Abstract
The molecular mechanisms through which alternative splicing and histone modifications regulate gene expression are now understood in considerable detail. Here, we discuss recent studies that connect these two previously separate avenues of investigation, beginning with the unexpected discoveries that nucleosomes are preferentially positioned over exons and DNA methylation and certain histone modifications also show exonic enrichment. These findings have profound implications linking chromatin structure, histone modification and splicing regulation. Complementary single gene studies provided insight into the mechanisms through which DNA methylation and histones modifications modulate alternative splicing patterns. Here, we review an emerging theme resulting from these studies: RNA-guided mechanisms integrating chromatin modification and splicing. Several groundbreaking papers reported that small noncoding RNAs affect alternative exon usage by targeting histone methyltransferase complexes to form localized facultative heterochromatin. More recent studies provided evidence that pre-messenger RNA itself can serve as a guide to enable precise alternative splicing regulation via local recruitment of histone-modifying enzymes, and emerging evidence points to a similar role for long noncoding RNAs. An exciting challenge for the future is to understand the impact of local modulation of transcription elongation rates on the dynamic interplay between histone modifications, alternative splicing and other processes occurring on chromatin.
INTRODUCTION
Alternative splicing is a versatile mechanism that explains both, how the vast complexity of the human proteome is generated from a limited number of genes and serves as a key target for the regulation of gene expression (1–3). The advent of high-throughput technologies paved the way for genome-wide analyses indicating that transcripts from up to 95% of multiple exon-containing human genes undergo alternative splicing (4–6). This review will focus on the most common form of alternative splicing in mammals, which involves differential selection of exons within primary RNA transcripts for inclusion in the mature mRNA (1). As the majority of the resulting isoforms are variably expressed at different times in development and/or in different cell and tissue types, alternative splicing must be precisely and robustly regulated (4–8). The importance of pre-mRNA splicing in mediating proper temporal and spatial expression of the human genome is underscored by the large number of genetic disorders associated with alterations in this process. Extensive surveys of disease-causing mutations in human genes revealed that the primary effect in up to 50% of the known examples is to disrupt constitutive splicing or perturb alternative splicing patterns (9–12).
Removal of introns from pre-mRNAs is carried out by a large macromolecular machine known as the spliceosome, which is comprised of five snRNAs and ∼300 proteins (13). Counter to the original view that only the earliest events in spliceosome assembly (formation of the E-complex containing the U1 snRNP and U2AF) are targeted by regulatory mechanisms, it is now understood that control over alternative splicing can be exerted at multiple later stages of the process including the transition from the pre-spliceosome containing the U1 and U2 snRNPs to the mature but pre-catalytic spliceosome [for comprehensive reviews, see (8,14)].
Mammalian gene architecture, in which exons comprise relatively small islands amidst a sea of intronic sequences, necessitates an initial recognition process in which cis-acting sequences recruit trans-acting factors to form exon definition complexes (15). Studies carried out over the past several decades have demonstrated that tissue- or developmental stage-specific alternative splicing is regulated at two different levels. At the most fundamental, local level, trans-acting splicing regulators, usually RNA-binding proteins, recognize cis-acting regulatory elements to control the formation of spliceosomes on pre-mRNAs (16). At a higher, more integrated level, transcriptional parameters, which in turn are influenced by chromatin structure and/or histone modifications, are key factors that ensure proper regulation of alternative splicing (17). Importantly, the two levels of alternative splicing regulation are connected by virtue of cotranscriptional deposition of protein factors including the serine–arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) onto nascent transcripts. Depending on the context, these factors may either antagonize or promote the recruitment of spliceosomal components to yield distinct alternative splicing patterns (17).
The mechanisms through which precise temporal and spatial regulation of spliceosome assembly ensures proper patterns of alternative splicing have been the subject of numerous recent reviews, several of which are cited above. In this article, we will focus on alternative splicing regulation at the chromatin level. This is a rapidly developing area of research that has benefited enormously from recent advances in genome-wide epigenetic analyses. These strategies have uncovered new mechanisms of alternative splicing regulation as well as adding a new dimension to our understanding of this process. We will first discuss exon definition in the context of the genome, emphasizing specific types of histone modifications that mark exons. Next, we will summarize how specific histone marks regulate alternative splicing through two proposed mechanisms. Last and most importantly, we will discuss an emerging theme in the epigenetic regulation of alternative splicing: possible roles of RNA-guided changes in histone modifications that lead to differential alternative splicing outcomes.
Exon definition at the chromatin level
Nucleosome positioning
The packaging of eukaryotic nuclear DNA into nucleosomes, in which 147 bp are wrapped around a histone octamer, limits accessibility and thus impinges on molecular processes that utilize DNA as a template including transcription and recombination (18). In addition to nucleosome positioning, the patterns of histone modifications demarcate functional genomic regions and influence the recruitment of trans-acting factors (19). Recently, it has become apparent that chromosomal features also influence processes that do not directly involve DNA yet occur cotranscriptionally such as pre-messenger RNA splicing. In 2009, four groups independently reanalyzed the genome-wide nucleosome positioning data obtained using activated human T cells (20) and discovered that nucleosome density is higher over exons than introns (21–24). A similar observation was also made using human sperm cells and Medaka (Japanese rice fish) early embryonic cells (25). Moreover, preferential association of nucleosomes with exons appears to be conserved throughout metazoans including Caenorhabditis elegans and Drosophila (22–24). By correlating exon inclusion levels and nucleosome distribution patterns, these studies suggested that nucleosome positioning defines exons at the chromatin level. Thus, the similarity between the average size of a vertebrate exon, 170 nt, and a single nucleosome + associated linker DNA may not be coincidental (26).
Given that nucleosomes serve as barriers to transcription, it is not surprising that RNA polymerase II (Pol II) binding in metazoans is also higher on exons than introns (22,27). Presumably, pausing of Pol II at exons allows more time for the splicing machinery to recognize and define exons. Consistent with this idea, it has been found that polymerase density is higher on alternative exons than on constitutive exons (28–31). Interestingly, for intron-containing genes in fission yeast, nucleosomes also appear to be enriched on exons, whereas Pol II preferentially accumulates over introns (32). This inverse distribution is nevertheless consistent with a role for polymerase speed in determining the locations of spliceosome assembly, as splice sites in fission yeast are initially paired across the relatively small introns (average length 81 nt) rather than the much larger exons (33).
Histone modification
These and additional studies also examined the patterns of histone posttranslational modifications (PTMs) in relation to exon–intron architecture. Histone PTMs, which can occur at multiple positions within a given polypeptide chain but are most frequently found at amino acids in the N-terminal tails of these proteins, play important roles in modulating chromatin structure. Of the 13 types of histone PTMs described to date, the most common and also most extensively studied are acetylation and methylation (34). Historically, these have been classified as activating or repressing, respectively, but more recent work indicates that the impact of a given PTM on transcription is context dependent. The popular histone code hypothesis, which states that the recognition of specific combinations of histone marks by protein complexes leads to distinct downstream events (35), can now be extended from DNA-level to RNA-level events.
Several studies analyzed genome-wide chromatin immunoprecipitation-sequencing (ChIP-seq) data for 38 histone PTMs and found a pronounced enrichment of the H3K36me3 (histone H3 Lysine 36 trimethyl) mark on exons (21–24,36). Remarkably, this histone mark appears to be associated more significantly with constitutive exons than with alternative exons (21,36,37). Several other histone PTMs are enriched over exons, but to a lesser extent, including H3K27me1/2/3 (21,23), H3K79me1 (22,23), H4K20me1 (22,23) and H2BK5me1 (22,23). Again, this phenomenon is conserved among metazoans from flies and worms to humans (22,23,36). Interestingly, a more recent study indicated that the H3K36me3 mark is also enriched over exons in fission yeast (32), which in this organism may facilitate intron definition. Thus, the overall picture is that exon/intron boundaries are marked by transitions between regions enriched in and depleted of H3K36me3. The mechanisms by which these differential markings of exons and introns by histone modifications define positions of spliceosome assembly remain to be elucidated.
DNA methylation
DNA methylation is a key epigenetic modification in the mammalian genome that serves as an important regulator of gene transcription [reviewed in (38)]. This modification is catalyzed by enzymes that add a methyl group to the cytosine at CpG dinucleotides (5-methylcytosine, 5-mC), the majority of which are methylated in most tissues and developmental stages that have been examined. Recently, three groups performed high-definition profiling of DNA methylation by single-molecule bisulfite sequencing technology and found that DNA methylation correlates with intron–exon junctions and is enriched in exonic regions in humans, honey bees and Arabidopsis (39–41). These results provide strong evidence that DNA methylation is another evolutionarily conserved mark for exon definition. Interestingly, in honey bees, a strong correlation was also found between methylation patterns on alternative exons and splicing patterns of these exons in queens and workers (40).
Less is known about the role of 5-hydroxymethylcytosine (5-hmC), a 5-mC derivative generated by the oxidation of 5-mC, which is abundant in the brain. Traditional bisulfite methods for assaying DNA modifications cannot differentiate between 5-mC and 5-hmC. Recently, Petronis and colleagues assayed 5-hmC by using glucosylation coupled with restriction-enzyme digestion and microarray analysis (42). They found that 5-hmC is highly enriched at the exon–intron boundary in the brain, whereas 5-mC is highly enriched at the exon–intron boundary in nonneuronal cells. Moreover, in the human frontal cortex, the DNA encoding constitutive exons that are included in 100% of gene transcripts contained higher levels of 5-hmC than alternatively spliced exons that are included in ≤80% of gene transcripts (42).
These studies indicate that modifications of DNA as well as histones serve as tissue-specific markers that help to differentiate exons from introns.
Epigenetic regulation of alternative splicing: two modes of action
The basis of epigenetic regulation of alternative splicing is that, in eukaryotes, splicing generally proceeds cotranscriptionally (43–47). Although a few recent studies indicated that some introns are removed posttranscriptionally, it is notable that, in these cases, the partially spliced transcripts remain tethered to chromatin until splicing is completed (48,49). These findings imply that chromatin structure and histone modifications can still influence splicing events that occur more slowly or involve removal of introns near the 3′-end of the transcript. Chromatin structure and histone modifications have been inferred to affect alternative splicing through two mechanisms, kinetic coupling and chromatin-splicing adaptor systems, which have been discussed extensively by Kornblihtt, Misteli and colleagues (17) and are briefly described below.
Kinetic coupling
For splicing events that occur cotranscriptionally, the kinetic coupling model states that there is a kinetic competition between transcription elongation and splicing (17). As alternative exons generally have suboptimal splicing signals that require more time to be recognized by the splicing machinery (50–52), it was predicted that faster transcriptional elongation speed of Pol II leads to increased skipping of these exons (17). Kornblihtt and colleagues directly tested this hypothesis in a study using an engineered mutant Pol II that mimics the Drosophila C4 mutant, which exhibits a slow rate of transcriptional elongation. In human cells, the mutant polymerase increased inclusion of the ED1 exon in fibronectin and promoted the use of a downstream alternative 5′-splice site in the Adenovirus pre-mRNA (51). In Drosophila, the C4 Pol II mutant was shown to affect splicing of the endogenous ultrobithorax pre-mRNA, suggesting a physiological role for the coupling between the kinetics of transcription and alternative splicing regulation (51).
The rate of transcriptional elongation is modulated by the dynamic cycle of acetylation and deacetylation of histones during transcription. A variety of histone acetyltransferases (HATs) and deacetylases (HDACs) mediate the addition and removal of acetyl groups (53). Rapid histone acetylation/deacetylation is very important for nucleosome dynamics during transcriptional elongation and is coordinated by the C-terminal domain (CTD) of Pol II (54–57). When pre-mRNA is transcribed by Pol II, acetylation of the nucleosomes in front of the elongation complex by HATs is required to destabilize histone–DNA interactions (55). The passage of Pol II causes displacement of histones, which are subsequently redeposited onto the DNA behind Pol II. These newly deposited nucleosomes are hyperacetylated, but only transiently. In order to maintain a normal chromatin configuration, histone deacetylase complexes remove the acetyl marks from histones (58–60). Overall, reversible histone acetylation serves as a mechanism to modulate local chromatin structure rapidly during transcription.
Recent global assays have indicated that the activity of HDACs also influences the selection of specific pairs of splice sites in many genes. Using splicing-sensitive microarrays, HDAC inhibitors were shown to affect the splicing patterns of ∼700 human genes (61). In particular, it was found that inhibition or depletion of HDAC1 led to increased histone H4 acetylation surrounding an alternative exon, which in turn enhanced the rate of elongation by Pol II, thereby decreasing inclusion of the exon (61).
Chromatin-splicing adaptor systems
During cotranscriptional splicing, chromatin remodeling proteins that recognize and bind specific histone marks recruit splicing factors to the site of transcription, an action that achieves one of two goals. First, core components of the spliceosome are recruited to ensure efficient and accurate removal of introns. For example, acetylation mediated by the HAT Gcn5 is required for cotranscriptional recruitment of the U2 snRNP to the branchpoint (62). In another example, the chromodomain protein CHD1 forms a bridge between H3K4me3 and the growing spliceosome via specific interactions with the U2 snRNP (19). Second, and are more relevant to the subject matter of this review, splicing regulators are recruited through chromatin remodeling proteins to directly affect the inclusion or exclusion of alternative exons. In the following section, we describe several examples to illustrate how histone marks dictate alternative splicing outcomes, focusing in particular on the impact of localized changes in histone modifications. It should be noted that kinetic coupling mechanisms and chromatin-splicing adaptor systems are not mutually exclusive; thus, in some examples, both are involved.
H3K9me
The marking of chromatin by histone H3 lysine 9 methylation including dimethylation (H3K9me2) and trimethylation (H3K9me3) is correlated with transcriptional repression. In mammalian genomes, H3K9me3, which serves as a specific binding site for the chromodomain protein HP1 (63), is generally enriched in heterochromatin and across transposons and other repetitive DNA elements. However, it has also been shown that H3K9me3 is associated with the coding regions of transcribed genes (64–66). Interestingly, multiple recent studies indicate that the H3K9me3 mark can influence alternative splicing.
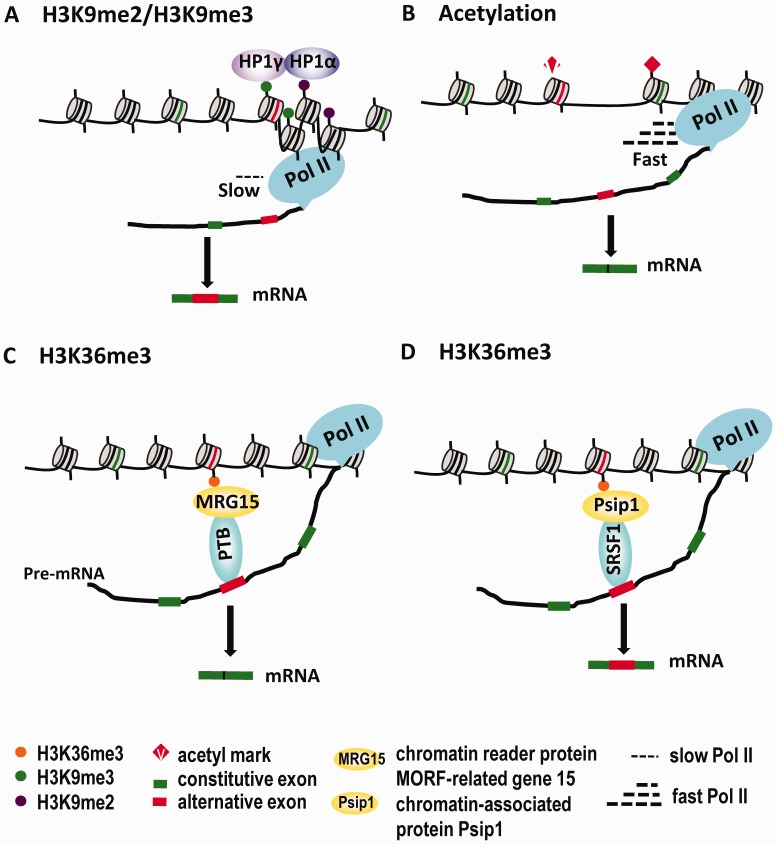
The first such study, by Muchardt and colleagues, carefully examined the distribution of a number of histone modifications including H3K4me2, H3K4me3, H3K36me3, H3K9me2 and H3K9me3 across the CD44 gene (29). Human CD44 exists in numerous isoforms generated by alternative splicing involving the variable region of the extracellular domain of CD44. The 10 variable exons (v1–v10) are located between two series of constitutive exons (C1–C5 and C16–C19) (67,68). An earlier study had demonstrated that Brm, one of the components of the chromatin remodeling complex Swi/Snf, associated with Pol II and reduced transcriptional elongation of the variable region of CD44, resulting in inclusion of the variable exons (28). In the recent study, it was found that the chromosomal region encompassing the CD44 variable exons is highly enriched in H3K9me3 marks, and the presence of this mark correlates with increased inclusion of the alternative exons in the mature CD44 mRNA. H3K9me3 marks in this localized region are recognized by HP1γ, which facilitates inclusion of the alternative exons by reducing the local transcriptional elongation rate (Figure 1A). This study provided the first evidence that local H3K9me3 modifications can impact alternative splicing by marking the chromosomal regions surrounding alternative exons (29).
Figure 1.
Mechanisms through which histone modifications affect alternative splicing outcomes. (A) H3K9me2 and H3K9me3 are recognized and bound by HP1α and HP1γ, respectively. As a result, Pol II slows down at the HP1-bound chromosomal region, leading to increased inclusion of a nearby alternative exon. (B) Hyperacetylation of H3 and H4 promotes a more relaxed chromatin structure, increased Pol II elongation rate and skipping of alternative exons. (C) The chromodomain protein MRG15 binds to H3K36me3 and recruits the splicing silencer protein PTB to its target RNA, thereby promoting skipping of an alternative exon. (D) Another H3K36me3-binding protein, Psip1, affects inclusion of alternative exons by recruiting the splicing regulator SRSF1.
In a second recent study, H3K9me2 was found in association with alternative exons in the Fibronectin (FN1) gene encoding a protein that mediates a wide variety of cellular interactions with the extracellular matrix and plays important roles in cell adhesion, migration, growth and differentiation (69). FN1 produces different protein isoforms through alternative splicing of exon EDI. The recent study indicated that local H3K9me2 marks induced by targeting of siRNAs to a region close to exon EDI influences splicing of exon EDI. Heterochromatin-associated protein HP1α recognizes the H3K9me2 marks and slows down transcriptional elongation, resulting in inclusion of exon EDI (70) (Figure 1A). The mechanism through which siRNAs mediate deposition of the H3K9me2 mark will be discussed below.
H3K9ac
The influence of changes in local histone acetylation patterns on splice site selection was first demonstrated in depolarized neuronal cells (71). The gene targeted by this mechanism encodes the neural cell adhesion protein (NCAM), which is involved in the development of the central nervous system. NCAM specifies two isoforms that contain different portions of the cytosolic region due to selective inclusion of exon 18 (72). Depolarization in neuronal cells induces H3K9 hyperacetylation restricted to an internal region of the NCAM gene surrounding exon 18, resulting in skipping of this exon. The hyperacetylation was also shown to increase the local transcriptional elongation rate (Figure 1B). These effects were fully reversible when the depolarizing agent was removed. Inhibition of HDAC activity by trichostatin A resulted in a similar molecular phenotype, indicating that depolarization most likely exerts its effects by increasing histone acetylation (71).
H3ac and H4ac
Studies from our laboratory provided two additional examples of how local histone hyperacetylation affects inclusion of alternative exons. The first target we uncovered is neurofibromatosis type 1 (NF1), a tumor-suppressor gene that contains alternative exon 23a, which encodes 21 amino acids within the domain related to GTPase-activating proteins (73), while the second target gene encodes the apoptosis-promoting receptor Fas, which produces a soluble isoform through exclusion of alternative exon 6 (74). Recently, we found that localized histone hyperacetylation surrounding these two alternative exons was significantly higher in neuronal than in nonneuronal cells. Histone hyperacetylation leads to an increased local transcriptional elongation rate and decreased inclusion of these exons (Figure 1B). This study provided direct evidence to support the proposal that local histone acetylation surrounding alternative exons influences splice site selection (30). Interestingly, we found that Hu proteins are among the major regulatory factors that repress inclusion of NF1 exon 23a and Fas exon 6 in neuronal cells (73); the mechanism of Hu-mediated histone acetylation will be discussed below.
H3K36me3
Histone H3 lysine 36 trimethylation (H3K36me3) is a hallmark of transcription elongation. Accordingly, H3K36me3 levels are lowest at the promoter and highest in the transcribed regions of genes (75). In metazoans, this histone modification marks exons in genes undergoing active transcription including alternative exons (76,77). In fission yeast, where alternative splicing via exon skipping is rare (33), H3K36me3 is nevertheless enriched over exons, particularly in poorly expressed genes (32). Thus, this mark may facilitate proper splice site pairing via intron definition as well as exon definition by modulating the rate of Pol II elongation. Although a kinetic coupling mechanism could in principle apply to metazoans, where excluded exons are excised together with the adjacent introns, the two examples discussed below involve chromatin-splicing adaptor systems.
A well-documented example in which H3K36me3 influences alternative splicing of a mammalian transcript is fibroblast growth factor receptor (FGFR2), which contains two mutually exclusive exons (IIIb and IIIc) that encode a region within the extracellular immunoglobulin-like domain responsible for distinct differences in ligand-binding specificity (78). The FGFR2-IIIb splice variant is expressed in epithelial cells, while FGFR2-IIIc is expressed in mesenchymal cells (79,80). Analysis of histone modifications throughout FGFR2 indicated that H3K36me3 is enriched over the FGFR2 gene in mesenchymal cells. This mark is recognized by the MORF-related gene 15 (MRG15) protein, which directly recruits the polypyrimidine tract-binding protein (PTB) to the intronic splicing silencer element surrounding exon IIIb to repress its inclusion in mesenchymal cells (Figure 1C). These results exemplify how local deposition of H3K36me3 can affect alternative splicing by recruiting the splicing regulator PTB to pre-mRNA via the chromatin-binding protein MRG15 (76).
The H3K36me3 mark can also be recognized by the chromatin-associated protein Psip1 to regulate alternative splicing (77). P52, the short isoform of Psip1 (Psip1/p52), is enriched in expressed genes, often at the downstream exons, and its distribution correlates with H3K36me3 patterns. A ChIP assay indicated a strong congruence in the presence of H3K36me3, Psip1/p52 and the SR protein SRSF1 on specific gene regions surrounding alternative exons such as exon 7 of Vcan and exon 5 of Diap2. Using biochemical and knockout assays, the authors found that Psip1/p52 binds the H3K36me3 marks through its PWWP domain, thereby recruiting SRSF1 to pre-mRNA to regulate alternative splicing (77) (Figure 1D).
DNA methylation (5-mC)
Multiple studies revealed a strong correlation between DNA methylation and alternative splicing, although evidence for a direct causal relationship between the two processes remained elusive. However, two recent studies established a mechanistic link between DNA methylation and alternative splicing outcomes (81,82). First, Oberdoerffer and colleagues found that a DNA-binding protein, CCCTC-binding factor (CTCF), can promote inclusion of many alternative exons containing weak upstream splice sites by mediating local RNA polymerase II pausing (81). They used alternative exon 5 of the CD45 gene as a model system to investigate the mechanism. CD45 is a hematopoietic receptor-like tyrosine phosphatase. Alternative splicing of three exons (4, 5 and 6) in CD45 produces multiple isoforms characterized by differential inclusion of heavily glycosylated protein segments A, B and C, respectively. In naive T cells, exons 4, 5 and 6 are included. Upon differentiation to memory T cells, exons 4, 5 and 6 are skipped (83). The authors found that 5-mC of CD45 exon 5 was increased during the transition from naive to mature T lymphocytes, which is correlated with exclusion of exon 5. In mature T lymphocytes, high levels of 5-mC promote exclusion of exon 5 by blocking CTCF binding to DNA. Second, a very recent study showed that DNA methylation is enriched in the DNA corresponding to included alternative exons in human cells, and that inhibition of DNA methylation with drug treatment disrupts regulation of alternative exons. Methyl-CpG-binding protein MeCP2 promoted the inclusion of alternative exons by recognizing DNA methylation and maintaining local histone hypoacetylation through the subsequent recruitment of HDACs (82).
Although the emerging evidence for chromatin-level regulation of alternative splicing is certainly exciting, it should be noted that it remains unclear whether these contributions are general. For example, modulation of splice site choice by transcriptional coupling mediated by a chromatin binding protein applies only to a subset of exons (75). Thus, the impact of histone and DNA modifications on exon inclusion may be critical in some genes and contexts but not in others.
Alternative splicing mediated by RNA and potential roles of RNA-guided local chromatin modifications in pre-mRNA processing
Selection of alternative exons is a complex process that responds to cell type-specific, developmental stage-specific and environmental signals. To accurately regulate alternative splicing at the chromatin level, it is necessary to modulate histone modifications at the chromosomal regions corresponding to alternative exons. However, histone modifications are created and removed by ubiquitously expressed histone-modifying complexes that appear to have no sequence specificity. Thus, a very important question is what guides these histone-modifying complexes to distinct and specific sites in different cellular contexts. From a number of recent studies, a theme has begun to emerge, namely that RNA molecules can recruit histone-modifying complexes to particular chromatin sites and regulate their activity to establish and maintain the proper local chromatin structure. In the following sections, we will first discuss the roles of siRNA/sRNA and pre-mRNA in regulating histone modifications needed for the accurate control of alternative splicing. Next, we will discuss the role of lncRNA in alternative splicing. Although numerous studies have implicated IncRNAs in chromatin modification, transcription and alternative splicing separately, no clear examples that link the two functions have yet been described. Many excellent reviews have already discussed the roles of lncRNAs in regulating local chromatin modification to regulate gene expression (84–87). Thus, in this section, we will focus on the impact of lncRNAs on alternative splicing. Lastly, we will briefly discuss the role of snoRNA in alternative splicing regulation.
Small noncoding RNA guides local chromatin remodeling to regulate alternative splicing
RNA interference (RNAi) is a silencing mechanism in which small (20–25 nt) RNA molecules suppress gene expression by pairing to complementary targets (88). The process generally begins with cleavage of long double-stranded RNA (dsRNA) precursor molecules by the enzyme Dicer to form short double-stranded siRNAs containing one passenger strand and one guide strand. These siRNAs mediate silencing in the cytoplasm (posttranscriptional gene silencing, PTGS) and in the nucleus (chromatin-dependent gene silencing, CDGS) by distinct mechanisms that nevertheless share certain components and features. In PTGS, the RNA-induced silencing complex (RISC) directs the guide strand siRNA to a complementary target sequence in an mRNA. The RISC subunit Argonaute (AGO) is responsible for cleaving the siRNA–mRNA duplex, rendering the mRNA vulnerable to degradation by exonucleases. More relevant to the subject at hand is CDGS, encompassing both transcriptional and cotranscriptional gene silencing, which is best understood in the context of heterochromatin formation in fission yeast but also occurs in plants and animals (88).
CDGS is a remarkably complex process in which the RNA-induced transcriptional silencing complex (RITS), the nuclear version of RISC that also contains a member of the Argonaute family, binds to the guide strand siRNA and directs it to the homologous region in the DNA by base pairing with nascent transcripts (89). In addition to RNA–RNA interactions, RITS is tethered to chromatin via binding of its chromodomain-containing subunit Chp1 to H3K9-methylated nucleosomes. Efficient heterochromatic silencing also requires a positive feedback loop triggered by two additional chromodomain proteins, Swi6 (the fission yeast ortholog of HP1), which binds to H3K9me, and Chp2, (a paralog of Swi6), which recruits a deacetylase complex (88). Thus, in organisms ranging from Schizosaccharomyces pombe to humans, silent chromatin is characterized by the presence of H3K9 methylation and hypoacetylated histones.
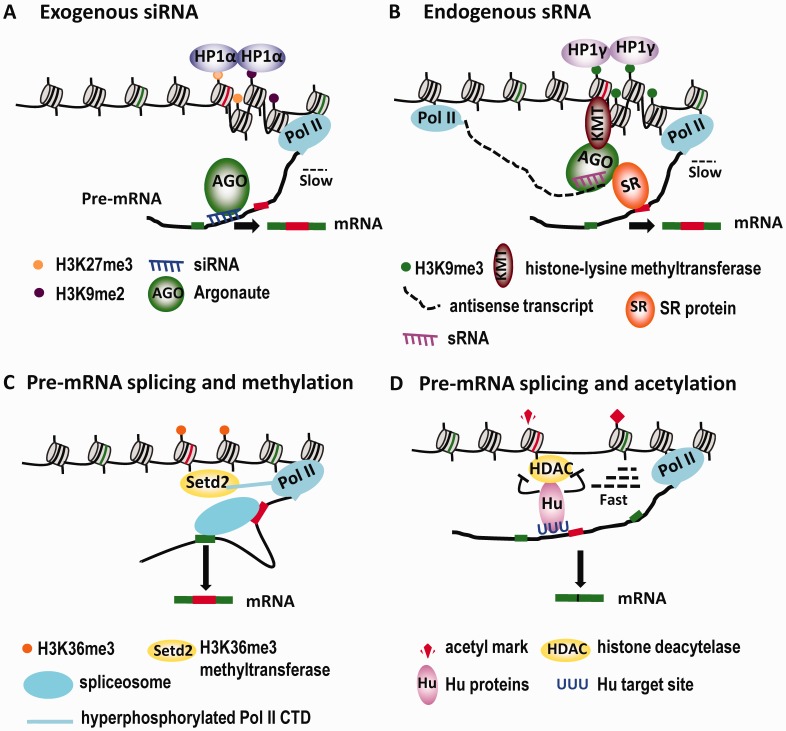
Whereas heterochromatin is present constitutively at centromeres and certain other chromosomal sites, it can also be formed facultatively to allow reversible transcriptional gene silencing (64,90). Interestingly, two recent studies have shown that facultative heterochromatin nucleated by small noncoding RNAs can regulate alternative splicing by inducing transient local chromatin remodeling (31,70). In a ‘proof of principle’ series of experiments, Kornblihtt and colleagues demonstrated that targeting of exogenously applied siRNA to intronic or exonic sequences close to the alternative exon EDI of the FN1 gene modulated the pattern of ED1 inclusion in an AGO1-dependent manner in human cells. Recently, the same group found another example in which alternative exon 18 of NCAM is also regulated by exogenously applied siRNA directed against an intron (91). These investigators further demonstrated that the siRNAs increased the levels of two heterochromatin marks, H3K9me2 and H3K27me3, at the target site. Recognition of these local heterochromatin marks by the human counterpart of fission yeast Swi6, HP1α, promotes EDI inclusion by reducing the elongation rate of RNA polymerase II (Figure 2A) (70).
Figure 2.
RNA-guided mechanisms that link chromatin modification and alternative splicing. (A) Treatment of cells with siRNAs complementary to regions surrounding an alternative exon induces deposition of the heterochromatin marks H3K9me2/H3K27me3 and H3K9me3 at the target site, which are bound by HP1α and HP1γ, respectively. This results in a reduction of the Pol II elongation rate and increased exon inclusion. (B) An intragenic antisense transcript generates sRNAs that target the intron–exon junction region close to the variable exons of CD44, which leads to enrichment of the H3K9me3 mark and recruitment of HP1γ. Consequently, the Pol II elongation rate is reduced and variable exons are included at a higher level. In addition, the AGO protein that is associated with the sRNA likely recruits multiple splicing factors through direct interactions, which further contributes to the increased inclusion of variable exons (see text for details). (C) The process of pre-mRNA splicing reinforces the coincident distribution of Setd2 (the mammalian ortholog of yeast Set2), the H3K36me3 methyltransferase and H3K36me3 marks on exons in genes undergoing active transcription. (D) Hu proteins bound at their target pre-mRNA sites interact with and inhibit the enzymatic activity of HDAC2, leading to localized H3 and H4 hyperacetylation in the corresponding chromosomal region. The resulting increase in the Pol II elongation rate through this region promotes skipping of the Hu-regulated alternative exons.
A subsequent study by Harel-Bellan and colleagues implicated endogenous small RNAs (sRNAs) in widespread regulation of alternative splicing (31). These investigators identified many splicing factors in the complexes of chromatin-embedded AGO1 and AGO2. Deep sequencing of sRNAs from the chromatin fraction containing AGO2 indicated that one-third of the population ranged in size from 19 to 30 nt and was specifically enriched near the 3′-ends of introns. The presumptive targeting of endogenous sRNAs to the neighborhood of intron–exon junctions provides an additional link between the RNAi pathway and splicing (Figure 2B). The authors of this study went on to use the variable exons of CD44 as a model to investigate the underlying mechanism. They found that upon treatment of cultured mammalian cells with phorbol-12-myristate-13-acetate (PMA), sRNAs recruited AGO1 and AGO2 to the transcribed regions of CD44, which increased local H3K9me3 levels at the chromosomal region corresponding to the variable exons (31). It is known that treatment with PMA increases inclusion of a cluster of nine variable exons of CD44 in many cell types by stimulating protein kinase C activity (92). Accumulation of H3K9me3 on the CD44 variable exon region resulted in recruitment of the chromodomain protein HP1γ, reduced Pol II elongation and promoted inclusion of CD44 variant exons. In addition, depletion of AGO1 or AGO2 in PMA-treated cells consistently decreased H3K9me3 levels over the variable exon region and reduced recruitment of the H3K9me3-binding protein HP1γ to this region (31). This study suggests that endogenously expressed sRNAs influence local H3K9me3 levels to regulate alternative splicing (Figure 2B).
Pre-mRNA guides local chromatin remodeling to regulate alternative splicing
During pre-mRNA splicing, the nascent transcript remains associated with both chromatin and RNA Pol II to ensure highly efficient and accurate splicing (49,93). This tight connection among pre-mRNA, RNA Pol II and chromatin provides the basis for pre-mRNA-induced chromatin remodeling. Recently, growing evidence has indicated that pre-mRNA splicing ‘reaches back’ to influence transcription and chromatin structure (94). The model that pre-mRNA splicing affects chromatin remodeling was first suggested by Kjems and colleagues, who found that the presence of a functional 5′ splice site can produce a significant enrichment of the histone marks H3K9ac and H3K4me3, which are indicative of active transcription (95).
In light of this finding, it is important to note that, as discussed in the previous section on exon definition, the exonic enrichment of the H3K36me3 mark is influenced by pre-mRNA splicing (Figure 2C) (96,97). Two studies using mammalian cells suggested that cotranscriptional splicing activity is at least one determinant of the establishment and maintenance of the H3K36me3 mark. In the first, Bentley and colleagues showed that inhibition of splicing specifically by splicing signal mutations, or globally by treatment with spliceostatin A, shifted the distribution of H3K36me3 away from the 5′-end and towards the 3′-end (96). In the second study, Carmo-Fonseca and colleagues showed that H3K36me3 distribution is dynamic, fluctuating with changes in local transcription activity (97). In addition, treatment of cells with the splicing inhibitor meayamycin or siRNA-mediated depletion of the splicing factor SAP130 decreased the enrichment of H3K36me3 on internal exons of at least three genes. More importantly, the distribution of the Huntingtin-interacting protein HYPB/Setd2 histone methyltransferase, which catalyzes H3K36 trimethylation, was also shifted in a pattern similar to H3K36me3 when splicing was inhibited (97). Taken together, these observations suggest that a self-reinforcing splicing/histone modification loop facilitates exon recognition in metazoans.
While the above findings strongly imply the existence of a mechanism through which splicing affects chromatin modifications, specifically the H3K36me3 distribution pattern, they do not rule out potential contributions of a reciprocal mechanism through which chromatin affects splicing (96). Support for the existence of such a mechanism is provided by a recent study from Guthrie, Yamamoto and colleagues. Interestingly, these investigators found that the histone marks H3K79me2 and H3K36me3 on alternative exons remained stable before and after treatments that resulted in significant inclusion of alternative exons for two different genes (98). Clearly, additional studies are needed to delineate the role of splicing in establishing and maintaining the patterns of histone marks.
Studies from our group established a distinct mechanism through which pre-mRNA serves as a guide to re-program the local histone acetylation state surrounding alternative exon 23a of the NF1 gene and exon 6 of the FAS gene to regulate alternative splicing of these exons. In this case, Hu proteins are recruited to the chromatin regions that contain alternative exons through binding to their target sequences on the pre-mRNA. Overexpression of Hu proteins in mouse embryonic stem (ES) cells, where Hu protein expression is low, promotes local histone hyperacetylation surrounding the two alternative exons. Conversely, knockdown of Hu proteins in neurons differentiated from mouse ES cells reduces localized histone hyperacetylation. Hu proteins interact with HDAC2 in neurons, and inhibit deacetylation activity of HDAC2 in an in vitro assay. Taken together, these results support a model in which Hu proteins are guided by pre-mRNA to induce histone hyperacetylation surrounding alternative exons by inhibiting HDAC activity (Figure 2D), leading to an increase in the local transcriptional elongation rate and decreased inclusion of the targeted exons (30).
Long noncoding RNAs
Long noncoding RNAs (lncRNAs) are a group of transcripts longer than 200 nt that lack significant protein-coding capacity (99). A subset, designated long intergenic noncoding RNAs (lincRNAs), is transcribed from and located within the stretches of DNA between genes, but the majority of lncRNAs are transcribed from complex loci that contain interlaced networks of noncoding and protein-coding transcripts including cis-antisense, intronic, bidirectional and promoter-associated lncRNAs (100,101).
A role for lncRNAs in the control of pre-mRNA alternative splicing was first suggested by studies of two natural antisense transcripts (NATs). The nuclear thyroid hormone receptor (TR) gene encodes two protein isoforms, NR1A1a and NR1Ab, which are generated by the selective usage of the NR1A1b-specific 5′-splice site. As NR1A1b antagonizes the nuclear thyroid hormone-activated NR1A1a, the TRalpha1:TRalpha2 ratio is a critical determinant of nuclear thyroid hormone action. Lazar and colleagues showed that two antisense lncRNAs (RevErbAα and TRα), which are transcribed from opposite directions at the same locus and overlap the TRα2 coding region, modulate the TRalpha1:TRα2 ratio in intact cells (102). A mechanistically distinct example, in which splicing regulation takes the form of intron retention, is found in the Zeb2 gene, encoding a transcriptional repressor of E-cadherin during an epithelial–mesenchymal transition (EMT) (103). A large intron located in the 5′-untranslated region of Zeb2 contains an internal ribosome entry site (IRES) that is necessary for expression of the protein. During EMT, an antisense lncRNA that includes sequences complementary to the 5′-splice site is transcribed from this region and prevents splicing of the IRES-containing intron from the pre-mRNA, resulting in efficient translation of the Zeb2 protein (103).
While the two examples described above involve lncRNAs acting in cis, recent studies conducted by several independent laboratories showed that the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) regulates alternative splicing by controlling the activity of trans-acting factors, specifically members of the SR protein family (104–107). MALAT1, which is among the most abundant nuclear lncRNAs, was originally identified in a screen for transcripts whose expression is altered in early-stage nonsmall cell lung cancers (108). In normal cells, MALAT1 is associated with SR proteins in both the nucleoplasm and nuclear speckles, and blocks the recruitment of these splicing factors to the pre-mRNA. However, in MALAT1-depleted cells, the levels of active SR proteins are increased, resulting in perturbed patterns of pre-mRNA alternative splicing. This study highlights a novel role for an lncRNA in the regulation of alternative splicing (104). A growing body of evidence indicates that, like siRNAs, lncRNAs can modulate local histone modifications by directing chromatin-remodeling complexes to specific regions of the genome. Thus, it will be interesting to determine whether lncRNAs can also regulate alternative splicing through modulation of chromatin modification.
Small nucleolar RNAs
Small nucleolar RNAs (snoRNAs) are a specific group of small nonprotein coding RNAs that are best known for their roles as guides for 2′-O-methylation or pseudouridylation of other nonprotein coding RNAs such as ribosomal, small nuclear and transfer RNAs (rRNA, snRNA, tRNA). Interestingly, a study by Kishore and Stamm implicated the snoRNA HBII-52 (a.k.a. SNORD115), which lacks significant complementarity to ribosomal RNAs, in promoting inclusion of Exon Vb of the serotonin receptor 5-HT2CR mRNA by binding to an 18-nt region of conserved complementarity that serves as a splicing silencer (109). Exon V of the serotonin receptor can produce exon Va– or Vb–containing isoforms through the use of two alternative 5′-splice sites. Skipping of exon Vb, encoding the second intracellular loop of the receptor, which is important for G protein binding, results in a truncated receptor due to a frame shift (110). The snoRNA HBII-52 is derived from an imprinted chromosomal domain containing two large arrays of tandemly repeated, paternally expressed box C/D snoRNA genes; the other product of this region, snoRNA H/MBII-85 (a.k.a. SNORD116), has been implicated in the etiology of Prader–Willi Syndrome (PWS) syndrome. Despite the controversy over whether these snoRNAs are processed into smaller RNAs (111), the proposal that defective pre-mRNA processing due to the absence of HBII-52 contributes to PWS disease development is tantalizing. It will be interesting to learn whether other snoRNAs modulate alternative pre-mRNA splicing and, if so, through what mechanism(s).
Concluding remarks
In this article, we have discussed a number of studies showing that local histone modifications including H3K36me3, H3K9me3 and acetylation, as well as DNA methylation, specifically tag chromosomal regions surrounding alternative exons to influence their inclusion or exclusion from the mature mRNA. Highlighted in particular were the two distinct modes by which specific local chromatin modifications within the coding regions of individual genes are established and/or maintained, presumably to ensure proper coordination of cotranscriptional events including RNA splicing. On the one hand, histone modifications can be established in the conventional transcription-coupled manner, whereas on the other hand, local alterations of this global scheme can be implemented or perhaps maintained/reinforced by additional mechanisms including those which are guided in cis by the nascent transcript, or in trans by siRNAs/sRNAs or lncRNAs.
While the role of histone modifications in the regulation of transcription initiation is well established, the new discoveries reviewed above have extended the role of histone modifications in regulating gene expression to the elongation phase of transcription. Interestingly, recent studies have revealed that both the rate of transcription elongation and the nature of the histone modifications at individual genes can be regulated in a cell type-specific as well as stimulus-specific manner (112–115). Perhaps, these new modes of regulation during transcription elongation also make critical contributions to increasing the complexity of the functional output from a given genome by influencing alternative splicing. Intriguingly, in addition to methylation and acetylation, histones can be modified by the addition of other chemical moieties including phosphorylation, ubiquitination, sumoylation and ADP-ribosylation (116). It will be interesting to determine whether these other histone modifications also regulate alternative splicing in metazoans.
Our growing appreciation for the importance of the elongation phase of transcription in regulating gene expression creates new challenges to building a unified framework of genome function. In particular, the pace of transcription must be highly regulated since inappropriate Pol II stalling can result in collisions with the DNA replication machinery and consequently genome instability or even cell death (117). It follows that any mis-steps in coordinating transcription elongation with other processes that utilize chromatin as a template, potentially including recombination and DNA repair as well as replication, could initiate such a disastrous sequence of events. In the context of this review, it is important to note that, like alternative pre-mRNA splicing, DNA-level transactions are modulated by chromatin modifications (75). Remarkably, recent studies have unexpectedly revealed that abnormal expression of factors pertinent to transcriptional elongation and splicing constitutes a major source of chromosomal breakage events in human cells (118). In this regard, it is tempting to speculate that defects in the dynamic balance of chromatin modifications engendered by perturbing the rate of Pol II progression and patterns of alternative splicing could potentially contribute to the etiology of some important human diseases such as cancer.
FUNDING
The National Institutes of Health [NS049103 to H.L. and GM073217 to J.A.W.]; National Science Foundation [MCB-1330788 to J.A.W.]; a pilot and feasibility grant from the Case Western Reserve University Skin Diseases Research Center [NIH-NIAMS; P30-AR-39750 to G.L.]; National Basic Research Program of China [973 Program, 2013CB917800 to H.-L.Z.]; a open research fund of the State Key Laboratory of Cognitive Neuroscience and Learning from Institute of Biophysics, Chinese Academy of Sciences [7Y1SNY7007 to H.-L.Z.]; ACES+ Opportunity Program (to J.A.W.). Funding for open access charge: The open access publication charge for this paper has been waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2011;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 6.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez SE, Petrillo E, Kornblihtt AR, Yanovsky MJ. Alternative splicing at the right time. RNA Biol. 2011;8:954–959. doi: 10.4161/rna.8.6.17336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J. Biol. Chem. 2007;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 9.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 10.Orengo JP, Cooper TA. Alternative splicing in disease. Adv. Exp. Med. Biol. 2007;623:212–223. doi: 10.1007/978-0-387-77374-2_13. [DOI] [PubMed] [Google Scholar]
- 11.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem. Sci. 2012;37:179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 15.Berget SM. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 16.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell. Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr. Opin. Genet. Dev. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 23.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 25.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8:3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 26.Sakharkar MK, Perumal BS, Sakharkar KR, Kangueane P. An analysis on gene architecture in human and mouse genomes. In Silico Biol. 2005;5:347–365. [PubMed] [Google Scholar]
- 27.Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 29.Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat. Struct. Mol. Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 30.Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc. Natl Acad. Sci. USA. 2011;108:E627–E635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 32.Wilhelm BT, Marguerat S, Aligianni S, Codlin S, Watt S, Bahler J. Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast. Genome Biol. 2011;12:R82. doi: 10.1186/gb-2011-12-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romfo CM, Alvarez CJ, van Heeckeren WJ, Webb CJ, Wise JA. Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe. Mol. Cell. Biol. 2000;20:7955–7970. doi: 10.1128/mcb.20.21.7955-7970.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 36.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput. Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu J. Epigenetics: unfinished symphony. Nature. 2006;441:143–145. doi: 10.1038/441143a. [DOI] [PubMed] [Google Scholar]
- 39.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT, 2nd, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25:2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 47.Osheim YN, Miller OL, Jr, Beyer AL. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985;43:143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- 48.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 52.Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 54.Svejstrup JQ. Transcription. Histones face the FACT. Science. 2003;301:1053–1055. doi: 10.1126/science.1088901. [DOI] [PubMed] [Google Scholar]
- 55.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 56.Spain MM, Govind CK. A role for phosphorylated Pol II CTD in modulating transcription coupled histone dynamics. Transcription. 2011;2:78–81. doi: 10.4161/trns.2.2.14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 59.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 60.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 61.Hnilicova J, Hozeifi S, Duskova E, Icha J, Tomankova T, Stanek D. Histone deacetylase activity modulates alternative splicing. PLoS One. 2011;6:e16727. doi: 10.1371/journal.pone.0016727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Black JC, Whetstine JR. Chromatin landscape: methylation beyond transcription. Epigenetics. 2011;6:9–15. doi: 10.4161/epi.6.1.13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 65.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brinkman AB, Roelofsen T, Pennings SW, Martens JH, Jenuwein T, Stunnenberg HG. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7:628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Screaton GR, Bell MV, Bell JI, Jackson DG. The identification of a new alternative exon with highly restricted tissue expression in transcripts encoding the mouse Pgp-1 (CD44) homing receptor. Comparison of all 10 variable exons between mouse, human, and rat. J. Biol. Chem. 1993;268:12235–12238. [PubMed] [Google Scholar]
- 68.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell. Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 69.White ES, Baralle FE, Muro AF. New insights into form and function of fibronectin splice variants. J. Pathol. 2008;216:1–14. doi: 10.1002/path.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 71.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl Acad. Sci. USA. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pollerberg GE, Schachner M, Davoust J. Differentiation state-dependent surface mobilities of two forms of the neural cell adhesion molecule. Nature. 1986;324:462–465. doi: 10.1038/324462a0. [DOI] [PubMed] [Google Scholar]
- 73.Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol. Cell Biol. 2008;28:1240–1251. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- 75.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell. Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Warzecha CC, Hovhannisyan R, Carstens RP. Dynamic fluorescent and luminescent reporters for cell-based splicing screens. Methods Mol. Biol. 2012;867:273–287. doi: 10.1007/978-1-61779-767-5_18. [DOI] [PubMed] [Google Scholar]
- 79.Orr-Urtreger A, Bedford MT, Burakova T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2) Dev. Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 80.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zikherman J, Weiss A. Alternative splicing of CD45: the tip of the iceberg. Immunity. 2008;29:839–841. doi: 10.1016/j.immuni.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 84.Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. Bioessays. 2011;33:830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kanhere A, Jenner RG. Noncoding RNA localisation mechanisms in chromatin regulation. Silence. 2012;3:2. doi: 10.1186/1758-907X-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nguyen TT, Cho K, Stratton SA, Barton MC. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol. Cell. Biol. 2005;25:2147–2157. doi: 10.1128/MCB.25.6.2147-2157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schor IE, Fiszbein A, Petrillo E, Kornblihtt AR. Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J. 2013;32:2264–2274. doi: 10.1038/emboj.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Konig H, Ponta H, Herrlich P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998;17:2904–2913. doi: 10.1093/emboj/17.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 96.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc. Natl Acad. Sci. USA. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 98.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat. Struct. Mol. Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Batista PJ, Chang HY. Cytotopic localization by long noncoding RNAs. Curr. Opin. Cell Biol. 2013;25:195–199. doi: 10.1016/j.ceb.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 101.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Munroe SH, Lazar MA. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J. Biol. Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- 103.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 108.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 109.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 110.Wang Q, O'Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J. Neurochem. 2000;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- 111.Bortolin-Cavaille ML, Cavaille J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader-Willi locus generate canonical box C/D snoRNAs. Nucleic Acids Res. 2012;40:6800–6807. doi: 10.1093/nar/gks321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell. 2013;50:212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fowler T, Sen R, Roy AL. Regulation of primary response genes. Mol. Cell. 2011;44:348–360. doi: 10.1016/j.molcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 117.Helmrich A, Ballarino M, Nudler E, Tora L. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013;20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- 118.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]