Abstract
The Polycomb-group protein, Ezh2, is required for epigenetic gene silencing in the adult heart by unknown mechanism. We investigated the role of Ezh2 and non-coding RNAs in a mouse model of pressure overload using transverse aortic constriction (TAC) attenuated by the prototypical histone deacetylase inhibitor, trichostatin A (TSA). Chromatin immunoprecipitation of TAC and TAC+TSA hearts suggests interaction of Ezh2 and primary microRNA-208b (pri-miR-208b) in the regulation of hypertrophic gene expression. RNAi silencing of pri-miR-208b and Ezh2 validate pri-miR-208b-mediated transcriptional silencing of genes implicated in cardiac hypertrophy including the suppression of the bi-directional promoter (bdP) of the cardiac myosin heavy chain genes. In TAC mouse heart, TSA attenuated Ezh2 binding to bdP and restored antisense β-MHC and α-MHC gene expression. RNA-chromatin immunoprecipitation experiments in TAC hearts also show increased pri-miR-208b dependent-chromatin binding. These results are the first description by which primary miR interactions serve to integrate chromatin modifications and the transcriptional response to distinct signaling cues in the heart. These studies provide a framework for MHC expression and regulation of genes implicated in pathological remodeling of ventricular hypertrophy.
INTRODUCTION
The heart undergoes dramatic remodeling under physiological and pathological stress that is associated with changes in gene function (1). Pressure overload by transverse aortic constriction (TAC) upregulates the expression of hypertrophy markers such as Anp (Nppa) and Bnp (Nppb), whereas it downregulates Serca2a gene expression (2). Cardiac hypertrophy is associated with a shift in myosin heavy chain (MHC) gene expression (3). Chromatin-modifying enzymes and non-coding RNAs (ncRNAs) are thought to mediate gene regulatory functions in cardiac hypertrophy (4,5). Chromatin remodeling complexes such as Brg1 and HDAC enzymes are known to regulate genes implicated in hypertrophy by directly associating to the intergenic bi-directional promoter (bdP) of the α- and β-MHC genes (6). Expression of antisense (AS) RNA (AS β-MHC) from the bdP is associated with β-MHC gene expression in hypertrophic and hypothyroid animals (7,8). Together with the evolutionary conserved Myh7b gene, these three myosin genes are regulated by MyomiRs, a family of intronic miRNAs (miR-208a, miR-208b and miR-499) that govern cardiac hypertrophy, growth and muscle performance (9,10).
The histone methyltransferase enzyme, Ezh2, regulates gene expression in mature cardiomyocytes (11). Although the functional relevance of the polycomb-group (PcG) histone methyltransferase enzyme, Ezh2, in the healthy heart was recently shown, its role in heart disease remains poorly characterized. Ezh2-deficient mice show increased cardiac growth with upregulated expression of hypertrophic and fibrotic genes such as Anp, Bnp, β-MHC and Tgfb3 (12). How these genes are precisely regulated by Ezh2 as well as the underlying mechanisms in the hypertrophied heart is not well understood. To study the role of Ezh2, we induced pressure overload by TAC in mice and attenuated the hypertrophy-associated gene expression using histone deacetylase (HDAC) inhibitors. HDAC inhibitors such as trichostatin A (TSA) can reverse pathological hypertrophy and ameliorate cardiac function (13). Structurally diverse HDAC inhibitors can attenuate the induction of hypertrophy markers such as Anp, Bnp and the embryonic β-MHC genes (14–16).
In this study, we examined the expression of a number of genes implicated in cardiac hypertrophy and assessed chromatin interaction of long ncRNAs in complex with PcG silencing proteins such as Ezh2. Results from TAC animals as well as in vitro loss-of-function studies suggest a novel role for Ezh2 and the primary microRNA-208b (pri-miR-208b) transcript. Administration of TSA in TAC mice attenuated the expression of pressure overload-induced hypertrophic and fibrotic genes. These results describe for the first time the interaction of pri-miR-208b targeting specific chromatin modifications and regulation of genes implicated in pathological remodeling of ventricular hypertrophy. These studies provide new insights for specific long non-coding RNAs (lncRNAs) underlying the exchange of regulatory complexes involved with chromatin modification and the control of gene transcription in cardiac hypertrophy.
MATERIALS AND METHODS
TAC surgery
Chronic pressure overload was induced by transverse aorta constriction (TAC) performed on C57BL6 male mice at 10–12 weeks of age. TAC mice were injected subcutaneously with either dimethyl sulfoxide (DMSO) or TSA (dissolved in 50% DMSO), twice daily at 0.6 mg/kg/day for a period of 4 weeks. Establishment of hypertrophy and attenuation by TSA was confirmed by echocardiography by measuring left ventricular (LV) wall thickness and dimensions end-diastole and end-systole (17). Sham control animals underwent the same surgical procedures without constriction of the aorta and exhibited no hypertrophy. Non-invasive echocardiographic test was performed at the end of the 4-week study period. Mice were anesthetized with isoflurane (4% for induction and 1.7% for maintenance). Using iE33 ultrasound system (Philips) and a 15-MHz liner-array transducer, 2D short-axis view of the left ventricle (LV) was obtained and M-mode traces were acquired as we previously described (18). LV diameters at diastole and systole (LVDd, LVDs) or wall thickness at diastole was measured, and fractional shortening and LV mass were calculated. Results are presented as mean ±SEM.
Sca1+ cell fractionation and isolation
Stem cell antigen-1 positive (Sca-1+) progenitor cells were isolated by magnetic activated cell sorting system using anti-Sca-1 microbeads (Miltenyi Biotech) from differentiated mouse embryonic stem cells by the removal of 103 U/ml leukemia inhibitory factor (ESGRO, Millapore), as previously described (19). In all, 80–95% of isolated cells positive for Sca1 antigen were evaluated by flow cytometry and immunostaining. Sca-1+ and mES cells were cultured on 1% gelatin-coated dishes with Dulbecco’s modified Eagle’s medium supplemented with ES-qualified fetal bovine serum (FBS), knockout serum replacement, non-essential amino acids, antibiotics (penicillin/streptomycin) and β-mercaptoethanol (Gibco) at 37°C in humid air with 5% CO2. To obtain higher numbers of Sca-1+ cells, these cells were allowed to grow in culture without leukemia inhibitory factor for two to eight population doublings.
Mouse neonatal ventricular cardiomyocyte isolation, culture and treatment
Day 1—C57BL/6 pups were sacrificed by single cut decapitation procedure and their chests were opened. Hearts were suspended in Hanks solution. Ventricles were carefully dissected and suspended into Hanks + Trypsin solution followed by 4°C incubation overnight on the orbital shaker. Day 2—Tissues were suspended in Hanks + Collagenase medium for tissue dissociation, and resulting cell suspension was collected. This step was repeated until complete tissue dissociation was achieved. The digested extract was pelleted, and adding Dulbecco’s modified Eagle's medium/10% fetal calf serum (FCS) deactivated the collagenase. The cells were plated in P150 culture dish and incubated for 50 min at 37°C to allow fibroblasts to adhere to the bottom of the dish. Cell suspension was collected and incubated again for 45 min at 37°C to allow the remaining fibroblasts to adhere to the dish. After second incubation, cell suspension was collected into a sterile container. Bromodeoxyuridine (Brdu) (3.15 mg/ml) 100 ul/10 ml media was added, and cells were plated at the density of 0.5 × 106/ml, 2 ml/well (6-well plate). Cardiomyocytes were allowed to settle for 2–3 days followed by serum starvation before TSA treatment was initiated. The serum-starved neonatal ventricular cardiomyocytes were incubated for 48 h in media containing DMSO (vehicle) or TSA (100 nM).
Total RNA preparation
For in vivo studies, mice heart LV tissue was homogenized in Trizol reagent (Invitrogen) in the presence of RNase inhibitor (SUPERase-In, Ambion). For in vitro studies, cultured mouse cardiac progenitor cells were trypsinized and pelleted by centrifugation, and then resuspended with Trizol reagent. Vigorous phenol–chloroform treatment to obtain RNA-containing aqueous phase was followed by purification of RNA using RNeasy Mini Kit preparation columns (Qiagen). DNase treatment ensured removal of residual DNA, and total RNA was accurately quantified using Qubit fluorometer (Invitrogen).
RNA quantification by quantitative reverse transcriptase-polymerase chain reaction
For gene expression studies, total RNA (1–2 µg) was reverse transcribed using high capacity cDNA synthesis kit (Applied Biosystems) in a 20-µl reaction volume containing random primers. For strand-specific quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) of MHC genes, forward and reverse primers were included in separate reactions, and cDNA synthesis was performed at 60°C using Thermoscript cDNA preparation system (Invitrogen) and real-time quantification was performed using Fast SyBr-Green qPCR system (Applied Biosystems). Gene expression levels are expressed as fold difference where relative RNA levels between the control and treatment groups were normalized. The specificity of template amplification was assessed by melt curve analysis. To ensure the RNA strand-specific cDNA synthesis, negative primer controls and negative enzyme controls were included and assessed for negligible non-specific amplification. Oligonucleotide sequences are listed in Supplementary Data. For miR detection, ∼10 ng of RNA was included in Taqman MicroRNA Reverse transcription kit (PN 4366596). miR-specific RT reactions were carried out for miR-208a (RT000511), miR-208b (RT002290) and control small ncRNA snoRNA135 (RT001230). Relative levels were amplified in PCR using Taqman small RNA Assays for miR-208a (TM000511), miR-208b (TM002290) and snoRNA135 (TM001230). The pri-miR-208b transcript was detected using strand-specific amplification as well as Taqman primary microRNA assays (Applied Biosystems).
Chromatin immunoprecipitation
For in vivo studies, LV tissue was carefully dissected and finely diced. Formaldehyde fixation (1%) was carried out in phosphate buffered saline (w/o Ca2+/Mg2+) by rotation for 10 min at room temperature, and quenching of excessive formaldehyde was achieved using glycine (0.125 M) (17,20). Fixed tissues were washed with ice-cold phosphate buffered saline and homogenized to a clear solution in warm sodium dodecyl sulphate (SDS) lysis buffer containing 1% SDS, 10 mM ethylenediaminetetraacetic acid (EDTA), 50 mM Tris–HCl, pH 8, containing protease inhibitor cocktail (Cayman). Homogenate was placed on ice to ensure proper cell lysis. For in vitro studies, cultured cells (∼80% confluent) were formaldehyde (1%) fixed and quenched by adding glycine. Collected cell pellet was lysed in warm SDS lysis buffer. Cell lysates prepared from LV as well as from cultured cells were separated into 300-µl aliquots, and sonication of chromatin was achieved using the bioruptor (Diagnode) with constant power settings. Sheared soluble chromatin was size-fractionated using MultiNA (Shimadzu) to ensure proper sonication of chromatin (∼500 bp). Soluble chromatin (5-µg) was resuspended in chromatin immunoprecipitation (ChIP) dilution buffer containing 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl, pH 8.0, and 167 mM NaCl. Immunopurification of soluble chromatin was performed using antibodies for specific histone modifications, H3K9/14ac (Millipore 06-599), H3K4m3 (Abcam ab8580), H3K9m3 (Abcam ab8898) and H3K27m3 (Millipore 07-449). Antibodies for histone-modifying enzymes are Ezh2 (Abnova PAB0648), HDAC2 (Sigma H3159) and HDAC1 (Upstate 06-720). The antibody-bound chromatin fraction was precipitated using dynabeads coated with protein A/G (Invitrogen). Conjugates were washed with increasing salt conditions followed by LiCl and SDS washes. Input and antibody-bound isolates were heated at 64°C/2 h in elution buffer containing proteinase K, and the reverse cross-linked DNA was purified by column fractionation (Nucleospin). ChIP-enriched DNA sequences were detected by amplification using real-time qPCR system. Percentage input (% input) was calculated for each ChIP experiment, and results are expressed as relative fold enrichment/ratio for the target sequences compared between the treated versus control groups. The non-specific IgG immunoprecipitation served as control for non-specific background enrichment.
Chromatin RNA immunoprecipitation (RNA-ChIP)
Mice heart LV tissue and cardiac stem cells were formaldehyde fixed, quenched with glycine and resuspended with SDS lysis buffer as described above. To ensure RNA integrity and to protect from RNase degradation, only freshly prepared buffers containing RNase inhibitors were used. Cell lysates were sonicated in the size optimized for RNA (∼1000-bp long), and residual DNA from sheared chromatin was removed by mild DNase treatment. Antibodies that recognize histone H3 (Abcam ab1791) and Ezh2 (Abnova PAB0648) were used in the immunoprecipitation procedure (4°C/4–5 h) to purify RNA associated with chromatin. Conjugates were reverse cross-linked, and RNA was purified using Qiagen RNeasy columns followed by removal of DNA (Roche). Strand-specific reverse transcription of chromatin-associated RNAs and input RNA was performed using Thermoscript reverse transcription protocol (Invitrogen). Relative enrichment of long ncRNAs such as AS β-MHC and pri-miR-208b was assessed using qRT-PCR. Strand-specific amplification was ensured using controls as described above.
short-hairpin RNA-mediated knockdown strategy
The knockdown of Ezh2 protein, ncRNAs AS β-MHC and pri-miR-208b in Sca1+ vascular progenitors was achieved by MISSION short-hairpin RNA (shRNA) expressing lentivirus vectors (Sigma) as described previously (21). Target sequences are as follows: Ezh2, 5′-GCTAGGCTAATTGGGACC-3′ (TRCN0000039041), shRNA for AS β RNA 5′-CTGCTAGTTAGTATCCTACGC-3′ and shRNA for miR-208b 5′-CCGAATATAAGACGAACAAAA-3′. The knockdown of Ezh2 in the cells was verified by qRT-PCR and immunoblots. The knockdown of AS β-MHC and pri-miR-208b was verified by strand-specific qRT-PCR assays, as described above. Cells transduced with the MISSION Non-target shRNA control vector (Sigma) served as controls.
Nuclear and cytoplasmic cellular fractionation
Preparation of cytosolic and nuclear fractions was performed as described previously (21). Sca-1+ cells (1 × 107) were resuspended in hypotonic buffer containing 5 mM Tris HCl, pH 7.5, 20 mM KCl, 2 mM MgCl2, 0.25 mM EDTA, 0.125 mM ethylene glycol tetraacetic acid (EGTA), 1 mM Dithiothreitol (DTT), 0.5 mM Phenylmethanesulfonylfluoride (PMSF), 0.05% NP-40 and protease inhibitor (Cayman) for 10 min at 4°C. The cytosolic fraction (supernatant) and the nuclear pellet were separated by centrifugation at 600 g for 5 min. The pellet was extracted with buffer containing 20 mM Hepes KOH, pH 7.9, 25% glycerol, 520 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 0.2% NP-40 and protease inhibitor for 15 min at 4 °C. The nuclear fraction was recovered by centrifugation at 15 000 g for 15 min. The nuclear and cytosolic fractions were verified by immunoblot procedure using anti-Brm and anti-MeCP2 (nuclear markers) as well as anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (cytoplasmic marker) antibodies.
In vitro dsDNA/RNA binding assay
The intergenic bdP fragment (∼2 kb) corresponding to positions −726 to −2756 relative to α-MHC transcription start site was amplified using PCR. One picomole of intergenic bdP fragment was incubated at 37°C for 30 min with 100 pmol of pri-miR-208b (CCUCUCAGGGAAGCUUUUUGCUCGCGUUAUGUUU-BiotinTg) or non-specific (UCCCCUGUGGGUGGGGGUGGGGGGUCUUU-BiotinTg) RNA oligonucleotides (Sigma). The RNA/DNA hybrids were immunoprecipitated using streptavidin-coated beads (Life technologies) in the presence or absence of RNase H or RNase V1. The bdP sequence was amplified using real-time qPCR, and the fold enrichment was determined between the specific and non-specific oligo-enriched samples.
Statistical analysis
Data were presented as ±SEM in each group. All data were evaluated with a two-tailed, unpaired Student’s t-test using QuickCalcs (GraphPad). Statistical significance was obtained by comparing groups viz Sham versus TAC and TAC versus TAC+TSA. For in vitro studies, data compared viz non-targets versus shRNA and shRNA versus shRNA+TSA. A value of P < 0.05 was considered statistically significant.
RESULTS
TSA regulates the hypertrophy-induced expression of MHC ncRNAs
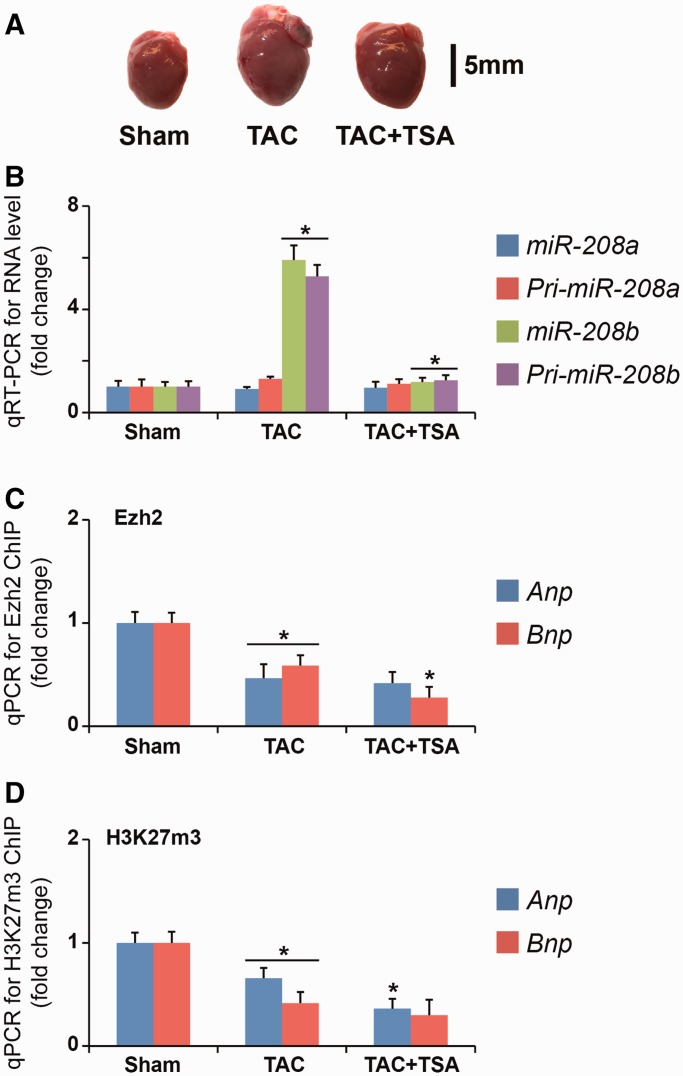
The mouse pressure overload model by TAC accelerated LV hypertrophy and heart failure (Table 1). Administration of TSA following TAC surgery reduced the severity of cardiac hypertrophy (Figure 1A). In TAC animals, we examined the expression of hypertrophy-associated genes, and demonstrated that increased Anp, Bnp and reduced Serca2a mRNA expression were attenuated by TSA (Supplementary Figure S1) (13). We confirmed in TAC animals that induction of hypertrophy altered MHC gene expression (elevated β-MHC as well as reduced α-MHC gene expression) was attenuated by TSA. Consistent with previous reports (7), expression of AS β-MHC from the bdP of the MHC intergenic region was reduced in TAC animals and TSA prevented this reduction. We quantified the expression of miR and report the induction of pri-miR-208b and miR-208b transcripts but not miR-208a in TAC animals were attenuated by TSA administration (Figure 1B) (22). The epigenetic regulation of Anp, Bnp and MHC genes by Ezh2 enzyme in wild-type animals was recently reported (11,12). The PcG protein, Ezh2, tri-methylates histone H3 at Lysine 27 (H3K27m3) and mediates gene suppression (23). In TAC animals, we examined the interaction of Ezh2 as well as H3K27m3 modification on Anp, Bnp and MHC gene promoters by ChIP-qPCR. Ezh2 binding was reduced in hypertrophic animals on the Anp and Bnp genes and this was not recovered in TSA-administered mice (Figure 1C). We also assessed H3K27m3 on Anp and Bnp promoters by ChIP and observed consistent reduction in TAC mice as well as in TSA-administered animals (Figure 1D). These results suggest that the induction of Anp and Bnp genes in TAC animals is associated with reduced Ezh2 binding and H3K27m3 modification.
Table 1.
Echocardiographic parameters and LV mass in TAC exposed TSA hearts
| ECG parameters | Sham | TAC + vehicle | TAC + TSA |
|---|---|---|---|
| Number | 10 | 12 | 12 |
| Heart rate (beats/min) | 578 ± 12 | 565 ± 11 | 552 ± 8 |
| LVDd (mm) | 4.1 ± 0.05 | 4.5 ± 0.1* | 3.9 ± 0.07*,† |
| LVDs (mm) | 2.7 ± 0.08 | 3.6 ± 0.2* | 2.6 ± 0.08† |
| Fractional shortening (%) | 34 ± 1 | 20 ± 1* | 32 ± 1† |
| Wall thickness (mm) | 0.72 ± 0.02 | 1.01 ± 0.04* | 0.90 ± 0.02* |
| LV mass (mg) | 116 ± 4 | 208 ± 13* | 147 ± 6*,† |
TAC, transverse aortic constriction; LVDd and LVDs, left ventricular diameter at diastole or systole; *P < 0.01 versus Sham group; †P < 0.05 versus TAC + vehicle group.
Figure 1.
TAC-induced hypertrophy is associated with the release of Ezh2 at ANP and BNP genes. (A) Pressure overload-induced cardiac hypertrophy in mice was attenuated by TSA administration. Image of Sham (control) and TAC hearts as well as reduced left arterial thrombus following TSA administration (black bar = 5 mm). (B) Hypertrophy-induced expression of miR-208b and pri-miR-208b transcripts in the LV of TAC mice was attenuated by TSA administration; *P < 0.001. (C) ChIP for Ezh2 enzyme followed by real-time qPCR for Anp and Bnp promoters shows reduced binding of Ezh2 in TAC mice; *P < 0.02. (D) ChIP for H3K27m3 followed by real-time qPCR for Anp and Bnp promoters shows reduced H3K27m3 enrichment in TAC mice. (n = 4); *P < 0.02.
Expression of AS β-MHC is determined by the binding of Ezh2 at the bdP
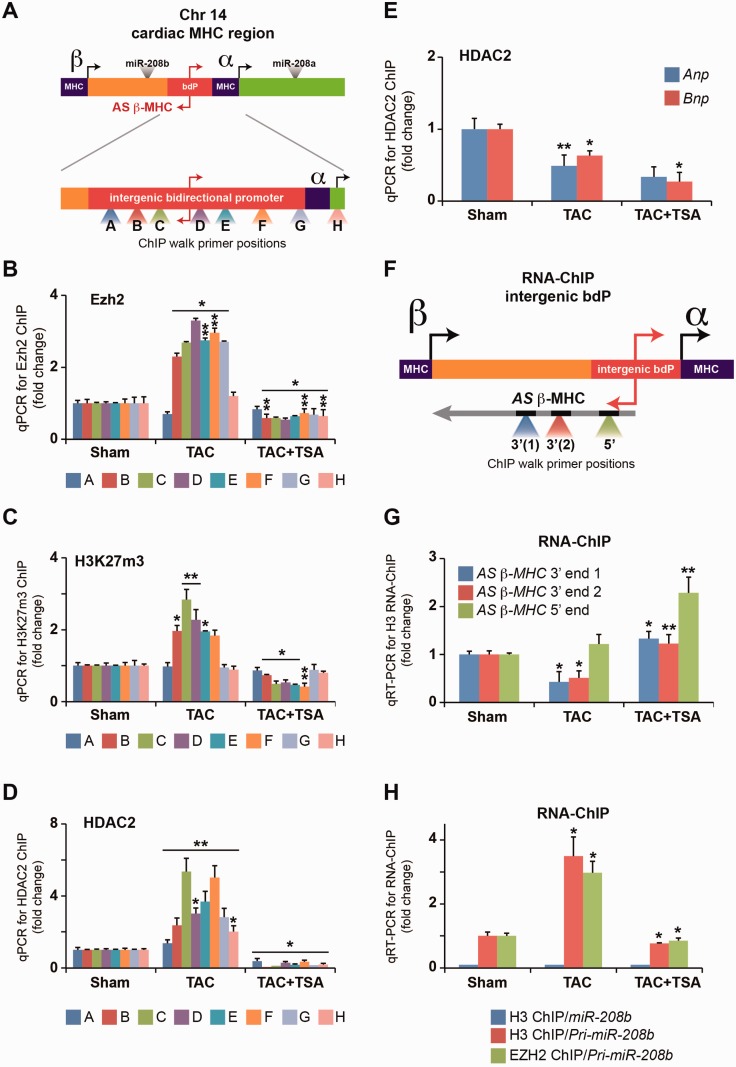
We screened the intergenic bdP region of the MHC genes by ChIP-qPCR (Figure 2A). We report increased Ezh2 interaction on the intergenic bdP in TAC animals and this was attenuated in TSA-administered mice (Figure 2B). This binding pattern was consistent with H3K27m3 enrichment (Figure 2C). We determined additional repressive histone marks. We report unremarkable enrichment of H3K9m3 on the bdP in TAC mice (Supplementary Figure S2). Ezh2 was recently demonstrated to interact with HDAC complexes on suppressed genes (24,25). To test the interaction of HDACs on the bdP sequence, LV chromatin was immunopurified from TAC mice using antibodies that recognize HDACs. The intergenic bdP was enriched for HDAC2 binding and not HDAC1 in TAC mice, which was abrogated by TSA (Figure 2D). Consistent with the changes in HDAC2 binding to the bdP, reduced binding on Anp and Bnp promoters in TAC and TSA mice was observed (Figure 2E). However, neither Ezh2 nor HDAC2 mRNA expression were altered in these mice (Supplementary Figure S3). In summary, these results suggest the reduced binding of Ezh2 and HDAC2 correlates with the induction of Anp and Bnp genes, whereas, the increased binding of these determinants participate in the suppression of AS β-MHC and α-MHC gene expression in the hypertrophic heart.
Figure 2.
Pathological hypertrophy induced α- to β-MHC shift is associated with the interaction of Ezh2 with the intergenic bdP. (A) Schematic representation of the cardiac MHC genomic region on chromosome 14 showing primer positions (A–H) used for ChIP walk. (B) Enrichment of intergenic bdP sequence was determined by real-time qPCR after Ezh2-ChIP, (C) H3K27m3-ChIP and (D) HDAC2-ChIP. *P < 0.01; **P < 0.03. (E) Real-time qPCR for Anp and Bnp promoters was performed after HDAC2-ChIP. *P < 0.008; **P < 0.03. (F) Schematic representation of the chromatin binding of AS β-MHC transcript and primer positions (3' and 5') used in qRT-PCR after RNA-ChIP. (G) Chromatin-dependent association of AS β-MHC transcript after histone H3 RNA-ChIP. *P < 0.006; **P < 0.01. (H) Interaction of pri-miR-208b with chromatin by RNA-ChIP using antibodies that recognize histone H3 and Ezh2 in the LV of TAC mice. *P < 0.01. All experiments independently performed n = 4.
The AS β-MHC transcript associates with chromatin
Recent evidence suggests that ncRNAs serve to integrate transcriptional responses conferred by chromatin modification (26). Notably, Ezh2 is thought to associate with long ncRNAs to regulate gene expression (27,28). To determine whether ncRNA expression in the hypertrophied heart could be directing Ezh2 methyltransferase to the bdP, soluble chromatin from LVs of TAC mice was immunoprecipitated using histone H3 antibody. Specific interaction of the AS β-MHC with chromatin was distinguishable from the sense β-MHC transcript (Supplementary Figure S4). The AS β-MHC transcript is >5 kb in length; therefore, we designed primers targeting the 5′- and 3′-ends of the transcript to examine chromatin-dependent association in TAC animals as well as in animals treated with TSA (Figure 2F). We detected two regions (end 1 and end 2) at the 3′ sequence of AS β-MHC transcript interacting with chromatin and this was reduced in LV chromatin isolated from TAC animals (Figure 2G). To extend these studies to the 5′-end, we validated the interaction of the AS β-MHC transcript with chromatin isolated from LV tissue. Increased association of the AS β-MHC transcript with chromatin in TSA-administered TAC mice was consistent with β-MHC expression. These results suggest that the AS β-MHC transcript could be interacting with the β-MHC gene to regulate histone modifications and gene expression. The β-MHC gene contains sense strand specificity, which is thought to also regulate its gene expression, so we examined two ncRNA transcripts that originate from the sense strand, a long pri-miR-208b as well as the short miR-208b. The interaction of the pri-miR-208b with chromatin was determined by RNA-ChIP using histone H3 antibody. TSA administration in TAC mice attenuated pri-miR-208b interaction with chromatin (Figure 2H). In contrast, the mature 22-nt miR-208b sequence was undetectable in chromatin isolated from the LV tissue of these animals. Next, we examined whether pri-miR-208b binds to Ezh2 enzyme in the heart. The immunopurification of soluble chromatin using antibody that recognizes Ezh2 shows specific interaction of pri-miR-208b transcript in the hypertrophic heart and this could be attenuated by the HDAC inhibitor, TSA. Taken together, these data suggest several scenarios that could mediate the expression of genes associated with hypertrophy. First, the Ezh2 methyltransferase directly suppresses the expression of target genes including AS β-MHC and α-MHC in response to hypertrophy. Alternatively, the expression of ncRNAs such as AS β-MHC and/or pri-miR-208b transcript could direct Ezh2 interaction to distinct gene promoters such as the intergenic bdP.
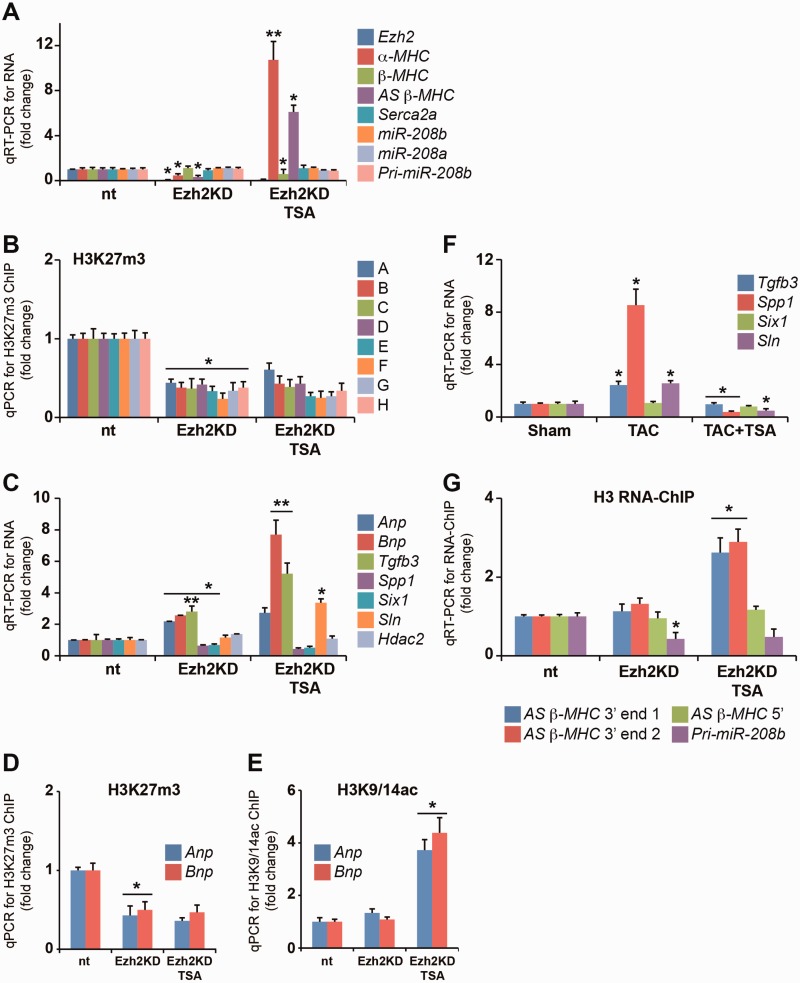
Interaction of AS β-MHC at the bdP is independent of Ezh2
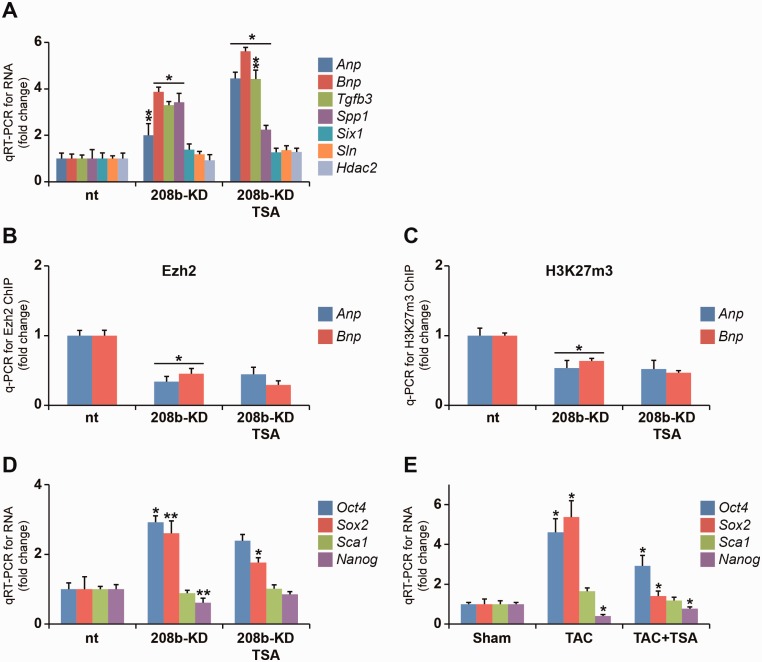
We explored chromatin-associated RNAs in Sca1+ progenitor cells because the immunoprecipitation procedure requires significant numbers of adult heart cells and the integrity of soluble chromatin was reduced because of collagenase treatments in cardiomyocyte isolation procedures (29). Ezh2 loss-of-function experiments using shRNA (Ezh2KD) were performed in mouse Sca1+ progenitor cells (Supplementary Figure S5). Ezh2 knockdown efficiency (93%) was determined using qRT-PCR (Figure 3A). ChIP results showed significant binding of Ezh2 at the intergenic bdP in non-target cells. We observed reduced AS β-MHC and α-MHC expression in Ezh2KD cells (Figure 3A). In Ezh2KD cells, HDAC inhibition by TSA increased the expression of AS β-MHC and α-MHC genes and this was consistent with the enrichment of histone H3K9/14ac and H3K4m3 at the intergenic bdP (Supplementary Figures S6 and S7). This observation corresponds with Ezh2 binding to the bdP suppressing AS β-MHC, whereas the activation of AS β-MHC requires H3K9/14ac modification in Ezh2KD cells. Ezh2 knockdown significantly reduced H3K27m3 enrichment at the intergenic bdP (Figure 3B). Consistent with our in vivo experimental results, we observed increased expression of Anp and Bnp genes in Ezh2KD cells (Figure 3C). ChIP experiments show reduced H3K27m3 on the Anp and Bnp genes in these cells (Figure 3D). Stimulation by TSA increased H3K9/14ac (Figure 3E). Taken together, these experimental results suggest Ezh2 confers chromatin silencing events by H3K27m3 to regulate transcriptional responses. Expression of profibrotic genes such as Tgfb3, Spp1 as well as Sln and Six1 is thought to be regulated by Ezh2 in the adult mouse heart (12). We report in Ezh2KD cells, Tgfb3 was upregulated, whereas Spp1 and Six1 genes were suppressed. In TAC animals, the expression of Tgfb3, Spp1 and Sln was increased while TSA administration attenuated gene expression (Figure 3F). These results in the mouse are consistent with in vitro observations and suggest Ezh2 could play a role in regulating H3K27m3-mediated gene expression in the hypertrophied heart. We hypothesized that Ezh2 binding confers the expression of hypertrophic as well as profibrotic genes through interaction with ncRNAs, such as AS β-MHC and pri-miR-208b. To test this, we examined the interaction of both transcripts associated with immunoprecipitated soluble chromatin prepared from Ezh2-deficient cells. The specific interaction of AS β-MHC transcript at the 3′-end remained unchanged on chromatin (Figure 3G). Interestingly, TSA increased AS β-MHC expression as well as its interaction with chromatin which was correlated with reduced β-MHC gene expression. We observed a strong reduction of interacting pri-miR-208b transcript on chromatin, which was not changed by TSA treatment. These results suggest that the interaction of AS β-MHC transcript with chromatin inversely correlates with β-MHC gene expression in TAC+TSA animals. In addition, experiments in Ezh2KD cells also indicate the interaction of pri-miR-208b transcript with chromatin was dependent on Ezh2 enzyme. In conclusion, the AS β-MHC interaction with chromatin was associated with β-MHC gene expression in mice and this was regulated by Ezh2 binding at the intergenic bdP in pathological hypertrophy.
Figure 3.
Chromatin association of AS β-MHC transcript is independent of Ezh2 and regulates β-MHC gene expression. (A) Gene expression was assessed by qRT-PCR in non-target (nt), Ezh2-deficient (Ezh2KD) and Ezh2KD cells stimulated with TSA. *P < 0.001; **P < 0.01. (B) H3K27m3-ChIP for intergenic bdP in Ezh2KD cells. *P < 0.001; **P < 0.01. (C) qRT-PCR detection of mRNA for Ezh2-regulated genes. *P < 0.001; **P < 0.015. (D) H3K27m3-ChIP for Anp and Bnp promoters in non-target (nt), Ezh2-deficient (Ezh2KD) and Ezh2KD cells stimulated with TSA; *P < 0.015. (E) H3K9/14ac-ChIP for Anp and Bnp promoters in non-target (nt), Ezh2-deficient (Ezh2KD) and Ezh2KD cells stimulated with TSA; *P < 0.005. (F) mRNA expression for Ezh2 cardiac gene targets determined by qRT-PCR in mice LV. *P < 0.005. (G) Loss of Ezh2 reduces the binding of pri-miR-208b but not AS β-MHC on chromatin binding of pri-miR-208b, whereas the binding of AS β-MHC transcript remained unaffected. *P < 0.01. All experiments independently performed n = 5.
Because AS β-MHC transcript binding to chromatin is reduced in the hypertrophic heart, we hypothesized that knockdown of AS β-MHC (AS β-MHC KD) could regulate the binding of Ezh2 at key cardiac genes. To do this, we used shRNA-mediated knockdown strategy to achieve efficient reduction of the AS β-MHC (86%) transcript. Knockdown of AS β-MHC did not alter Ezh2 binding nor its determinant H3K27m3 on the intergenic bdP as determined by ChIP-qPCR (Supplementary Figures S8 and S9). These results indicate that Ezh2 binding to intergenic bdP as well as to Anp and Bnp genes occurs independently of AS β-MHC transcript. Compatible with our hypothesis that AS β-MHC transcript is not guiding the interaction of Ezh2 with chromatin, we show reduced association of the AS β-MHC transcript with chromatin in AS β-MHC KD cells (Supplementary Figure S10). Interestingly, the interaction of the pri-miR-208b transcript with chromatin remained unaltered in AS β-MHC KD cells, whereas TSA exposure reduced this association. These data suggest that the AS β-MHC transcript does not guide Ezh2 to the bdP of the MHC gene.
Interaction of Ezh2 with chromatin is determined by pri-miR-208b
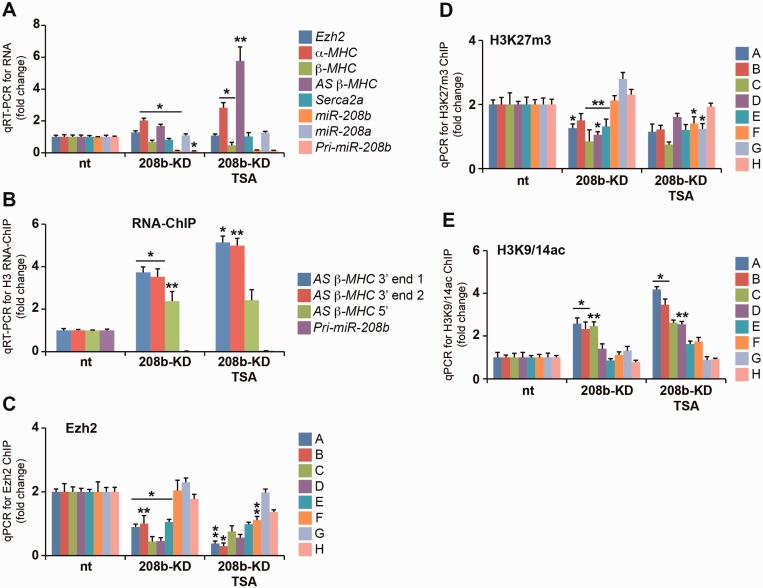
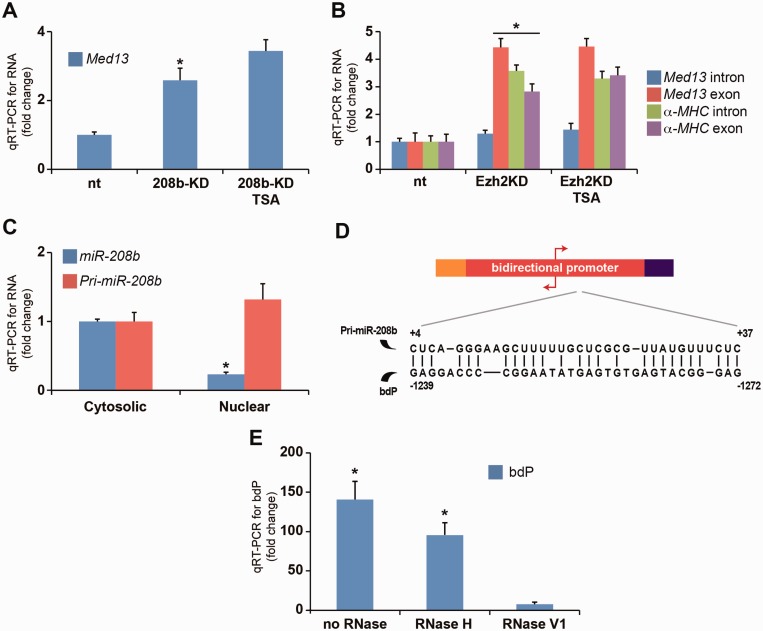
Next, we examined whether pri-miR-208b could direct Ezh2-mediated H3K27m3 modification on key cardiac genes. Primary miR transcripts are processed to mature miRNAs processed to mature miRNAs (∼22 nt in length) by RNase III family enzymes such as Drosha and Dicer (30). Using shRNA construct, we targeted the knockdown of the pri-miR-208b and miR-208b transcripts (90% reduction) in 208b-KD cells (Figure 4A). We used RNA-ChIP to determine the association of pri-miR-208b transcript with chromatin and show significant reduction in 208b-KD cells (Figure 4B). We then examined gene expression by qRT-PCR. While miR-208b deletion in mice show no changes in MHC gene expression (10), the loss of pri-miR-208b in 208b-KD cells increased the expression of bdP (AS β-MHC and α-MHC genes) and downregulated β-MHC (Figure 4A). We examined whether the chromatin interaction of AS β-MHC transcript resulted in β-MHC suppression in these cells. Association of the AS β-MHC transcript with chromatin was increased and inversely correlated with β-MHC gene expression in 208b-KD cells (Figure 4B). These changes to MHC gene expression suggest that the chromatin interaction of pri-miR-208b may be critical for epigenetic regulation of the intergenic bdP. Next, we assessed whether the loss of pri-miR-208b transcript altered Ezh2 and H3K27m3 association with the intergenic bdP region. ChIP results confirmed reduced Ezh2 binding at the bdP (Figure 4C) and this was consistent with reduced H3K27m3 modification in 208b-KD cells (Figure 4D). This was associated with H3K27m3 marks in these cells (Figure 4D). Release of Ezh2 from the intergenic region was consistent with H3K9/14ac increase in TSA-stimulated cells and increased AS β-MHC and α-MHC gene expression (Figure 4E). These results suggest the interaction of Ezh2 with the intergenic bdP requires pri-miR-208b transcript for gene silencing. We then determined whether the loss of pri-miR-208b transcript could deregulate other Ezh2 gene targets, and show increased Anp and Bnp gene expression (Figure 5A). Furthermore, additional Ezh2-regulated genes including the expression of Tgfb3 and Spp1 genes were increased, whereas Six1 and Sln remained unchanged in 208b-KD cells. Taken together, these experimental results support the idea that the pri-miR-208b transcript regulates gene expression by its interaction with Ezh2 and chromatin. Consistent with this idea, binding of Ezh2 and its determinant, H3K27m3, was reduced on the Anp and Bnp promoters in 208b-KD cells (Figure 5B and C). Therefore, the loss of pri-miR-208b results in changes to expression of Ezh2-regulated genes such as Anp, Bnp, Tgfb3, Spp1 as well as AS β-MHC and α-MHC genes. We examined whether pri-miR-208b-dependent chromatin binding of Ezh2 could regulate the expression of cardiac stem cell markers in TAC animals because an essential role for Ezh2 in adult muscle and neuronal regeneration after injury has been recently documented (31,32). Increased expression of endogenous cardiac stem cell (eCSC) genes such as Oct4 and Sox2 was observed, whereas Nanog was downregulated in 208b-KD cells (Figure 5D). TSA administration downregulated the expression of Oct4 and Sox2 while derepressing Nanog expression in 208b-KD cells. We extended these observations in LVs of TAC mice and report increased Oct4, Sox2 and decreased Nanog expression consistent with our in vitro observations (Figure 5E). Release of Ezh2 from gene targets in TAC animals as well as the activation of Ezh2-regulated genes in 208b-KD cells suggests that the pri-miR-208b transcript could regulate the chromatin binding of the Ezh2 enzyme. The precise role of pri-miR-208b in regulating eCSC marker expression remains to be investigated.
Figure 4.
Loss of pri-miR-208b transcript reduces the interaction of Ezh2 binding at the intergenic bdP. (A) qRT-PCR for RNA levels in 208b-KD cells shows activation of AS β-MHC and α-MHC genes. *P < 0.01; **P < 0.02. (B) RNA-ChIP in 208b-KD cells shows increased chromatin binding for AS β-MHC transcript. *P < 0.01; **P < 0.02. (C) Ezh2-ChIP in 208b-KD cells was assessed for intergenic bdP. *P < 0.001; **P < 0.004. (D) H3K27m3-ChIP showing reduced enrichment for bdP sequence. *P < 0.01; **P < 0.02. (E) H3K9/14ac-ChIP for intergenic bdP in 208b-KD cells. *P < 0.001; **P < 0.01. All experiments independently performed n = 5.
Figure 5.
Loss of pri-miR-208b is associated with the expression of Ezh2 gene targets. (A) Relative mRNA expression of Ezh2 gene targets was assessed by qRT-PCR in 208b-KD cells. *P < 0.003; **P < 0.015. (B) Ezh2 binding to Anp and Bnp promoters was reduced in 208b-KD cells; *P < 0.002. (C) H3K27m3-ChIP in 208b-KD cells followed by real-time qPCR detection of Anp and Bnp promoters; *P < 0.01. (D) Loss of pri-miR-208b is associated with the expression of stem cell genes. *P < 0.007. (E) mRNA expression for eCSC markers in Sham, TAC and TAC+TSA mice. *P < 0.001; **P < 0.01. All experiments independently performed n = 5.
Regulation by pri-miR-208b transcript
Our results suggest a regulatory function for the pri-miR-208b at the transcriptional level mediated by the PcG protein, Ezh2 as well as post-transcriptional regulation in the hypertrophied heart. To further characterize the functions of pri-miR-208b, we studied gene expression changes in 208b-KD cells because mature miRNAs suppress mRNA expression by binding to 3′-UTR sequences (30). Because miR-208b is a repressive determinant of Med13 (THRAP1) genes (9,10), we examined gene expression in 208b-KD cells. Figure 6A shows elevated Med13 expression in 208b-KD cells suggesting regulation is dependent on mature miR-208b. We also assessed the expression of exonic and intronic Med13 gene sequences. The expression of intronic Med13 does not change in 208b-KD cells (Figure 6B). These data suggest that miR-208b silences Med13 expression post-transcriptionally. In contrast, we observed elevated expression of intronic and exonic sequences of the α-MHC gene in 208b-KD cells (Figure 6B) suggesting that pri-miR-208b transcriptionally regulates gene expression. These data are also consistent with reduced Ezh2 interaction at the bdP in 208b-KD cells (Figure 4C). The expression of Ezh2 remains unchanged in 208b-KD cells (Figure 4A). To determine whether gene targets of pri-miR-208b are regulated transcriptionally, we isolated nuclear and cytosolic RNAs (21). We observe predominant expression of miR-208b in cytosolic fractions of Sca1+ cells, whereas the pri-miR-208b transcript was detected in both the nuclear and cytosolic preparations (Figure 6C). As expected, the expression of snoRNA was enriched in the nuclear compartment (Supplementary Figure S11), whereas the expression of 18s rRNA was cytosolic (Supplementary Figure S12). As controls for the nuclear and cytosolic fractions, we show by protein immunoblotting the isolation of Brm and GAPDH, respectively (Supplementary Figure S13). Taken together, these results suggest the mature miR-208b transcript is less likely to regulate gene silencing in the nucleus. We then examined whether pri-miR-208b transcript could directly interact with the intergenic bdP sequence. First, in silico analysis indicates specific 5′-end complementarity between the pri-miR-208b transcript and the intergenic bdP at sequences –1227 to –1330 relative to the α-MHC transcription start site (Figure 6D). Second, we designed a synthetic RNA oligomer using the pri-miR-208b sequence to determine its binding efficiency to the bdP using in vitro dsDNA/RNA immunoprecipitation assay. This procedure involves hybridization of biotin-incorporated RNA oligonucleotides with DNA, which are immunoprecipitated with streptavidin beads (33). Following immunoprecipitation, the isolates are treated with ribonucleases, RNase H or RNase V1. The endonuclease RNase H specifically catalyzes the cleavage of RNA in RNA/DNA hybrids, whereas the RNase V1 enzyme does not distinguish base-paired RNA/DNA hybrids. To quantify enrichment of the bdP, we used qPCR. Immunoprecipitation indicates the pri-miR-208b sequence interacts specifically with the bdP, which was confirmed by RNase H cleavage, as shown in Figure 6E. Similarly, treatment with the RNase V1 ribonuclease abolished bdP enrichment. Taken together, these results suggest the pri-miR-208b transcript interacts with the bdP template.
Figure 6.
Regulation by the pri-miR-208b transcript. (A) Increased expression of Med13 gene in 208b-KD cells; *P < 0.0038 (B) qRT-PCR of exonic and intronic sequences shows transcriptional activation of α-MHC gene in 208b-KD cells; *P < 0.0005 (C) Wild-type Sca1+ cells were assessed for miR expression in cytosolic and nuclear fractions. *P < 0.0007. (D) Pri-miR-208b recognition sequence within the bdP. (E) In vitro dsDNA/RNA binding assay using biotin-tagged RNA oligomers followed by real-time qPCR of the bdP sequence. Fold enrichment was calculated relative to non-specific oligonucleotide binding. *P < 0.0038. All experiments independently performed n = 4.
These results show for the first time that the association of the pri-miR-208b transcript with Ezh2 enzyme is linked to changes in gene function in the hypertrophied LV. This concept is supported by the observation that loss of pri-miR-208b transcript is associated with altered chromatin binding of Ezh2 and derepression of Ezh2-regulated genes. However, the chromatin interaction of AS β-MHC occurs independent of pri-miR-208b and Ezh2 enzyme. To date, there is little information regarding the chromatin interaction of primary miRNAs regulating epigenetic states and gene expression. Besides functioning as precursor transcripts for miR processing, the current data support novel chromatin regulatory roles for primary miR transcripts in the hypertrophied heart.
While our laboratory is intensely refining the techniques to isolate heart-derived cardiomyocytes for the specific purpose to study chromatin-associated RNAs from small cell numbers, many of the current observations published use considerably larger numbers of cultured cells (34,35). To explore the mechanism of gene regulation, we studied the mouse neonatal ventricular cardiomyocytes to show that gene expression changes were consistent with our observations with Sca1+ cells. For example, we observed the upregulation of α-MHC and AS β-MHC in TSA-stimulated cells as well as the downregulation of Anp genes (Supplementary Figure S14). TSA stimulation did not change the expression of pri-miR-208a and pri-miR-208b transcripts. While our studies highlight similarities between terminally differentiated muscle and Sca1+ progenitor cells, we cannot rule out other regulatory differences mediating chromatin-associated ncRNAs.
DISCUSSION
The PcG protein, Ezh2, is a key regulatory enzyme required for tissue-specific gene silencing. We observed binding of Ezh2 to the Anp and Bnp genes in LV tissues isolated from adult hearts. Pressure overload by TAC surgery results in the activation of fetal genes such as Anp and Bnp consistent with reduced binding of Ezh2 on these genes. Conversely, the repression of adult genes such as α-MHC and AS β-MHC in TAC animals is associated with increased Ezh2 binding and H3K27m3 modification at the bdP (Figure 7). Although HDAC inhibition by TSA is known to attenuate the induction of fetal genes and repression of adult-specific genes such as α-MHC, the precise molecular regulators targeted by TSA still remain poorly understood. We show that derepression of α-MHC and AS β-MHC genes by TSA is associated with the release of Ezh2 and H3K27m3 modification at MHC genes. The chromatin binding of Ezh2 in the adult heart is essential for maintaining the homeostatic gene expression.
Figure 7.
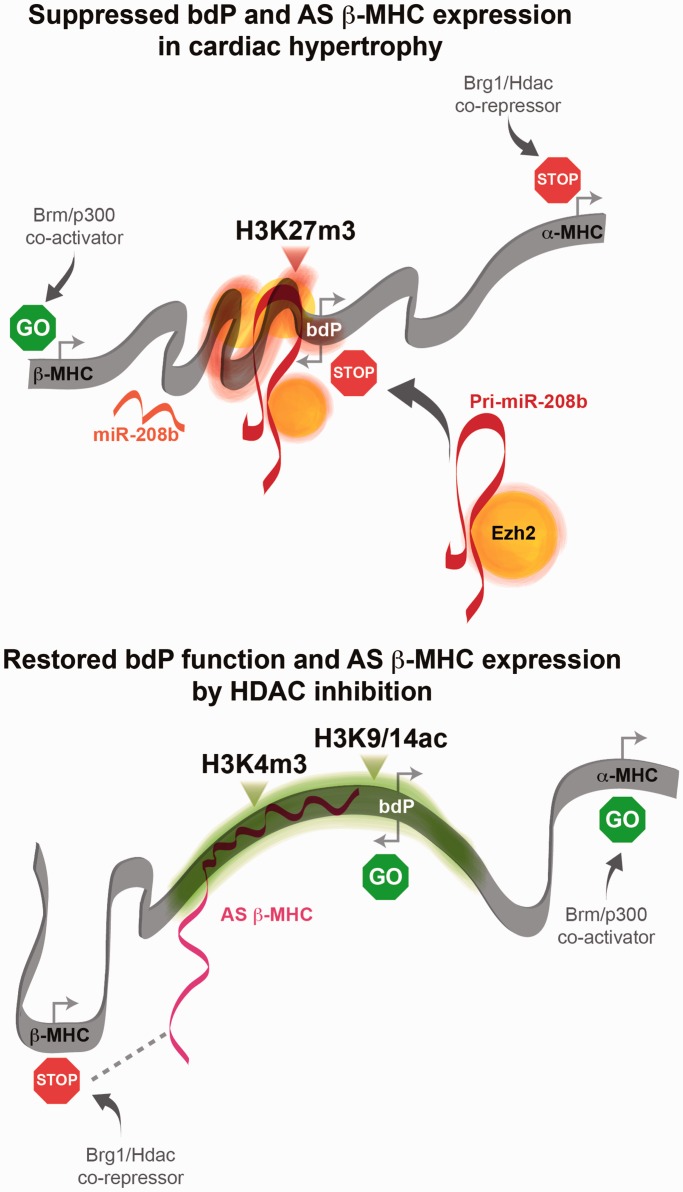
Schematic representation of the α/β MHC gene regulating events at the intergenic bdP mediated by ncRNA. Cardiac hypertrophy is associated with H3K27m3 of intergenic bdP by the PcG methyltransferase protein, Ezh2 and suppression of coding α-MHC and non-coding AS β-MHC gene expression. Increased expression of pri-miR-208b is associated with Ezh2 interaction at the bdP in cardiac hypertrophy. HDAC inhibition by TSA attenuates cardiac hypertrophy by restoring the expression of α-MHC and AS β-MHC genes and this is associated with hyperacetylation of the intergenic bdP as well as the release of Ezh2/pri-miR-208b complex. Restoration of AS β-MHC expression results in chromatin binding and inhibition of sense β-MHC transcription.
As shown in Figure 7, MHC chromatin content as well as specific co-regulatory complexes serve to integrate ncRNAs to distinct transcriptional responses in the hypertrophied heart. We propose that the pri-miR-208b transcript as a substrate for histone modification (36). We observed the pri-miR-208b transcript mediates the α- to β-MHC switch in the hypertrophied heart. HDAC inhibition by TSA attenuates pri-miR-208b dependent-chromatin binding in TAC animals. In the hypertrophied heart, recruitment of Ezh2 is mediated by pri-miR-208b chromatin interaction. In vitro silencing of either the pri-miR-208b transcript or Ezh2 enzyme are associated with gene expression changes that were comparable with pri-miR-208b and Ezh2 knockdown. Specifically, the reduced chromatin interaction of pri-miR-208b transcript in miR-208b-KD cells alters Ezh2 binding to target genes. This was associated with the derepression of Ezh2-regulated genes in pri-miR-208b knockdown cells. The loss of AS β-MHC transcript in cardiac progenitors did not change the expression of Ezh2-regulated genes. This suggests that pri-miR-208b could regulate the binding of Ezh2 and H3K27m3 at genes. Experimental results derived from TAC animals suggest increased chromatin interaction of pri-miR-208b transcript could mediate the release of Ezh2 from Anp and Bnp genes, while increase Ezh2 binding to the bdP in the hypertrophied heart.
Recent evidence suggests that antisense transcription is critical for H3K27m3-mediated silencing (37). The long ncRNA, HOTAIR, interacts with Ezh2 (34,35,38). While, a mouse ortholog to the HOTAIR transcript (mHOTAIR) has been reported, its the function in the heart remains uncharacterized (39). We report no appreciable expression of the HOTAIR transcript in our experiments. The molecular basis of ncRNA-dependent chromatin interactions conferring gene-regulating epigenetic changes in the heart remains incomplete (40,41). The experimental results presented in this study suggest that ncRNA could guide chromatin-modifying complexes to regulate hypertrophy-associated gene expression (42). Recent studies also indicate that TSA attenuates pathological hypertrophy in mice as well as regulates the co-repressive Brg1-HDAC complexes interacting with the bdP of the intergenic region (6). The mechanism underlying the induction of stem cell population within the myocardium in response to increased pressure overload is poorly understood; however, it is thought to contribute negatively to ventricular function in the pathological heart (43–45). Our results also suggest a novel role for HDAC inhibition in regulating cardiac stem cell genes by mechanisms that involve ncRNA–chromatin interaction. Not only is this work of direct therapeutic relevance to hypertrophy, but will also be important for our understanding of new mechanisms of gene regulation conferred by interacting ncRNAs that influence chromatin structure and function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Victorian Government’s Operational Infrastructure Support program (in part). Funding for open access charge: National Health and Medical Research Council and National Heart Foundation of Australia.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge grant and fellowship support from the National Health and Medical Research Council (NHMRC) and the National Heart Foundation of Australia (NHF). P.M. was awarded a Monash Graduate Scholarship, and A.E-O. and X-J. D. are NHMRC Senior Research Fellows.
REFERENCES
- 1.Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, Epstein JA. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Invest. 2003;112:863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 3.Gupta MP. Factors controlling cardiac myosin-isoform shift during hypertrophy and heart failure. J. Mol. Cell Cardiol. 2007;43:388–403. doi: 10.1016/j.yjmcc.2007.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luther HP. Role of endogenous antisense RNA in cardiac gene regulation. J. Mol. Med. (Berl.) 2005;83:26–32. doi: 10.1007/s00109-004-0613-5. [DOI] [PubMed] [Google Scholar]
- 5.Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circ. Res. 2011;108:378–396. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haddad F, Qin AX, Bodell PW, Zhang LY, Guo H, Giger JM, Baldwin KM. Regulation of antisense RNA expression during cardiac MHC gene switching in response to pressure overload. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H2351–H2361. doi: 10.1152/ajpheart.01111.2005. [DOI] [PubMed] [Google Scholar]
- 8.Giger J, Qin AX, Bodell PW, Baldwin KM, Haddad F. Activity of the beta-myosin heavy chain antisense promoter responds to diabetes and hypothyroidism. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H3065–H3071. doi: 10.1152/ajpheart.01224.2006. [DOI] [PubMed] [Google Scholar]
- 9.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, et al. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ. Res. 2012;110:406–415. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat. Genet. 2012;44:343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 14.Glenn DJ, Wang F, Chen S, Nishimoto M, Gardner DG. Endothelin-stimulated human B-type natriuretic peptide gene expression is mediated by Yin Yang 1 in association with histone deacetylase 2. Hypertension. 2009;53:549–555. doi: 10.1161/HYPERTENSIONAHA.108.125088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat. Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 16.Davis FJ, Pillai JB, Gupta M, Gupta MP. Concurrent opposite effects of trichostatin A, an inhibitor of histone deacetylases, on expression of alpha-MHC and cardiac tubulins: implication for gain in cardiac muscle contractility. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H1477–H1490. doi: 10.1152/ajpheart.00789.2004. [DOI] [PubMed] [Google Scholar]
- 17.Chang L, Kiriazis H, Gao XM, Du XJ, El-Osta A. Cardiac genes show contextual SWI/SNF interactions with distinguishable gene activities. Epigenetics. 2011;6:760–768. doi: 10.4161/epi.6.6.16007. [DOI] [PubMed] [Google Scholar]
- 18.Gao XM, Kiriazis H, Moore XL, Feng XH, Sheppard K, Dart A, Du XJ. Regression of pressure overload-induced left ventricular hypertrophy in mice. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H2702–H2707. doi: 10.1152/ajpheart.00836.2004. [DOI] [PubMed] [Google Scholar]
- 19.Xiao Q, Zeng L, Zhang Z, Margariti A, Ali ZA, Channon KM, Xu Q, Hu Y. Sca-1+ progenitors derived from embryonic stem cells differentiate into endothelial cells capable of vascular repair after arterial injury. Arterioscler Thromb. Vasc. Biol. 2006;26:2244–2251. doi: 10.1161/01.ATV.0000240251.50215.50. [DOI] [PubMed] [Google Scholar]
- 20.Mathiyalagan P, Chang L, Du XJ, El-Osta A. Cardiac ventricular chambers are epigenetically distinguishable. Cell Cycle. 2010;9:612–617. doi: 10.4161/cc.9.3.10612. [DOI] [PubMed] [Google Scholar]
- 21.Okabe J, Orlowski C, Balcerczyk A, Tikellis C, Thomas MC, Cooper ME, El-Osta A. Distinguishing hyperglycemic changes by Set7 in vascular endothelial cells. Circ. Res. 2012;110:1067–1076. doi: 10.1161/CIRCRESAHA.112.266171. [DOI] [PubMed] [Google Scholar]
- 22.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, Wagner DR, Staessen JA, Heymans S, Schroen B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 23.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 24.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 25.Chng KR, Chang CW, Tan SK, Yang C, Hong SZ, Sng NY, Cheung E. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 2012;31:2810–2823. doi: 10.1038/emboj.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 29.Bolli P, Vardabasso C, Bernstein E, Chaudhry HW. Chromatin immunoprecipitation of adult murine cardiomyocytes. In: Bonifacino JS, et al., editors. Current Protocols in Cell Biology/Editorial Board. Wiley, Hoboken, New Jersey: 2013. Chapter 17, Unit17 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell. Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 31.Juan AH, Derfoul A, Feng X, Ryall JG, Dell'Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sher F, Boddeke E, Olah M, Copray S. Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS One. 2012;7:e40399. doi: 10.1371/journal.pone.0040399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattick JS. RNA as the substrate for epigenome-environment interactions: RNA guidance of epigenetic processes and the expansion of RNA editing in animals underpins development, phenotypic plasticity, learning, and cognition. Bioessays. 2010;32:548–552. doi: 10.1002/bies.201000028. [DOI] [PubMed] [Google Scholar]
- 37.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 38.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 41.Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ. Res. 2012;111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- 42.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsieh PC, Segers VF, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat. Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urbanek K, Quaini F, Tasca G, Torella D, Castaldo C, Nadal-Ginard B, Leri A, Kajstura J, Quaini E, Anversa P. Intense myocyte formation from cardiac stem cells in human cardiac hypertrophy. Proc. Natl Acad. Sci. USA. 2003;100:10440–10445. doi: 10.1073/pnas.1832855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.