and zero, 5 or 15 decoy sequences (randomly generated sequences lacking EREs) were added. Sequence lengths between 50 and 5000 bp were tested. For each sequence set, the P-values of the 108 Jaspar motifs, relative to shuffling the sequences, were recorded (Fig. 3). In the majority of cases the PPARγ motif has a P-value <0.01, and is also the most significantly over-represented motif. In the remaining cases PPARγ is usually among the most significant handful of motifs, and occasionally it is surpassed by one of the other ERE-like nuclear receptors. Only for long sequences with many decoys does detection become less robust.
Figure 3.
Detection by Clover of ERE motifs embedded in random DNA sequences of varying length. In all panels, the P-values of the 108 Jaspar motifs are plotted as dots. P-values of zero were increased to 0.001 to fit on the log scale. Crosses indicate the PPARγ motif, and circles indicate the six other ERE-like nuclear receptor motifs. (A) Results for 15 ERE-containing sequences with no decoy sequences. (B) Results for 15 ERE-containing sequences with five decoy sequences. (C) Results for 15 ERE-containing sequences with 15 decoy sequences.
Clover compares favorably with our alignment-based program GLAM, which was tested on the same datasets (3). GLAM returns completely random alignments for the 5000-bp sequence sets, and also for sequence lengths ≥1000 bp when decoys are present. In many of these cases Clover finds ERE-like motifs to be the most significant or among the top handful of motifs. Even when GLAM succeeds, it does not always attach a significant P-value to its alignment, meaning that we cannot tell that it has succeeded. Thus library-based motif finders are promising alternatives to alignment-based methods even when the library only contains a homolog of the motif to be found. When Clover is given the ERE matrix (which is constructed, in part, from the embedded EREs), it assigns it a P-value of zero in every single case.
Comparison with contingency table based methods
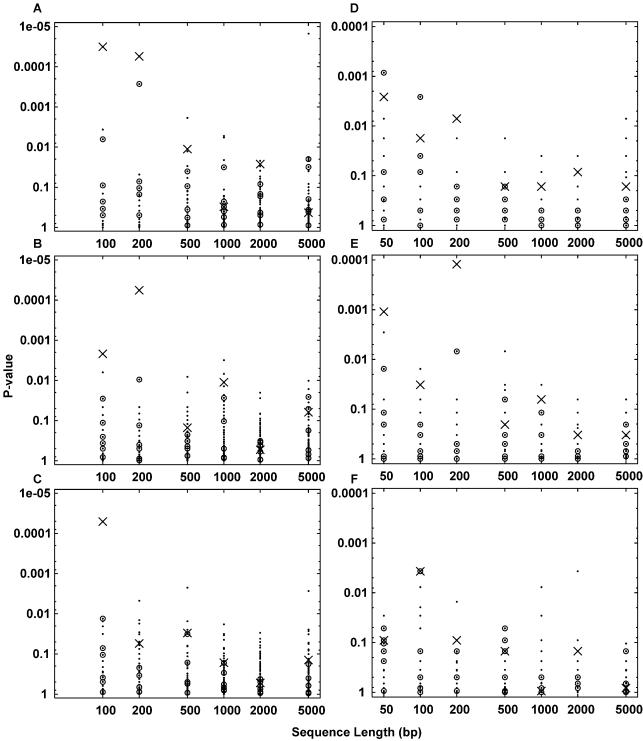
For comparison, two contingency table based methods, ‘motif counting’ and ‘sequence counting’, were also tested on these sequence sets. As described in the Introduction, these methods scan motif matrices across target and control sequences, recording matches with score greater than some arbitrary threshold. We used the same target sets as above (EREs embedded in random DNA), and for each target set a control set containing the same number of same-length random DNA sequences was constructed (without EREs). As the names imply, the sequence counting method counts the number of sequences with one or more motif matches in the target and control sets, whereas the motif counting method counts the total number of matches in each set. For the motif counting method we used score thresholds such that 0.1% of locations in the control set were deemed matches, as suggested by Haverty et al. (9). For the sequence counting method, we chose thresholds such that 10% of sequences in the control set contained one or more matches, similarly to Elkon et al. (7). P-values for over-representation of counts in the target set were calculated using Fisher’s exact test.
Although the contingency table based methods tend to rank PPARγ among the top handful of most over-represented motifs (Fig. 4), it is not ranked highest in most cases, and never when the sequence length is >200 bp. Moreover, in most cases motifs that do not resemble the ERE are ranked highest. Clover’s advantage in this comparison stems from incorporating motif scores rather than cutting them off at a threshold. It should be noted that these test sets are artificial in that they have only one motif per sequence which favors sequence counting, and they lack repetitive sequences that can interfere with the motif counting approach. The problem with real test sets, of course, is that the correct answer is not known for certain.
Figure 4.
Detection by contingency table based methods of EREs embedded in random DNA sequences of varying length. In all panels, the P-values of the 108 Jaspar motifs are plotted as dots. Crosses indicate the PPARγ motif, and circles indicate the six other ERE-like nuclear receptor motifs. (A, B, C) Motif counting method. Length 50 sequences were not analyzed because the number of possible locations is <1000 for some motifs, making the 0.1% threshold criterion impossible. (D, E, F) Sequence counting method. (A, D) Results for 15 ERE-containing sequences with no decoy sequences. (B, E) Results for 15 ERE-containing sequences with five decoy sequences. (C, F) Results for 15 ERE-containing sequences with 15 decoy sequences.
To further illustrate the method’s utility, Clover was used to predict functional DNA motifs in a diverse range of biological systems. We chose systems of special biological or medical interest (e.g. Drosophila segmentation enhancers, dopamine-responsive genes), where collections of functionally similar promoters are available, and some functional motifs are already known so that the predictions can be checked.
Analysis of dopamine responsive promoters
In order to find DNA motifs involved in transcriptional regulation by dopamine signaling, we collected 1500 bp human genomic sequences upstream of 23 dopamine- responsive genes (18). Clover was used to search for significant motifs from the Jaspar library in these sequences. P-values were obtained relative to three background datasets: human chromosome 20, 2000-bp sequences upstream of human genes and human CpG islands, the latter two being derived from annotations at the UCSC genome website (19). Several motifs are significantly over-represented relative to all backgrounds (Table 1A). The presence of the CREB motif is consistent with previous knowledge: dopamine signaling activates the cAMP pathway and induces transcription via the CREB protein (20). In addition, the program makes novel predictions that a Forkhead and a MADS-box factor may be involved in this signaling pathway. We reiterate that Clover makes direct predictions about binding motifs rather than specific proteins.
To strengthen these results, the analysis was repeated on sequences upstream of the orthologous genes in mouse. CREB, MADS and Forkhead motifs are again found to be over-represented (Table 1B), increasing our confidence in these predictions. Some motifs receive negative raw scores, indicating that the sequences lack good matches for these motifs, but nevertheless achieve low P-values relative to all background sequence sets (Table 1A and 1B). This result might be expected if the motif itself is not functionally present in the sequences (i.e. the corresponding transcription factor does not interact with the sequences), but a similar motif for a homologous factor is. Another explanation is that motifs derived from in vitro site selection (indicated with asterisks in all tables), including CREB, might exhibit less variability than the real in vivo motifs.
Table 1B. Significant Jaspar motifs in sequences upstream of mouse dopamine responsive genes.
| Motif | Raw score | P-value relative to | |
|---|---|---|---|
| Mouse chrom. 19 | Mouse promoters | ||
| *Pax-4 paired-homeo | 24.8 | 0.004 | 0.007 |
| Broad-complex_1 Zn-finger | 22.2 | 0 | 0.002 |
| *SQUA MADS | 17 | 0 | 0 |
| *SRF MADS | 15.9 | 0 | 0 |
| *MEF2 MADS | 12.7 | 0 | 0 |
| *AGL3 MADS | 11.9 | 0 | 0 |
| *Agamous MADS | 5.68 | 0 | 0 |
| *Brachyury T-BOX | 3.01 | 0.002 | 0.001 |
| Broad-complex_2 Zn-finger | 2.68 | 0.001 | 0.006 |
| *FREAC-7 Forkhead | 1.65 | 0.001 | 0.007 |
| *Athb-1 homeo-ZIP | 0.912 | 0.001 | 0.009 |
| cEBP bZIP | –0.185 | 0.002 | 0.005 |
| *CREB bZIP | –0.241 | 0 | 0 |
| *S8 homeo | –0.48 | 0 | 0.006 |
| *HLF bZIP | –1.8 | 0 | 0.009 |
| *bZIP910 bZIP | –1.82 | 0 | 0.001 |
Motifs indicated with an asterisk are derived from in vitro site selection experiments.
Analysis of sequences flanking LSF binding sites
In order to understand better transcriptional regulation involving the transcription factor LSF, we searched for significant motifs in 15 (mostly mammalian) sequences flanking LSF binding sites. No Jaspar motifs were found to be significant, but several Transfac motifs are significantly over-represented relative to three background datasets (Table 2). The LSF motif itself was not recovered, because Transfac lacked a high-quality matrix for LSF (we have since submitted an LSF matrix to Transfac) or a homolog with similar binding properties. However, the presence of the NFκB motif is consistent with previous knowledge: NFκB interacts physically with LSF and can synergize with it to activate transcription of the mouse serum amyloid A3 gene (21). This result provides further evidence that Clover detects functionally relevant motifs, and suggests that the LSF–NFκB interaction may be more widespread than previously thought, since the serum amyloid A3 gene was not among the 15 sequences that we collected. The discovery of SRF, AP-1 and Oct motifs in these sequences suggests that these factors may also be involved in regulation by LSF.
Table 2. Significant Transfac motifs in sequences flanking LSF binding sites.
| Motif | Raw score | P-value relative to | ||
|---|---|---|---|---|
| Human chrom. 20 | Human promoters | Human CpG islands | ||
| V$NFKB_C | 7.08 | 0.002 | 0.002 | 0.001 |
| V$NFKB_Q6 | 5.61 | 0.007 | 0.006 | 0.008 |
| V$OCT1_B | 4.96 | 0.007 | 0.002 | 0.008 |
| V$AP1_Q2 | 4.44 | 0.008 | 0.004 | 0.003 |
| V$SRF_C | 3.57 | 0.003 | 0.003 | 0.009 |
| V$SRF_01 | 2.98 | 0 | 0 | 0.007 |
| V$OLF1_01 | –3.4 | 0.997 | 0.993 | 0.992 |
| V$PAX5_01 | –3.41 | 0.996 | 0.994 | 0.993 |
Analysis of muscle regulatory regions
To validate Clover’s ability to detect functional motifs more comprehensively, in a more extensively studied system, we applied it to a well characterized set of 27 muscle regulatory regions from mammals and birds (22), using the Jaspar database. Four motifs known to function in these sequences, MEF2, Myf, TEF-1 and SRF (22), are found to be over-represented with P-values of zero relative to all backgrounds (Table 3). The Myf and TEF-1 motifs were constructed from in vivo binding site compilations, very likely including sites within these 27 sequences, and so their recovery is perhaps not surprising. On the other hand, the MEF2 and SRF motifs were constructed from in vitro site selection data, and so their recovery constitutes further evidence of Clover’s ability to detect functional motifs. Interestingly, Forkhead and SOX-family motifs are over-represented with equally strong significance, hinting at undiscovered regulatory influences on these sequences. Separate studies support the involvement of Fox and Sox factors in gene regulation in muscle: Sox15 can specifically inhibit activation of muscle-specific genes (23), and mice lacking myocyte nuclear factor/Foxk1, a Forkhead family member, exhibit atrophic skeletal muscles and impaired muscle regeneration after injury (24). Thus the method predicts previously known motifs and plausible novel candidates with high confidence.
Table 3. Significant Jaspar motifs in muscle regulatory regions.
| Motif | Raw score | P-value relative to | ||
|---|---|---|---|---|
| Human chrom. 20 | Human promoters | Human CpG islands | ||
| *MEF2 MADS | 30.1 | 0 | 0 | 0 |
| Myf bHLH | 29.5 | 0 | 0 | 0 |
| *AGL3 MADS | 20.3 | 0 | 0 | 0 |
| *SQUA MADS | 17.2 | 0 | 0 | 0 |
| TEF-1 TEA | 15.1 | 0 | 0 | 0 |
| *SRF MADS | 13 | 0 | 0 | 0 |
| *FREAC-7 Forkhead | 12 | 0 | 0 | 0 |
| *MZF_1-4 Zn-finger | 11.9 | 0.005 | 0.005 | 0.009 |
| *SRY HMG | 11.2 | 0 | 0 | 0.002 |
| *Agamous MADS | 9.4 | 0 | 0 | 0 |
| Broad-complex_2 Zn-finger | 6 | 0 | 0 | 0.006 |
| *SOX17 HMG | 5.91 | 0 | 0 | 0 |
| *Sox-5 HMG | 5.21 | 0 | 0.001 | 0.003 |
| TBP TATA-box | 4.33 | 0 | 0.001 | 0.001 |
Motifs indicated with an asterisk are derived from in vitro site selection experiments.
Analysis of liver regulatory regions
A compilation of 16 liver regulatory regions from mammals and birds (25) was also analyzed using the 428 Transfac motifs. Most of the over-represented motifs are hepatic nuclear factors (HNF1, 3, 4 and 6) (Table 4), which are known to be important for liver-specific gene regulation. The motif for the Xenopus Forkhead domain 3 (XFD3), which is the Xenopus homolog of HNF-3β, is constructed from in vitro site selection experiments, again providing independent evidence that Clover detects biologically relevant motifs. A number of motifs are very significantly under-represented in these liver sequences, with P-values of 1 (i.e. the raw scores were always exceeded in random fragments of the background sequences). We propose that the presence of these motifs in liver regulatory regions would be detrimental. Some of the under-represented motifs are GC rich (MZF1, SPZ1, AP2 and MAZR). Although the liver sequences do have a slightly lower GC content (42.7%) than any of the background sets, the difference in GC content from human chromosome 20 (44.1%) is minimal and unlikely to generate such under-representation. The significance of this unexpected under-representation of sites remains to be experimentally determined. In the case of the muscle initiator sequence, whose binding protein(s) has not yet been identified, we hypothesize that its absence in liver-specific regulatory regions (i) prevents inappropriate expression of these genes in muscle and/or (ii) prevents inappropriate repression of these genes in liver, if one role of the ‘muscle initiator element’ is actually to repress expression of muscle-specific genes in non-muscle cells.
Table 4. Significant Transfac motifs in liver regulatory regions.
| Motif | Raw score | P-value relative to | ||
|---|---|---|---|---|
| Human chrom. 20 | Human promoters | Human CpG islands | ||
| V$HNF1_01 | 32 | 0 | 0 | 0 |
| V$HNF1_C | 25.5 | 0 | 0 | 0 |
| V$HNF3ALPHA_Q6 | 8.38 | 0.001 | 0.002 | 0 |
| V$HNF4ALPHA_Q6 | 7.23 | 0.001 | 0 | 0.001 |
| V$HNF4_01 | 5.77 | 0.003 | 0.001 | 0.002 |
| V$HNF3B_01 | 5.76 | 0.001 | 0.004 | 0.003 |
| V$COUP_01 | 4.9 | 0.004 | 0.002 | 0.002 |
| *V$XFD3_01 | 4.05 | 0.004 | 0.004 | 0.009 |
| V$POLY_C | 4.03 | 0.005 | 0.002 | 0.004 |
| V$HNF6_Q6 | 3.66 | 0.001 | 0.001 | 0.005 |
| V$AP1_Q2 | 3.46 | 0.01 | 0.006 | 0.004 |
| V$IPF1_Q4 | 2.28 | 0.005 | 0.002 | 0.006 |
| *V$MZF1_01 | –3.03 | 0.995 | 0.999 | 1 |
| *V$SPZ1_01 | –3.78 | 1 | 1 | 1 |
| V$AP2_Q6 | –4.03 | 0.996 | 1 | 1 |
| V$MUSCLE_INI_B | –4.07 | 0.998 | 0.999 | 1 |
| V$CACCC_Q6 | –4.29 | 0.992 | 0.994 | 0.999 |
| V$MINI20_B | –4.31 | 1 | 1 | 1 |
| V$MINI19_B | –4.34 | 0.999 | 1 | 1 |
| *V$PAX4_01 | –4.66 | 0.997 | 0.997 | 0.999 |
| *V$MAZR_01 | –6.07 | 0.998 | 1 | 1 |
Motifs indicated with an asterisk are derived from in vitro site selection experiments.
Analysis of Drosophila segmentation enhancers
Finally, we analyzed 19 Drosophila regulatory regions active in the embryonic segmentation process (26). There is previous evidence that the transcription factors Bicoid, Caudal, Hunchback, Krüppel and Knirps regulate these sequences (26). Consistent with this evidence, the Hunchback motif is significantly over-represented, and although Krüppel is absent from Jaspar, the Krüppel-like factor Gklf is recovered (Table 5). We also discover that HMG-IY (recently renamed HMGA), Dorsal/REL and Ets motifs are highly over- represented. The relevant HMGA protein may well be Lilliputian, which appears to regulate fushi tarazu and huckebein (27), both segmentation genes although not among the 19 analyzed here. Dorsal and the Ets factors Pointed or Yan, which are active in the Drosophila embryo (28), are also good candidate factors for regulating these sequences. Furthermore, Snail, Engrailed and Forkhead motifs are under-represented. These crucial embryonic enhancers clearly undergo stringent selection to avoid unwanted regulatory interference from other transcription factors. For instance, it would seem evolutionarily advantageous to exclude binding motifs for the Snail repressor, since Snail is activated by Dorsal and would counteract the desired activation by Dorsal of these genes (29).
SUMMARY
We propose that our method for finding over-represented motifs opens a door to the reverse genetics of regulatory elements. In every regulatory system that we examined, some previously known motifs were recovered and a manageable number of novel candidate motifs were identified. Screening sequences against a precompiled motif library is superior to ab initio motif discovery algorithms in cases where functional motifs are likely to be present in the library: it has greater power to detect weak motifs, it is less prone to be misled by repetitive elements and accurate estimates of statistical significance are more readily available. Nonetheless, ab initio methods will always be useful for studying novel types of regulatory mechanism. The ability to find over-represented motifs in regulatory regions should greatly assist methods that predict regulatory regions by finding clusters of specific motifs in DNA sequences (26,30–36). Previously, the main bottleneck with such methods was ignorance of which motifs cluster with one another to form the various types of regulatory region. The power of the motif library screening approach obviously depends on the coverage and accuracy of the motif library that is used. The tools are now in hand to obtain accurate motif models for every transcription factor in a genome (37,38); our understanding of gene regulation is poised for a quantum leap when such comprehensive libraries become available.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Kim McCall, Peter Haverty, Dmitriy Leyfer, Anason Halees, Katya Ravid and Jun Lu for helpful discussions, and Wyeth Wasserman and Albin Sandelin for assistance with Jaspar. We thank Joseph Ryan and Li Li for beta-testing Clover. This work was partly supported by NSF Major Research Instrument grant DBI-0116574. U.H. and M.C.F. were supported in part by funding from the National Institutes of Health (R01-CA81157). Z.W. and M.C.F. were supported in part by NSF grant DBI-0078194. U.H. and Z.W. were partially funded by NIH grant 1P20GM066401–01. Y.F. and Z.W. were partially funded by NIH grant 1R01HG03110–01. L.Y. and J.C. were funded by NIH grant NS37403. M.C.F. is a Howard Hughes Medical Institute Predoctoral Fellow.
REFERENCES
- 1.Stormo G.D. (2000). DNA binding sites: representation and discovery. Bioinformatics, 16, 16–23. [DOI] [PubMed] [Google Scholar]
- 2.Pennacchio L.A. and Rubin,E.M. (2001). Genomic strategies to identify mammalian regulatory sequences. Nature Rev. Genet., 2, 100–109. [DOI] [PubMed] [Google Scholar]
- 3.Frith M.C., Hansen,U., Spouge,J.L. and Weng,Z. (2004). Finding functional sequence elements by multiple local alignment. Nucleic Acids Res., 32, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu R., McEachin,R.C. and States,D.J. (2003). Computationally identifying novel NF-kappa B-regulated immune genes in the human genome. Genome Res., 13, 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aerts S., Thijs,G., Coessens,B., Staes,M., Moreau,Y. and De Moor,B. (2003). Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res., 31, 1753–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng J., Wu,J. and Sun,Z. (2003). An approach to identify over-represented cis-elements in related sequences. Nucleic Acids Res., 31, 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elkon R., Linhart,C., Sharan,R., Shamir,R. and Shiloh,Y. (2003). Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res., 13, 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharan R., Ovcharenko,I., Ben-Hur,A. and Karp,R.M. (2003). CREME: a framework for identifying cis-regulatory modules in human-mouse conserved segments. Bioinformatics, 19 (Suppl 1), I283–I291. [DOI] [PubMed] [Google Scholar]
- 9.Haverty P.M., Hansen,U. and Weng,Z. (2004). Computational inference of transcriptional regulatory networks from expression profiling and transcription factor binding site identification. Nucleic Acids Res., 32, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000). Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 11.Papatsenko D.A., Makeev,V.J., Lifanov,A.P., Regnier,M., Nazina,A.G. and Desplan,C. (2002). Extraction of functional binding sites from unique regulatory regions: the Drosophila early developmental enhancers. Genome Res., 12, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dermitzakis E.T. and Clark,A.G. (2002). Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol., 19, 1114–1121. [DOI] [PubMed] [Google Scholar]
- 13.Schneider T.D., Stormo,G.D., Gold,L. and Ehrenfeucht,A. (1986). Information content of binding sites on nucleotide sequences. J. Mol. Biol., 188, 415–431. [DOI] [PubMed] [Google Scholar]
- 14.Berg O.G. and von Hippel,P.H. (1987). Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol., 193, 723–750. [DOI] [PubMed] [Google Scholar]
- 15.Kel-Margoulis O.V., Tchekmenev,D., Kel,A.E., Goessling,E., Hornischer,K., Lewicki-Potapov,B. and Wingender,E. (2003). Composition-sensitive analysis of the human genome for regulatory signals. In Silico Biol., 3, 145–171. [PubMed] [Google Scholar]
- 16.Sandelin A., Alkema,W., Engstrom,P., Wasserman,W.W. and Lenhard,B. (2004). JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res., 32, D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Pruss,M., Reuter,I. and Schacherer,F. (2000). TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berke J.D., Paletzki,R.F., Aronson,G.J., Hyman,S.E. and Gerfen,C.R. (1998). A complex program of striatal gene expression induced by dopaminergic stimulation. J. Neurosci., 18, 5301–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karolchik D., Baertsch,R., Diekhans,M., Furey,T.S., Hinrichs,A., Lu,Y.T., Roskin,K.M., Schwartz,M., Sugnet,C.W., Thomas,D.J. et al. (2003). The UCSC Genome Browser Database. Nucleic Acids Res., 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konradi C., Leveque,J.C. and Hyman,S.E. (1996). Amphetamine and dopamine-induced immediate early gene expression in striatal neurons depends on postsynaptic NMDA receptors and calcium. J. Neurosci., 16, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bing Z., Huang,J.H. and Liao,W.S. (2000). NFkappa B interacts with serum amyloid A3 enhancer factor to synergistically activate mouse serum amyloid A3 gene transcription. J. Biol. Chem., 275, 31616–31623. [DOI] [PubMed] [Google Scholar]
- 22.Wasserman W.W. and Fickett,J.W. (1998). Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol., 278, 167–181. [DOI] [PubMed] [Google Scholar]
- 23.Beranger F., Mejean,C., Moniot,B., Berta,P. and Vandromme,M. (2000). Muscle differentiation is antagonized by SOX15, a new member of the SOX protein family. J. Biol. Chem., 275, 16103–16109. [DOI] [PubMed] [Google Scholar]
- 24.Garry D.J., Meeson,A., Elterman,J., Zhao,Y., Yang,P., Bassel-Duby,R. and Williams,R.S. (2000). Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl Acad. Sci. USA, 97, 5416–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krivan W. and Wasserman,W.W. (2001). A predictive model for regulatory sequences directing liver-specific transcription. Genome Res., 11, 1559–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman B.P., Nibu,Y., Pfeiffer,B.D., Tomancak,P., Celniker,S.E., Levine,M., Rubin,G.M. and Eisen,M.B. (2002). Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA, 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang A.H., Neufeld,T.P., Rubin,G.M. and Muller,H.A. (2001). Transcriptional regulation of cytoskeletal functions and segmentation by a novel maternal pair-rule gene, lilliputian. Development, 128, 801–813. [DOI] [PubMed] [Google Scholar]
- 28.Hsu T. and Schulz,R.A. (2000). Sequence and functional properties of Ets genes in the model organism Drosophila. Oncogene, 19, 6409–6416. [DOI] [PubMed] [Google Scholar]
- 29.Stathopoulos A. and Levine,M. (2002). Dorsal gradient networks in the Drosophila embryo. Dev. Biol., 246, 57–67. [DOI] [PubMed] [Google Scholar]
- 30.Wagner A. (1999). Genes regulated cooperatively by one or more transcription factors and their identification in whole eukaryotic genomes. Bioinformatics, 15, 776–784. [DOI] [PubMed] [Google Scholar]
- 31.Frith M.C., Hansen,U. and Weng,Z. (2001). Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics, 17, 878–889. [DOI] [PubMed] [Google Scholar]
- 32.Frith M.C., Spouge,J.L., Hansen,U. and Weng,Z. (2002). Statistical significance of clusters of motifs represented by position specific scoring matrices in nucleotide sequences. Nucleic Acids Res., 30, 3214–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frith M.C., Li,M.C. and Weng,Z. (2003). Cluster-Buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res., 31, 3666–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson O., Alkema,W., Wasserman,W.W. and Lagergren,J. (2003). Identification of functional clusters of transcription factor binding motifs in genome sequences: the MSCAN algorithm. Bioinformatics, 19 (Suppl 1), I169–I176. [DOI] [PubMed] [Google Scholar]
- 35.Rajewsky N., Vergassola,M., Gaul,U. and Siggia,E.D. (2002). Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics, 3, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey T.L. and Noble,W.S. (2003). Searching for statistically significant regulatory modules. Bioinformatics, 19 (Suppl 2), II16–II25. [DOI] [PubMed] [Google Scholar]
- 37.Roulet E., Busso,S., Camargo,A.A., Simpson,A.J., Mermod,N. and Bucher,P. (2002). High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol., 20, 831–835. [DOI] [PubMed] [Google Scholar]
- 38.Bulyk M.L., Gentalen,E., Lockhart,D.J. and Church,G.M. (1999). Quantifying DNA–protein interactions by double-stranded DNA arrays. Nat. Biotechnol., 17, 573–577. [DOI] [PubMed] [Google Scholar]