Abstract
Thousands of somatic mutations accrue in most human cancers and causes are largely unknown. We recently showed that the DNA cytosine deaminase APOBEC3B accounts for up to half of the mutational load in breast carcinomas expressing this enzyme. Here, we address whether APOBEC3B is broadly responsible for mutagenesis in multiple tumor types. We analyzed gene expression data and mutation patterns, distributions, and loads for 19 different cancer types, totaling over 4,800 exomes and 1,000,000 somatic mutations. Remarkably, APOBEC3B is upregulated and its preferred target sequence is frequently mutated and clustered in at least 6 distinct cancers: bladder, cervix, lung (adeno- and squamous cell), head/neck, and breast. Interpreted in light of prior genetic, cellular, and biochemical studies, the most parsimonious conclusion based on these global analyses is that APOBEC3B catalyzed genomic uracil lesions are responsible for a large proportion of both dispersed and clustered mutations in multiple distinct cancers.
Somatic mutations are essential for normal cells to develop into cancers. Partial and full tumor genome sequences have revealed the existence of hundreds to thousands of mutations in most cancers1–10. The observed mutation spectrum is the result of DNA lesions that either escaped repair or were misrepaired. This spectrum can be used to help determine the cause or source of the initial damage. For instance, the C-to-T transition bias in skin cancers can be explained by a mechanism in which UV-induced lesions, cyclobutane pyrimidine dimers (C*C, C*T, T*C, or T*T), are bypassed by DNA polymerase-catalyzed insertion of two adenine bases opposite each unrepaired lesion11. A second round of DNA replication or excision and repair of the pyrimidine dimer results in C-to-T transitions. Notably, the nature of this type of DNA damage dictates that each resulting C-to-T transition occurs in a dipyrimidine context, with each mutated cytosine invariably flanked on the 5’ or the 3’ side by a cytosine or thymine. Similar rationale combining observed mutation spectra and knowledge of biochemical mechanisms may be used to delineate other sources of DNA damage and mutation in human cancers.
Non-random mutation patterns are also observed in other types of cancer, such as C/G base pairs being more frequently mutated than A/T pairs1–10 and the occurrence of strand-coordinated clusters of cytosine mutations9,12,13. Spontaneous hydrolytic deamination of cytosine to uracil (C-to-U) may explain a subset of these events, but not the majority because most occur outside of potentially methylatable CpG dinucleotide motifs (i.e., sites most prone to spontaneous deamination) and the occurrence of these mutations in clusters is highly non-random. Another possible source of these mutations is enzyme-catalyzed C-to-U deamination by one or more of the nine active DNA cytosine deaminases encoded by the human genome. Such a mechanism was originally hypothesized when the DNA deaminase activity of these enzymes was discovered14, and was recently highlighted with demonstrations of clustered mutations in breast, head/neck, and other cancers9,12,13. These clusters have been named kataegis, as their sporadic but concentrated nature bears likeness to rain showers9. Although enzymatic deamination has been implicated in this phenomenon, the actual enzyme responsible has not been determined.
Enzyme-catalyzed DNA C-to-U deamination is central to both adaptive and innate immune responses. B lymphocytes use activation-induced deaminase (AID) to create antibody diversity by inflicting uracil lesions in the variable regions of expressed immunoglobulin genes, which are ultimately processed into all six types of base substitution mutations15,16. AID also catalyzes uracil lesions in antibody gene switch regions that lead to DNA breaks and juxtaposition of the expressed, and often mutated, variable region next to a new constant region (i.e., isotype switch recombination)15,16. In humans, seven related enzymes, APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H, combine to provide innate immunity to a variety of DNA-based parasitic elements17,18. A well-studied example is the cDNA replication intermediate of HIV-1, which during reverse transcription is vulnerable to enzymatic deamination by at least 3 different APOBEC3 proteins19,20. APOBEC1 also has a similar capacity for viral cDNA deamination, and it is the only family member known to have a biological role in cellular mRNA editing21–24. More distantly related proteins, APOBEC2 and APOBEC4, have yet to elicit enzymatic activity. In total, nine of eleven APOBEC family members have demonstrated DNA deaminase activity in a variety of biochemical and biological assay systems14,25–29.
However, a possible drawback of encoding nine active DNA deaminases could be chromosomal DNA damage and, ultimately, mutations that lead to cancer14. AID has been linked to B cell tumorigenesis through off-target chromosomal deamination as well as triggering translocations between the expressed heavy chain locus and various oncogenes30. Transgenic expression of AID causes tumor formation in mice31, as does transgenic expression of APOBEC132. Most recently, we showed that APOBEC3B is upregulated in breast tumors and correlated with a doubling of both C-to-T and overall base substitution mutation loads33. Since AID and APOBEC1 are expressed tissue specifically and there is no reason to suspect developmental confinement of APOBEC3B, we hypothesized that APOBEC3B may be a general mutagenic factor impacting the genesis and evolution of many different cancers. This hypothesis is supported by studies indicating APOBEC3B expression in many different cancer cell lines33–35, in contrast to relatively low expression in 21 normal human tissues spanning all major organs33,35,36. This DNA mutator hypothesis is additionally supported by the fact that APOBEC3B is the only deaminase family member with constitutive nuclear localization33,37.
Here, we test this mutator hypothesis by performing a global analysis of all available DNA deaminase family member expression data and exomic mutation data from 19 different carcinomas, representing over 4,800 tumors and 1,000,000 somatic mutations. Mutation frequencies, local sequence contexts, and distributions including kataegis events were analyzed systematically for each tumor and cancer type. In addition, we calculated the hierarchical distances between the deamination signature of recombinant APOBEC3B derived from biochemical experiments33 and the observed frequencies of cytosine mutation spectra in all 19 cancer types. Taken together, these analyses converge upon APOBEC3B as the most likely cause of a large fraction of the both the dispersed and clustered cytosine mutations in six distinct cancers.
RESULTS
As a first test of the hypothesis that APOBEC3B is a general endogenous cancer mutagen, we performed a comprehensive analysis of the expression profiles of all eleven APOBEC family members across a panel of 19 distinct tumor types, including breast cancer as a positive control33 (Table 1 and Supplementary Fig. 1). The expression values for each target mRNA were normalized to those of the constitutive housekeeping gene, TATA-binding protein (TBP), to enable quantitative comparisons between RNAseq and RT-qPCR data sets and to provide controls for the few instances where RNAseq values for normal tissues were not available publicly (Online Methods).
Table 1.
Summary statistics for the 19 different tumor types in this study.
| A3B expression data1 | Exome mutation data2 | Clustered mutation data3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor type | TCGA ID |
n | Range | Median | n | Range | Median | Average | Total number of clusters |
Mean per tumor |
Percentage of total mutations |
| Low Grade Glioma | LGG | 174 | 0 – 0.69 | 0.06 | 170 | 5 – 15458 | 45 | 138 | 280 | 1.6 | 5.1 |
| Prostate adenocarcinoma | PRAD | 140 | 0 – 0.76 | 0.12 | 150 | 19 – 165 | 54 | 59 | 27 | 0.18 | 1.1 |
| Thyroid carcinoma | THCA | 384 | 0 – 4.1 | 0.18 | 326 | 3 – 98 | 20 | 22 | 25 | 0.08 | 1.2 |
| Glioblastoma multiforme | GBM | 169 | 0.014 – 2.0 | 0.22 | 167 | 1 – 173 | 28 | 34 | 114 | 0.68 | 7.9 |
| Kidney renal papillary cell carcinoma | KIRP | 76 | 0.0079 – 3.0 | 0.24 | 100 | 15 – 214 | 64 | 69 | 18 | 0.18 | 1.0 |
| Kidney renal clear cell carcinoma | KIRC | 480 | 0.011 – 4.5 | 0.29 | 244 | 6 – 696 | 73 | 92 | 42 | 0.17 | 0.67 |
| Acute myeloid leukemia | LAML | 179 | 0.027 – 2.3 | 0.44 | 74 | 1 – 151 | 12 | 17 | 1 | 0.010 | 0.21 |
| Ovarian serous cystadenocarcinoma | OV | 266 | 0.0015 – 8.6 | 0.48 | 469 | 1 – 145 | 39 | 55 | 1 | 0.0021 | 0.010 |
| Breast invasive carcinoma | BRCA | 849 | 0.0012 – 39 | 0.67 | 777 | 2 – 443 | 45 | 59 | 122 | 0.16 | 0.86 |
| Stomach adenocarcinoma | STAD | 57 | 0.18 – 3.6 | 0.68 | 156 | 6 – 8849 | 172 | 551 | 66 | 0.42 | 0.32 |
| Lung adenocarcinoma | LUAD | 355 | 0.0041 – 9.6 | 0.68 | 392 | 12 – 2547 | 259 | 355 | 310 | 0.79 | 0.73 |
| Rectum adenocarcinoma | READ | 72 | 0.082 – 3.2 | 0.81 | 88 | 28 – 7204 | 136 | 227 | 44 | 0.50 | 1.2 |
| Colon adenocarcinoma | COAD | 192 | 0.017 – 3.7 | 0.85 | 266 | 27 – 8459 | 250 | 487 | 133 | 0.50 | 0.39 |
| Uterine corpus endometrioid carcinoma | UCEC | 370 | 0.012 – 12 | 0.94 | 248 | 1 – 14687 | 68 | 722 | 1093 | 4.4 | 2.9 |
| Skin cutaneous melanoma | SKCM | 267 | 0.0011 – 10 | 1.1 | 255 | 6 – 6174 | 389 | 697 | 353 | 1.4 | 0.68 |
| Bladder urotheilal carconoma | BLCA | 122 | 0.0050 – 24 | 1.6 | 99 | 45 – 1802 | 226 | 291 | 293 | 3.0 | 3.5 |
| Head & neck squamous cell carcinoma | HNSC | 303 | 0.0038 – 20 | 1.7 | 306 | 7 – 2070 | 138 | 180 | 203 | 0.66 | 1.4 |
| Lung squamous cell carcinoma | LUSC | 259 | 0.094 – 15 | 1.7 | 177 | 1 – 3910 | 299 | 363 | 144 | 0.81 | 0.77 |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | CESC | 97 | 0.0010 – 20 | 2.4 | 39 | 30 – 1779 | 138 | 233 | 98 | 2.5 | 3.4 |
A3B expression values relative to those of the housekeeping gene TBP by RNAseq.
Somatic mutations in each exome, spanning aproximately 38 Mb of the human genome.
Kataegis events from exome mutation data are defined as ≥2 cytosine mutations within 10kb intervals which meet Gordenin significance (see Methods).
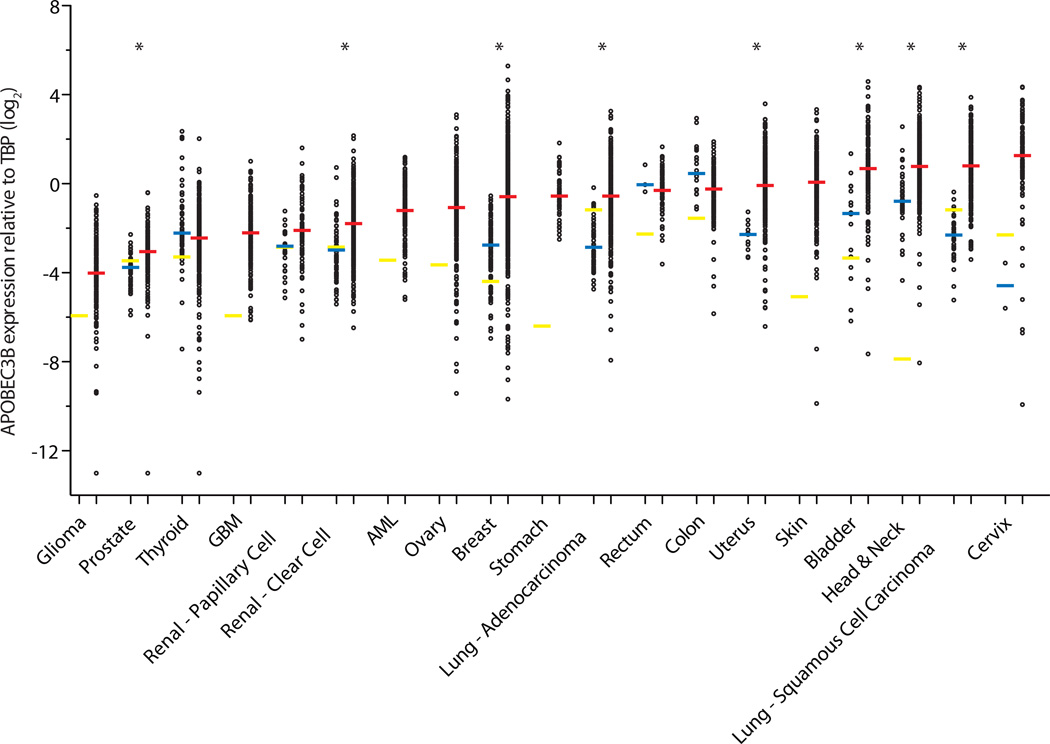
Several cancers showed APOBEC3B expression levels comparable to those in corresponding normal tissues (Fig. 1, Table 1, Supplementary Fig. 1, and Supplementary Table 1). Prostate and renal clear cell carcinomas showed statistically significant upregulation of APOBEC3B in the tumors, albeit with median expression values that are only a fraction of TBP. In contrast, 6 different cancers showed evidence for strong APOBEC3B upregulation in the majority of tumors of the breast, uterus, bladder, head & neck, and lung (adeno- and squamous cell carcinomas) (p<0.0001 by Mann-Whitney U-test). Other cancers such as cervical and skin also showed high APOBEC3B levels, but a lack of data for corresponding normal tissues precluded statistical analysis. Remarkably, a total of 10 cancers showed a median level of APOBEC3B upregulation greater than that of the intended positive control, breast cancer. This was particularly striking for bladder, head/neck, both lung, and cervical cancers.
Figure 1. APOBEC3B is upregulated in numerous cancer types.
Each data point represents one tumor or normal sample, and the Y-axis is log-transformed for better data visualization. Red, blue, and yellow horizontal lines indicate the median APOBEC3B/TBP value for each cancer type (Table 1), the median value for each set of normal tissue RNAseq data (Supplementary Table 1), and individual RT-qPCR data points, respectively. Asterisks indicate significant upregulation of APOBEC3B in the indicated tumor type relative to the corresponding normal tissues (p<0.0001 by Mann-Whitney U-test). P-values for negative or insignificant associations are not shown.
The second major prediction of the APOBEC mutator hypothesis is chromosomal DNA C-to-U deamination, which should result in strong biases toward mutations at C/G base pairs. Such mutational events may be either transitions or transversions because genomic uracils can directly template the insertion of adenines during DNA replication and, if converted to abasic sites by uracil DNA glycosylase, the lesions become non-instructional and error-prone polymerases may insert adenine, thymine, or cytosine opposite the abasic site (most often adenine following the A-rule). In both scenarios, an additional round of DNA synthesis or repair can yield either transitions or transversions at C/G base pairs (i.e., C/G-to-T/A, C/G-to-G/C, and C/G-to-A/T mutations; see Discussion for model).
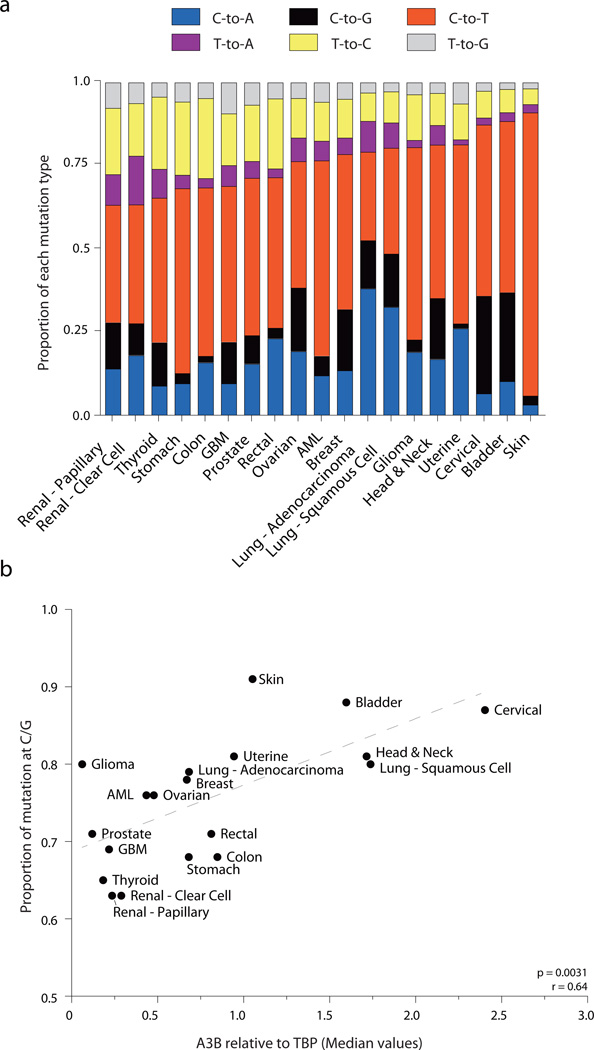
Interestingly, the fraction of mutations at C/G base pairs ranges considerably, from a low of 60% in renal cancers to a high of approximately 90% in skin, bladder, and cervical cancers (Fig. 2a). The massive bias in skin cancers is largely attributable to error-prone DNA synthesis (A insertion) opposite cyclobutane pyrimidine dimers caused by UV light11. However, the biases observed in urogenital carcinomas such as bladder and cervical cancers are probably not due to UV but more likely to an alternative mutagenic source such as enzymatic DNA deamination. Indeed, the top 5 tumor types with C/G dominated mutation spectra are among the top 6 tumors in terms of APOBEC3B expression (compare Fig. 1 and Fig. 2a). A possible mechanistic relationship is further supported by a positive correlation between overall proportion of mutations occurring at C/G base pairs and median APOBEC3B levels (p=0.0031, r=0.64 by Spearman’s correlation; Fig. 2b). The positive correlation is remarkable given the fact that all available data were included in the analysis and multiple variables could have undermined a positive correlation, such as known mutational sources (UV in skin cancer), undefined mutational sources (glioma with the 6th highest C/G mutation bias and lowest APOBEC3B levels), and differential DNA repair capabilities among the distinct tumor types (discussed further below).
Figure 2. Mutation types and signatures in 19 human cancers.
(a) Stacked bar graph summarizing the 6 types of base substitution mutations as proportions of the total mutations per cancer.
(b) Median APOBEC3B relative to TBP expression levels plotted against the proportion of mutations at C/G base pairs (Spearman p = 0.0031, r = 0.64). Dashed grey line is the best-fit for visualization.
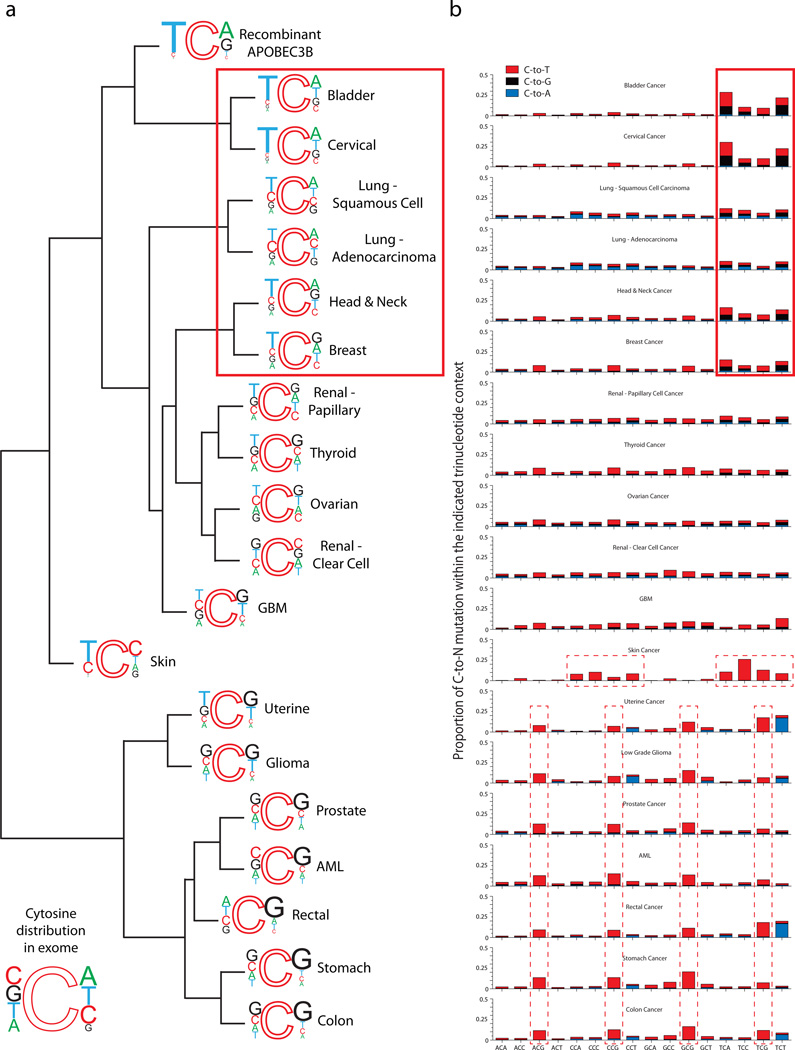
DNA deaminases such as APOBEC3B are strongly influenced by the bases adjacent to the target cytosine, particularly at the immediate 5’ position. For instance, AID prefers 5’ adenines or guanines, APOBEC3G prefers 5’ cytosines, and other family members prefer 5’ thymines38–40. We recently showed that recombinant APOBEC3B prefers 5’ thymines and strongly disfavors 5’ purines; on the 3’ side, it prefers adenines or guanines, and disfavors pyrimidines33 (Fig. 3a). Therefore, the third and possibly most important prediction of the APOBEC mutator hypothesis is that cancers impacted by enzymatic deamination should show non-random nucleotide distributions immediately 5’ and 3’ of mutated cytosines, and that these signatures can then be used with expression information (above), additional mutation data (below), and existing literature and biochemical constraints (below) to identify the enzyme responsible.
Figure 3. Cytosine mutation spectra for 19 cancers.
(a) Dendrogram with weblogos indicating the relationship among cancer types determined by the trinucleotide contexts of mutations occurring at C nucleotides for the top 50% APOBEC3B expressing samples within each cancer type. Font size of the bases at the 5’ and 3’ positions are proportional to their observed occurrence in exome mutation datasets. The preferred mutation context for recombinant APOBEC3B from Ref. 33 is included in the hierarchical clustering in order to determine how closely each cancers’ actual mutation spectrum matches the preferred motif for APOBEC3B in vitro. The pattern expected if the mutations were to occur at random C bases in the exome is included as an inset at the bottom left.
(b) Stacked bars indicate the observed proportion of cytosine mutations at each unique trinucleotide [5’-NCN-to-N(T/G/A)N]. Bar color indicates each mutation type: red: C-to-T, black: C-to-G, and blue: C-to-A. The top 6 cancer types (highlighted by solid line box) show clear biases toward mutations within 5’TCN motifs, at frequencies that resemble the preferences of recombinant APOBEC3B in vitro (Ref. 33). Skin cancer and the bottom 7 cancers (highlighted by dashed line boxes) have obviously different cytosine mutation spectra.
We therefore performed a global sequence signature analysis on all available cytosine mutation data from the upper 50% of APOBEC3B-expressing tumors for each tumor type (this cut-off was chosen to minimize the impact of unrelated mutational mechanisms). These mutation data were first compiled and subjected to a hierarchical cluster analysis to group tumors with similar cytosine mutation signatures (Fig. 3a). Short Euclidean distances (i.e., smaller measures) between the mutation signatures of different tumors indicate a high degree of concordance, i.e. similar mutational patterns (Supplementary Table 2 lists calculated values). Bladder and cervical cancers, two of the top APOBEC3B-expressing cancers, had cytosine mutation signatures remarkably similar to each other and to that of recombinant APOBEC3B. This is visually evidenced by strong mutation biases at 5’TCA motifs, which match the enzyme’s optimal in vitro substrate. The two lung cancers, breast cancer, and head/neck cancer also had cytosine mutation signatures that strongly resembled the preference of recombinant APOBEC3B (Fig. 3a and Supplementary Table 2). Several cancers had cytosine mutation signatures with an intermediate relatedness to recombinant APOBEC3B (renal papillary, thyroid, ovarian, renal clear cell, GBM, and skin). In further contrast, the seven remaining cancers had the largest separation from recombinant APOBEC3B ranging from uterine to colon cancer (Fig. 3a and Supplementary Table 2).
We next separated each composite mutation distribution into the 16 individual local trinucleotide contexts to further resolve cytosine-focused mutational mechanisms that may be influencing each cancer. Bladder, cervical, lung squamous, lung adeno, head/neck, and breast carcinomas all shared strong 5’TCN mutation signatures, with 5’TCA being strongest of the four possibilities (boxed in Fig. 3b). A background of other mutations was apparent in the two types of lung cancer, possibly associated with tobacco carcinogens or other mutational mechanisms. The next most obvious signature occurred in skin cancer, as expected, with C-to-T transitions predominating within dipyrimidine contexts (middle dashed boxes in Fig. 3b). Only two other obvious cytosine-focused mutation patterns were evident. C-to-T mutations at 5’CG contexts dominated at least seven types of cancer, consistent with a 5’CG targeted mechanism such as spontaneous deamination of methyl-cytosine (lower dashed boxes in Fig. 3b). Finally, uterine, low-grade glioma, rectal, and colon cancers had an inordinate number to C-to-A transversions in 5’(YCT contexts) consistent with at least one additional distinct cytosine-focused mutational mechanism (e.g., POLE proofreading domain variants have been implicated in a subset of colorectal tumors41).
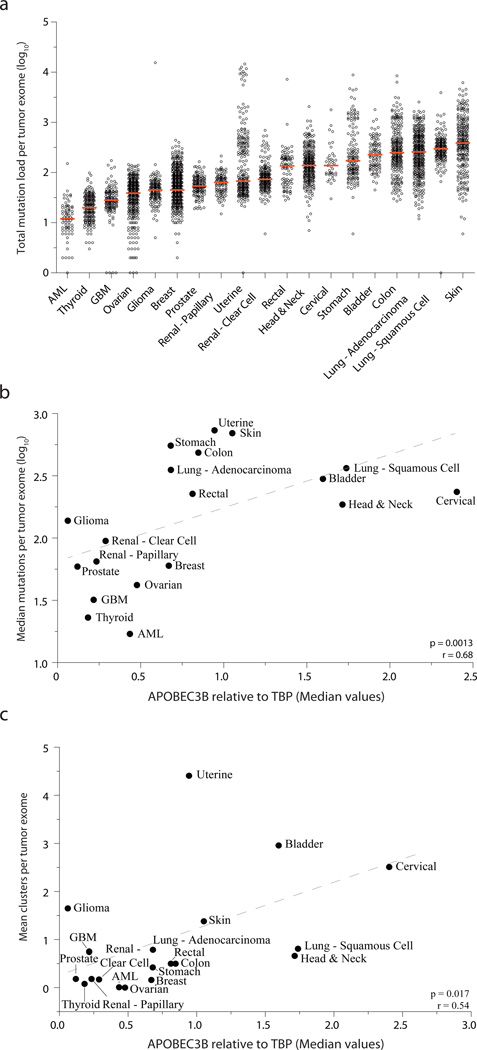
A fourth prediction of a general mutator hypothesis is that tumor mutation loads ought to correlate with APOBEC3B expression levels. To test this possibility on a global level, we used median mutation loads for each tumor type and median APOBEC3B expression values. Median values were chosen ensure the inclusion of all data, yet simultaneously minimize the impact of uncontrollable variables such as other mutational mechanisms, jackpot effects, bottlenecks, tumor ages, etc. As recently reviewed42, mutation loads vary considerably within each tumor type and between the different cancers with more than a full log difference from the bottom to the top of this range (AML to skin cancer in Fig. 4a). However, despite this incredible variation, a strong positive correlation was found between median mutation loads and APOBEC3B expression levels (p=0.0013, r=0.68 by Spearman’s correlation; Fig. 4b). This result is consistent with the possibility that APOBEC3B may be a general endogenous mutagen that contributes to most human cancers albeit, as outlined above, clearly much more to a subset of cancers. A dominant role for APOBEC3B in a subset of cancers is further evidenced by significant correlations between mutation loads and APOBEC3B expression levels when these analyses were performed for each cancer type on a tumor-by-tumor basis (Supplementary Fig. 2 and Supplementary Fig. 3).
Figure 4. APOBEC3B expression levels correlate with total mutation loads and kataegis events.
(a) A dot plot showing the total mutation loads for each tumor exome from each of the indicated cancers. Each data point represents one tumor, and the Y-axis is log-transformed for better visualization. A red horizontal line shows the median mutation load for each cancer type.
(b) Median mutation loads per tumor exome for each cancer type plotted against the median APOBEC3B relative to TBP expression values (Spearman p = 0.0013, r = 0.68). Dashed grey line is the best-fit for visualization.
(c) The mean number of cytosine mutation clusters per exome for each cancer type plotted against median APOBEC3B relative to TBP expression values (Spearman p = 0.0017, r = 0.54). Dashed grey line is the best-fit for visualization.
A final prediction of a general APOBEC mutator hypothesis is that impacted cancers should bear evidence for strand-coordinated clusters of cytosine mutations9,12,13. As proposed12, clusters can be defined as 2 or more mutation events within a 10 kbp window. By this criterion, every cancer showed evidence for cytosine mutation clustering with a large range between different cancer types (0.016 to 38 cytosine mutation clusters per tumor). However, it is necessary to apply an additional calculation to take into consideration the sequence length of each cluster, which also varies dramatically and can result in the inclusion of false-positives (see Roberts et al.12 and Online Methods). This additional filter yielded a much smaller number of likely kataegis events, ranging from 0.002 clusters per ovarian carcinoma to 4.4 clusters per uterine tumor (Table 1). Interestingly, the number of mutations grouped into kataegis was a relatively small percentage of the total number of cytosine mutations for each cancer (maximally 7.9%). However, the sheer existence of clustered cytosine mutation in nearly every cancer provides further evidence for APOBEC involvement. For most cancers this is likely to be APOBEC3B because average number of kataegis per tumor correlates positively with median APOBEC3B expression levels (p=0.017 and r=0.54 by Spearman correlation; Fig. 4c). The 6 cancer types with cytosine mutation signatures that grouped most closely with recombinant APOBEC3B, bladder, cervix, lung (adeno- and squamous cell), head/neck, and breast, all showed strong evidence for kataegis with a mean of 3.0, 2.5, 0.79, 0.81, 0.66, and 0.16 clusters per tumor, respectively. It is notable that breast cancer is at the low end of this range, but 50-fold higher frequencies would be expected if full genomic sequences had been available (concordant with analyses of Nik-Zainal et al.9). Interestingly, low-grade gliomas and uterine carcinomas are clear outliers in this analysis, consistent with the close hierarchical clustering of their cytosine mutation signatures (distant from recombinant APOBEC3B) and strongly suggesting another distinct mutational mechanism.
DISCUSSION
We performed an unbiased analysis of all available DNA deaminase expression profiles and cytosine mutation patterns in 19 different cancer types to try to explain the origin of the cytosine-biased mutation spectra and clustering observed in many different cancers1–10,13. The observed cytosine mutation patterns were compared using a hierarchical clustering method to group cancers with similar mutation patterns. Six distinct cancer types, bladder, cervical, lung squamous cell, lung adenocarcinoma, head/neck, and breast, clearly stood out, with elevated APOBEC3B expression in the majority of tumors, strong overall C/G mutation biases, cytosine mutation contexts that closely resemble the deamination signature of recombinant APOBEC3B, and evidence for kataegis events. The most parsimonious explanation for this convergence of independent data sets is that APOBEC3B-dependent genomic DNA deamination is the direct cause of most of these cytosine mutations in these types of cancers. These data are consistent with a general mutator hypothesis, in which APOBEC3B mutagenesis has the capacity to broadly shape the mutation landscapes of at least six distinct tumor types and possibly also those of several others, albeit to lesser extents.
The large data sets analyzed here support a model in which upregulated levels of APOBEC3B cause genomic C-to-U lesions, which may be processed into a variety of mutagenic outcomes33 (Supplementary Fig. 4). In most instances, uracil lesions are repaired faithfully by canonical base excision repair. However, in some instances, uracil lesions may template the insertion of adenines during DNA synthesis, which may result in C-to-T transitions (G-to-A on the opposing strand). In other instances, genomic uracils may be converted to abasic sites by uracil DNA glycosylase. These lesions are noninstructional such that DNA polymerases, in particular translesion DNA polymerases, may place any base opposite, with an A leading to a transition and a C or T leading to a transversion. In addition, uracil lesions that are processed into nicks through the concerted action of a uracil DNA glycosylase and an abasic site endonuclease, can result in single- or double-stranded DNA breaks, which are substrates for recombination repair and undoubtedly intermediates in the formation of cytosine mutation clusters (kataegis)9,12,13 and larger-scale chromosomal aberrations such as translocations.
The significant positive correlations between APOBEC3B expression levels and the percentage of mutations at C/G pairs, the overall mutation loads, and the number of kataegis events combine to suggest that most cancers are impacted by APOBEC3B-dependent mutagenesis, but unambiguous determinations were not possible for several cancers for a variety of reasons. Skin cancer, for example, has the fifth highest APOBEC3B expression rank and clear evidence for kataegis, but it also has a strong dipyrimidine-focused C-to-T mutation pattern that could easily eclipse an APOBEC3B deamination signature. APOBEC3B may help explain melanomas that occur with minimal UV exposure43. Several other cancers such as uterine, rectal, stomach, and ovarian also have significant APOBEC3B upregulation and evidence for kataegis, which combine to suggest direct involvement, but the trinucleotide cytosine mutation motifs were too distantly related to that of the recombinant enzyme to enable unambiguous associations. Therefore, additional large data sets such as high-depth full genome sequences will be required to distinguish an APOBEC3B-dependent mechanism unambiguously from the multiple other mechanisms contributing to these tumor types.
We note that we have not completely excluded the possibility of other DNA deaminase family members contributing to mutation in cancer but, apart from AID in B cell cancers30, roles for other APOBECs are unlikely to be as great as those of APOBEC3B for the following reasons: i) no reported enzymatic activity (APOBEC2 and APOBEC4), ii) tissue-restricted expression profiles (AID, APOBEC3A, APOBEC1, APOBEC2, and APOBEC4)33,35,36,44–48, iii) localization to the cytoplasmic compartment (APOBEC3A, APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H)29,37,49,50, and iv) in two instances, a completely different intrinsic preference for bases surrounding the target cytosine (AID and APOBEC3G prefer 5’RC and 5’CC, respectively)33,38–40. Thus, taken together with the comprehensive analyses presented here of expression data (Fig. 1), C/G mutation frequencies (Fig. 2), local cytosine mutation signatures (Fig. 3), overall mutation loads (Fig. 4), and kataegis (Fig. 4c and Table 1), all available data converge upon the conclusion that APOBEC3B is a major source of mutation in multiple human cancers. This knowledge provides foundations for future studies focused on each cancer type and sub-type to further delineate the impact of this potent DNA mutator on each cancer genome and on associated therapeutic responses and patient outcomes.
ONLINE METHODS
Data Analyses
A description of tumor types, tumor APOBEC3B expression data, and tumor exome mutation data is provided in Table 1. Information for the corresponding normal tissues is provided in Supplementary Table 1. Somatic mutations and RNAseq expression data were retrieved from the Cancer Genome Atlas Data Matrix on January 3rd, 2013. Gene expression data were mined from RNAseqV2 datasets for all cancers (normalized expression values) with the exception of LAML and STAD, which were from RNAseq datasets (RPKM values). Additional normal sample RNAseqV2 data were downloaded from TCGA on April 4th, 2013 to include recently released normal sample information for READ and COAD. APOBEC3B expression values were normalized to the expression of TBP for each patient sample. Comparisons between the normal RNAseq-derived gene expression values and the tumor expression values were performed using the Mann-Whitney U test to determine significance. All RT-qPCR values for normal tissues were reported previously based on data from pooled normal samples33,35, with the exception of salivary gland, stomach, skin, and rectal tissues, which are unique to this report. The primary tissue RNA was generated using published methods35 and total RNA obtained commercially (salivary gland RNA for head/neck and stomach RNA were obtained from Clontech and skin and rectal RNA were obtained from USBiological). Each A3B relative to TBP value from RTqPCR was multiplied by an experimentally derived factor of 2 to facilitate direct comparisons with RNAseq values (unpublished data).
Mutation data were taken from maf files downloaded from TCGA Somatic Mutation database (http://tcga-data.nci.nih.gov/tcga/). Insertions/deletions and adjacent multiple mutations (di- and trinucleotide variations) were removed and the remaining single nucleotide variations (SNVs) were converted to hg19 coordinates (Supplementary Table 3). Non-mutations with respect to the reference genome (e.g., C-to-C) were eliminated and duplicate entries were removed unless they were reported for different patient samples. Comparisons between mutation and gene expression were calculated using Spearman’s rank correlation.
Trinucleotides with cytosines in the center position were used to calculate the sequence context-dependence of mutations. There are a total of 16 unique trinucleotides containing C in the center position. The corresponding 16 reverse complements were also included in the analysis but, for simplicity, discussion was focused on the cytosine-containing strand. For each unique trinucleotide the observed C-to-T, C-to-G, and C-to-A mutations were counted and placed in a table and normalized to one to reflect the fraction of each mutation type. This table reflects the global mutation profile of cytosines for each cancer. These data were then used to hierarchically cluster the cancer mutation signatures. This was done using the hclust function of R using Euclidean distance and “complete” option (http://www.r-project.org). The Euclidean distance is the ordinary distance between two data points on a 2D plot (Supplementary Table 2 lists all calculated Euclidean distances).
A kataegis event is defined as two or more mutations within a 10,000 nucleotide genomic DNA window. The probability of each event occurring by chance is then calculated following the work of Gordenin and colleagues12. Briefly, the p-value of observing a given number of mutations within a given number of base pairs was calculated using a negative binomial distribution utilizing the genomic size of each event, the number of mutations in each event and the base probability of finding a random mutation in the exome (number of mutations in each cancer type divided by the number of patients and exome size). The significant kataegis events with p-values less than 10−4 for each cancer are reported in Table 1. “Gordenin significance” indicates that a given cluster of mutations has met the above criteria and attained significance. This approach minimizes false positive cluster-calls resulting by random chance.
Supplementary Material
ACKNOWLEDGEMENTS
We thank The Cancer Genome Atlas (TCGA) Network for generating the RNAseq and somatic mutation data and providing open access, and Harris lab members and S. Kaufmann for comments. M.B.B. was supported by a Department of Defense Breast Cancer Research Program Predoctoral Fellowship (BC101124). This work was supported by grants from the Jimmy V Foundation, Minnesota Ovarian Cancer Alliance, and National Institutes for Health (R01 AI064046 and P01 GM091743).
Footnotes
AUTHOR CONTRIBUTIONS
All authors contributed to the study designs, data analyses, and manuscript preparation. MBB and NAT analyzed data from TCGA. NAT performed mutation and cluster analysis.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Stephens P, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 2.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 5.Kumar A, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108:17087–17092. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makridakis NM, Reichardt JK. Translesion DNA polymerases and cancer. Front Genet. 2012;3:174. doi: 10.3389/fgene.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drier Y, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23:228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 15.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 16.Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaRue RS, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci. 2009;364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanc V, Davidson NO. C-to-U RNA editing: mechanisms leading to genetic diversity. J Biol Chem. 2003;278:1395–1398. doi: 10.1074/jbc.R200024200. [DOI] [PubMed] [Google Scholar]
- 22.Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 23.Petit V, et al. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J Mol Biol. 2009;385:65–78. doi: 10.1016/j.jmb.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda T, et al. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res. 2011;39:5538–5554. doi: 10.1093/nar/gkr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 26.Petersen-Mahrt SK, Neuberger MS. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1) J Biol Chem. 2003;278:19583–19586. doi: 10.1074/jbc.C300114200. [DOI] [PubMed] [Google Scholar]
- 27.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 28.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3' --> 5' on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 29.Hultquist JF, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu Rev Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- 31.Okazaki IM, et al. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamanaka S, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 35.Refsland EW, et al. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koning FA, et al. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lackey L, et al. APOBEC3B and AID have similar nuclear import mechanisms. J Mol Biol. 2012;419:301–314. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohli RM, et al. Local sequence targeting in the AID/APOBEC family differentially impacts retroviral restriction and antibody diversification. J Biol Chem. 2010;285:40956–40964. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M, Rada C, Neuberger MS. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J Exp Med. 2010;207:141–153. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujino T, Navaratnam N, Scott J. Human apolipoprotein B RNA editing deaminase gene (APOBEC1) Genomics. 1998;47:266–275. doi: 10.1006/geno.1997.5110. [DOI] [PubMed] [Google Scholar]
- 45.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 46.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato Y, et al. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J Biol Chem. 2010;285:7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 49.Rada C, Jarvis JM, Milstein C. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc Natl Acad Sci U S A. 2002;99:7003–7008. doi: 10.1073/pnas.092160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Land AM, et al. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and non-genotoxic. J Biol Chem. 2013 doi: 10.1074/jbc.M113.458661. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.