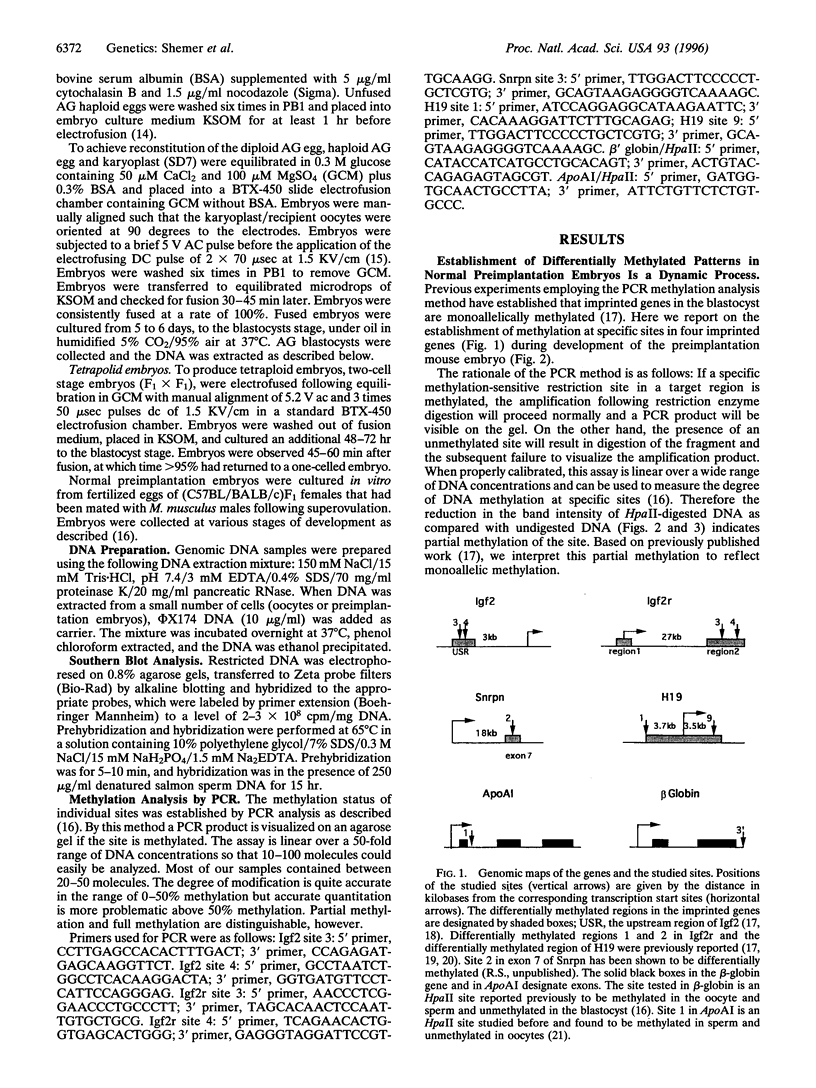
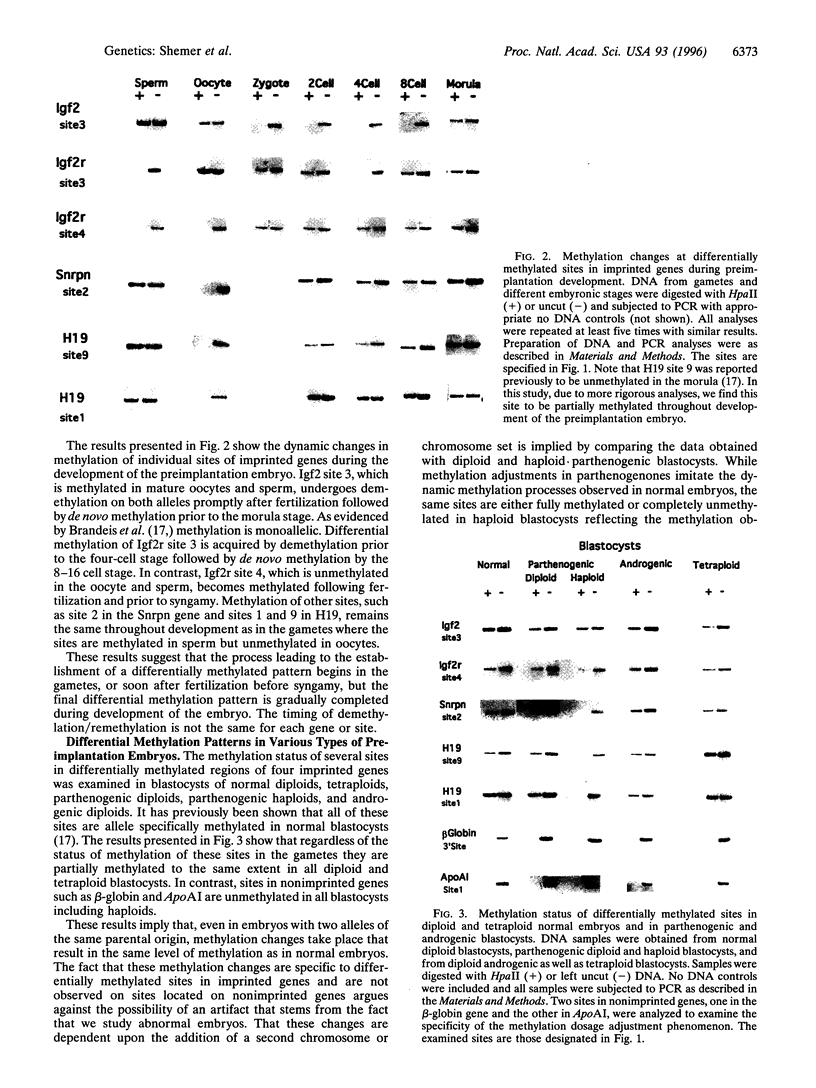
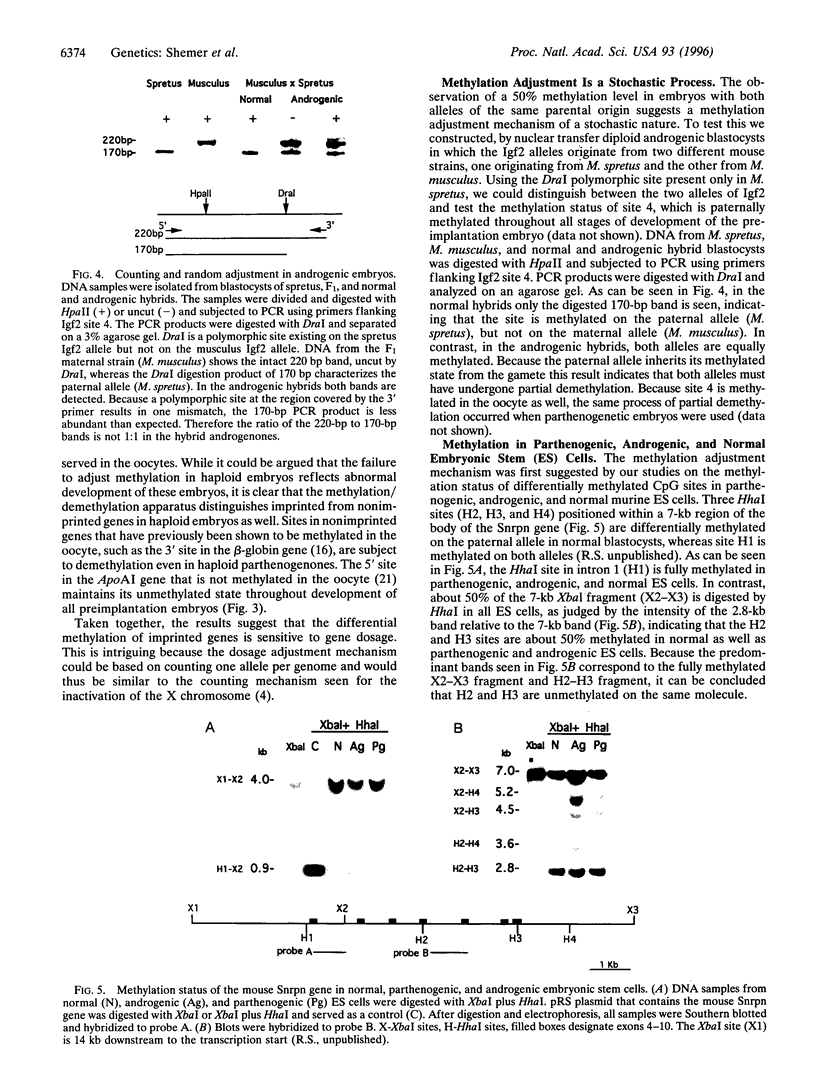
Abstract
Monoallelic expression in diploid mammalian cells appears to be a widespread phenomenon, with the most studied examples being X-chromosome inactivation in eutherian female cells and genomic imprinting in the mouse and human. Silencing and methylation of certain sites on one of the two alleles in somatic cells is specific with respect to parental source for imprinted genes and random for X-linked genes. We report here evidence indicating that: (i) differential methylation patterns of imprinted genes are not simply copied from the gametes, but rather established gradually after fertilization; (ii) very similar methylation patterns are observed for diploid, tetraploid, parthenogenic, and androgenic preimplantation mouse embryos, as well as parthenogenic and androgenic mouse embryonic stem cells; (iii) haploid parthenogenic embryos do not show methylation adjustment as seen in diploid or tetraploid embryos, but rather retain the maternal pattern. These observations suggest that differential methylation in imprinted genes is achieved by a dynamic process that senses gene dosage and adjusts methylation similar to X-chromosome inactivation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartolomei M. S., Webber A. L., Brunkow M. E., Tilghman S. M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993 Sep;7(9):1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- Brandeis M., Kafri T., Ariel M., Chaillet J. R., McCarrey J., Razin A., Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993 Sep;12(9):3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillet J. R., Vogt T. F., Beier D. R., Leder P. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell. 1991 Jul 12;66(1):77–83. doi: 10.1016/0092-8674(91)90140-t. [DOI] [PubMed] [Google Scholar]
- Cuthbertson K. S. Parthenogenetic activation of mouse oocytes in vitro with ethanol and benzyl alcohol. J Exp Zool. 1983 May;226(2):311–314. doi: 10.1002/jez.1402260217. [DOI] [PubMed] [Google Scholar]
- Driscoll D. J., Waters M. F., Williams C. A., Zori R. T., Glenn C. C., Avidano K. M., Nicholls R. D. A DNA methylation imprint, determined by the sex of the parent, distinguishes the Angelman and Prader-Willi syndromes. Genomics. 1992 Aug;13(4):917–924. doi: 10.1016/0888-7543(92)90001-9. [DOI] [PubMed] [Google Scholar]
- Feil R., Walter J., Allen N. D., Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994 Oct;120(10):2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- Glenn C. C., Porter K. A., Jong M. T., Nicholls R. D., Driscoll D. J. Functional imprinting and epigenetic modification of the human SNRPN gene. Hum Mol Genet. 1993 Dec;2(12):2001–2005. doi: 10.1093/hmg/2.12.2001. [DOI] [PubMed] [Google Scholar]
- Grant S. G., Chapman V. M. Mechanisms of X-chromosome regulation. Annu Rev Genet. 1988;22:199–233. doi: 10.1146/annurev.ge.22.120188.001215. [DOI] [PubMed] [Google Scholar]
- Hatada I., Sugama T., Mukai T. A new imprinted gene cloned by a methylation-sensitive genome scanning method. Nucleic Acids Res. 1993 Dec 11;21(24):5577–5582. doi: 10.1093/nar/21.24.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992 May;6(5):705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Kay G. F., Barton S. C., Surani M. A., Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell. 1994 Jun 3;77(5):639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Kitsberg D., Selig S., Brandeis M., Simon I., Keshet I., Driscoll D. J., Nicholls R. D., Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993 Jul 29;364(6436):459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Knoll J. H., Cheng S. D., Lalande M. Allele specificity of DNA replication timing in the Angelman/Prader-Willi syndrome imprinted chromosomal region. Nat Genet. 1994 Jan;6(1):41–46. doi: 10.1038/ng0194-41. [DOI] [PubMed] [Google Scholar]
- Lalande M. In and around SNRPN. Nat Genet. 1994 Sep;8(1):5–7. doi: 10.1038/ng0994-5. [DOI] [PubMed] [Google Scholar]
- Latham K. E., Doherty A. S., Scott C. D., Schultz R. M. Igf2r and Igf2 gene expression in androgenetic, gynogenetic, and parthenogenetic preimplantation mouse embryos: absence of regulation by genomic imprinting. Genes Dev. 1994 Feb 1;8(3):290–299. doi: 10.1101/gad.8.3.290. [DOI] [PubMed] [Google Scholar]
- Lawitts J. A., Biggers J. D. Culture of preimplantation embryos. Methods Enzymol. 1993;225:153–164. doi: 10.1016/0076-6879(93)25012-q. [DOI] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993 Nov 25;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Lyon M. F. Epigenetic inheritance in mammals. Trends Genet. 1993 Apr;9(4):123–128. doi: 10.1016/0168-9525(93)90206-w. [DOI] [PubMed] [Google Scholar]
- Migeon B. R. X-chromosome inactivation: molecular mechanisms and genetic consequences. Trends Genet. 1994 Jul;10(7):230–235. doi: 10.1016/0168-9525(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Mohandas T., Sparkes R. S., Shapiro L. J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981 Jan 23;211(4480):393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Mutter G. L., Stewart C. L., Chaponot M. L., Pomponio R. J. Oppositely imprinted genes H19 and insulin-like growth factor 2 are coexpressed in human androgenetic trophoblast. Am J Hum Genet. 1993 Nov;53(5):1096–1102. [PMC free article] [PubMed] [Google Scholar]
- Nicholls R. D. New insights reveal complex mechanisms involved in genomic imprinting. Am J Hum Genet. 1994 May;54(5):733–740. [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Tanguay R. L., Steigerwald S. D., Riggs A. D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990 Aug;4(8):1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- Rachmilewitz J., Goshen R., Ariel I., Schneider T., de Groot N., Hochberg A. Parental imprinting of the human H19 gene. FEBS Lett. 1992 Aug 31;309(1):25–28. doi: 10.1016/0014-5793(92)80731-u. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and genomic imprinting. Cell. 1994 May 20;77(4):473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- Reik W., Allen N. D. Genomic imprinting. Imprinting with and without methylation. Curr Biol. 1994 Feb 1;4(2):145–147. doi: 10.1016/s0960-9822(94)00034-5. [DOI] [PubMed] [Google Scholar]
- Reik W., Howlett S. K., Surani M. A. Imprinting by DNA methylation: from transgenes to endogenous gene sequences. Dev Suppl. 1990:99–106. [PubMed] [Google Scholar]
- Reis A., Dittrich B., Greger V., Buiting K., Lalande M., Gillessen-Kaesbach G., Anvret M., Horsthemke B. Imprinting mutations suggested by abnormal DNA methylation patterns in familial Angelman and Prader-Willi syndromes. Am J Hum Genet. 1994 May;54(5):741–747. [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Pfeifer G. P. X-chromosome inactivation and cell memory. Trends Genet. 1992 May;8(5):169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Jones P. A., Chaillet J. R., Ferguson-Smith A. C., Barton S. C., Reik W., Surani M. A. Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev. 1992 Oct;6(10):1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- Shemer R., Kafri T., O'Connell A., Eisenberg S., Breslow J. L., Razin A. Methylation changes in the apolipoprotein AI gene during embryonic development of the mouse. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11300–11304. doi: 10.1073/pnas.88.24.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solter D. Differential imprinting and expression of maternal and paternal genomes. Annu Rev Genet. 1988;22:127–146. doi: 10.1146/annurev.ge.22.120188.001015. [DOI] [PubMed] [Google Scholar]
- Stöger R., Kubicka P., Liu C. G., Kafri T., Razin A., Cedar H., Barlow D. P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993 Apr 9;73(1):61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Surani M. A. Genomic imprinting. Silence of the genes. Nature. 1993 Nov 25;366(6453):302–303. doi: 10.1038/366302a0. [DOI] [PubMed] [Google Scholar]
- Surani M. A., Kothary R., Allen N. D., Singh P. B., Fundele R., Ferguson-Smith A. C., Barton S. C. Genome imprinting and development in the mouse. Dev Suppl. 1990:89–98. [PubMed] [Google Scholar]
- Sutcliffe J. S., Nakao M., Christian S., Orstavik K. H., Tommerup N., Ledbetter D. H., Beaudet A. L. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994 Sep;8(1):52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- Szabó P., Mann J. R. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development. 1994 Jun;120(6):1651–1660. doi: 10.1242/dev.120.6.1651. [DOI] [PubMed] [Google Scholar]
- Takagi N., Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975 Aug 21;256(5519):640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Tremblay K. D., Saam J. R., Ingram R. S., Tilghman S. M., Bartolomei M. S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995 Apr;9(4):407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- Walsh C., Glaser A., Fundele R., Ferguson-Smith A., Barton S., Surani M. A., Ohlsson R. The non-viability of uniparental mouse conceptuses correlates with the loss of the products of imprinted genes. Mech Dev. 1994 Apr;46(1):55–62. doi: 10.1016/0925-4773(94)90037-x. [DOI] [PubMed] [Google Scholar]