Abstract
We introduce macromolecular crowding quantitatively into the model for kinetics of gene regulation in Escherichia coli. We analyse and compute the specific-site searching time for 180 known transcription factors (TFs) regulating 1300 operons. The time is between 160 s (e.g. for SoxS Mw = 12.91 kDa) and 1550 s (e.g. for PepA6 of Mw = 329.28 kDa). Diffusion coefficients for one-dimensional sliding are between 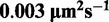 for large proteins up to
for large proteins up to  for small monomers or dimers. Three-dimensional diffusion coefficients in the cytoplasm are 2 orders of magnitude larger than 1D sliding coefficients, nevertheless the sliding enhances the binding rates of TF to specific sites by 1–2 orders of magnitude. The latter effect is due to ubiquitous non-specific binding. We compare the model to experimental data for LacI repressor and find that non-specific binding of the protein to DNA is activation- and not diffusion-limited. We show that the target location rate by LacI repressor is optimized with respect to microscopic rate constant for association to non-specific sites on DNA. We analyse the effect of oligomerization of TFs and DNA looping effects on searching kinetics. We show that optimal searching strategy depends on TF abundance.
for small monomers or dimers. Three-dimensional diffusion coefficients in the cytoplasm are 2 orders of magnitude larger than 1D sliding coefficients, nevertheless the sliding enhances the binding rates of TF to specific sites by 1–2 orders of magnitude. The latter effect is due to ubiquitous non-specific binding. We compare the model to experimental data for LacI repressor and find that non-specific binding of the protein to DNA is activation- and not diffusion-limited. We show that the target location rate by LacI repressor is optimized with respect to microscopic rate constant for association to non-specific sites on DNA. We analyse the effect of oligomerization of TFs and DNA looping effects on searching kinetics. We show that optimal searching strategy depends on TF abundance.
INTRODUCTION
For many decades, macromolecular crowding has been well advertised as a factor strongly affecting all biochemical reactions and other processes in living cells (1–4). Yet, rarely this effect has been quantitatively analysed. We follow classical works on facilitated target location (5–11), extended by Li et al. (12) to analyse gene regulations in Escherichia coli by transcription factors (TFs). We explicitly include new factors to the model: reduction of mobility of TFs in the cytoplasm and on DNA due to macromolecular crowding (13,14) and reduction of nucleoid volume caused by nucleoid-associated proteins (NAPs) covering DNA. We use the model of TF diffusion along DNA (sliding) of Blainey et al. (15,16). We calibrate the model using non-specific binding constant of LacI to DNA calculated from single-molecule experimental data (17,18). Next we apply the model to all known 180 TFs that bind specifically to cis-regulatory elements and regulate 1300 operons. In particular we determine 3D and 1D diffusion constants for TFs, enhancement rates due to the sliding and times for location of operators by TFs. We discuss various strategies used by TFs for rapid target location in vivo. We provide a large number of data, which can be used for analysis of genetic networks in systems biology. These data can also guide future experiments, since quantitative experimental data in vivo are available for only one repressor (LacI) regulating only one operon.
TFs use diffusion driven by thermal fluctuations to search for specific sites on DNA. The search process combines 3D diffusion of TF through a cytoplasm, subsequent non-specific binding of TF to DNA, next 1D diffusion along a DNA and finally unbinding into cytoplasm (and subsequent repeat of the previous steps) or target recognition when TF encounters an operator during sliding (Figure 1). This process is known as the facilitated target location (5–11). Typical bacterial genome consists of 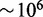 base pairs, and location of the specific site on DNA is through enormous amount of bindings to non-specific sites. One-dimensional diffusion along DNA increases effective target size on DNA and speeds the searching up to 2 orders of magnitude (8,12,18). Recent in vivo single-molecule experiment has shown that LacI repressor dimer needs
base pairs, and location of the specific site on DNA is through enormous amount of bindings to non-specific sites. One-dimensional diffusion along DNA increases effective target size on DNA and speeds the searching up to 2 orders of magnitude (8,12,18). Recent in vivo single-molecule experiment has shown that LacI repressor dimer needs 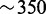 s to locate the operator (17) and on average slides along 36 bp when non-specifically bound (18) to DNA. The diffusion coefficient for sliding has been measured in the buffers only (16,17).
s to locate the operator (17) and on average slides along 36 bp when non-specifically bound (18) to DNA. The diffusion coefficient for sliding has been measured in the buffers only (16,17).
Figure 1.
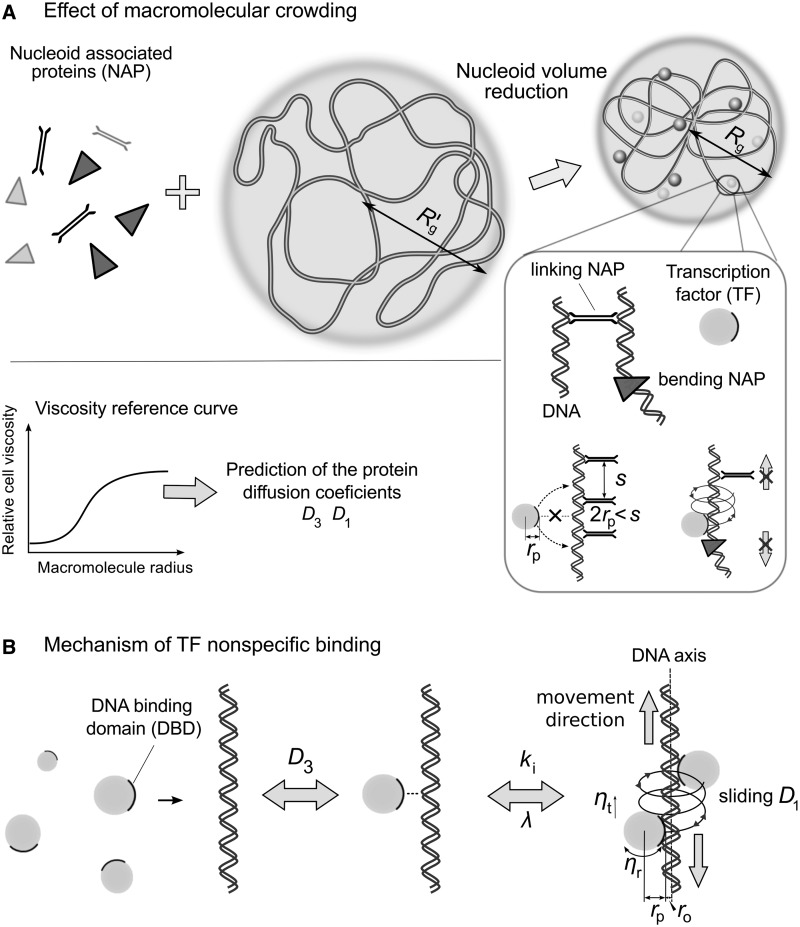
Influence of macromolecular crowding on gene regulation kinetics. (A) NAPs cause shrinking of available searching volume by linking and bending of DNA chains. NAPs when bound to DNA disturb non-specific association of TFs and 1D diffusion along DNA (12). Macromolecular crowding increases viscosity of the cytoplasm and diminishes TF 1D and 3D-mobility constants [Equations (1–2)]. The viscosity experienced by a translocating macromolecule is a stretched exponential function of size of the macromolecule (13,14). (B) TF searches for specific site on DNA through numerous DNA–non-specific associations. Three-dimensional diffusion brings TF close to DNA. Next, TF binds to DNA with microscopic association rate constant ki (8). Non-specifically bound TF slides performing curvilinear motion (16,19) along DNA and dissociates with rate constant λ.
The prokaryotic cytoplasm is a highly crowded environment (14,20–22). Two aspects of macromolecular crowding affect regulation of gene expression (Figure 1A): (i) cytoplasmic crowding reduces 3D and 1D diffusion constant of TF and (ii) crowding on DNA disturbs TF non-specific binding and 1D diffusion along DNA due to presence of NAPs (12,23,24). However, NAPs additionally shrink the nucleoid, thereby decrease the size of space where the process of facilitated target location occurs. Elowitz et al. (25) have shown that protein diffusion in the E. coli cytoplasm is the normal diffusion, but significantly reduced as compared with its mobility in water. This reduction is a non-linear function of protein molecular weight (21,22). We have introduced the phenomenological model of cytoplasm length-scale dependent viscosity (13,14) to describe all available experimental data for diffusion of small ligands, proteins and DNA plasmids in E. coli. We have previously tested the model (13) for motion of proteins in buffers crowded by polymers (13,26) or micelles (27). In this article, we apply the model to describe the influence of macromolecular crowding on 1D diffusion along DNA. There is a marked difference between diffusion in the cytoplasm and sliding. During sliding on DNA, the protein follows the path along major grooves and thus rotates around its own axis of rotation and also around the DNA. The friction for rotational motion is proportional to the volume of the protein (15,16), whereas for the translational diffusion in the cytoplasm, the friction is proportional to the radius of the protein. One-dimensional diffusion coefficients are substantially (2 orders of magnitude) smaller than 3D diffusion constants for the typical sizes of TFs.
MATERIALS AND METHODS
Macromolecular crowding
The prokaryotic cell is a densely packed structure with concentration of macromolecules between 200 and 400 g/l (20–30% of cellular volume) (20,28). This macromolecular crowding in the cytoplasm reduces in the first place diffusional mobilities of TFs. The reduction of mobility is proportional to the scale-dependent viscosity (13,26). This viscosity describes the mobility in complex liquids (13,26), in cytoplasm of the eukaryotic cells (HeLa and Swiss 3T3 fibroblasts) (13) and prokaryotic cells (E. coli) (14). This model based on the viscosity describes protein–protein association kinetics in Hela cytoplasm (29). The model is based on the Stokes-Sutherland-Einstein relation 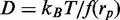 , where D is a diffusion constant of macromolecule,
, where D is a diffusion constant of macromolecule, 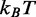 is the Boltzmann constant times the absolute temperature and f is the length-scale–dependent hydrodynamic drag friction. For 3D diffusion coefficient D3, this drag has the following form:
is the Boltzmann constant times the absolute temperature and f is the length-scale–dependent hydrodynamic drag friction. For 3D diffusion coefficient D3, this drag has the following form:
| (1) |
Thus 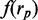 is proportional to the length-scale–dependent viscosity
is proportional to the length-scale–dependent viscosity 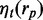 . At fixed temperature, the viscosity
. At fixed temperature, the viscosity 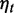 experienced by an object of hydrodynamic radius rp is described by the formula (13,26)
experienced by an object of hydrodynamic radius rp is described by the formula (13,26) 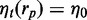
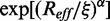 , where
, where 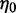 is the solvent viscosity, ξ is the correlation length,
is the solvent viscosity, ξ is the correlation length, 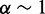 is a constant and effective radius Reff is defined as
is a constant and effective radius Reff is defined as 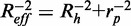 , where Rh is the effective hydrodynamic radius of macromolecular crowding agents. We find for E. coli these characteristic parameters by fit to experimental data:
, where Rh is the effective hydrodynamic radius of macromolecular crowding agents. We find for E. coli these characteristic parameters by fit to experimental data: 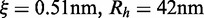 and
and 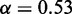 (14). The length-scale–dependent viscosity of E. coli cytoplasm change by a factor of 104 from 0.001 Pas for small solutes to 13 Pas for large plasmids (250 nm). The calculation of the 1D coefficient D1 is based on an observation that TFs spin while non-specifically bound to DNA (16). The friction experienced by TF of size rp during its curvilinear motion along DNA is given by the following expression (19):
(14). The length-scale–dependent viscosity of E. coli cytoplasm change by a factor of 104 from 0.001 Pas for small solutes to 13 Pas for large plasmids (250 nm). The calculation of the 1D coefficient D1 is based on an observation that TFs spin while non-specifically bound to DNA (16). The friction experienced by TF of size rp during its curvilinear motion along DNA is given by the following expression (19):
| (2) |
where the first term corresponds to frictional force experienced by TF bound to DNA during translational motion parallel to the DNA axis, the second term describes rotational friction that arises from TF rotation along its axis, and the last term is the rotational friction experienced by TFs that spin around DNA axis (see Figure 1B). TF makes full 360° rotation (2π rotation) around DNA after translating over the distance of 3.54 nm, i.e. 10.4 base pairs along DNA (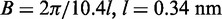 is a length between consecutive base pairs). ro accounts for the protein offset from the DNA axis (Figure 1B). We use the value of
is a length between consecutive base pairs). ro accounts for the protein offset from the DNA axis (Figure 1B). We use the value of 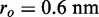 (19,30). Brownian dynamics simulations (21) have shown that rotational viscosity experienced by rotating proteins of molecular weights in the range
(19,30). Brownian dynamics simulations (21) have shown that rotational viscosity experienced by rotating proteins of molecular weights in the range 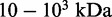 in E. coli cytoplasm is ∼10 times larger than water viscosity. Therefore, we assume that
in E. coli cytoplasm is ∼10 times larger than water viscosity. Therefore, we assume that 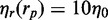 .
.
Nucleoid volume
NAPs reduce volume of the nucleoid (Figure 1A), by bending and linking DNA strands. There are two types of proteins belonging to NAP family that affect nucleoid volume: bending NAPs that bends DNA and change its Kuhn length and linking NAPs that connect neighbouring DNA chains. We describe the volume reduction using a model of a randomly cross-linked polymer chain of the Kuhn length κ, consisting of N base pairs subjected to Ml uncorrelated cross-linking constraints (31). The radius of gyration Rg of the polymer is (31):
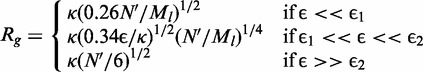 |
(3) |
where ε is a mean distance between monomers that form a cross-link, 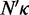 is a contour length of the polymer and
is a contour length of the polymer and 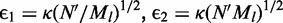 . The first case in Equation (3) concerns rigid cross-linking polymer regime where
. The first case in Equation (3) concerns rigid cross-linking polymer regime where 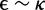 or smaller, second soft cross-linking regime and the last one describes free polymer chain regime. The volume occupied by nucleoid is calculated from
or smaller, second soft cross-linking regime and the last one describes free polymer chain regime. The volume occupied by nucleoid is calculated from 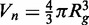 . The mean distance between bending proteins is
. The mean distance between bending proteins is 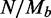 , where Mb is a number of bending NAPs. The Kuhn length is a non-linear function of the mean distance between bending proteins (32,33).
, where Mb is a number of bending NAPs. The Kuhn length is a non-linear function of the mean distance between bending proteins (32,33).
Model of facilitated target location
We follow the classical works of Berg et al. (6–8) with the extension by Li et al. (12). The TF association rate constant to a specific site on DNA is as follows:
| (4) |
The association rate constant is equal to the rate constant per binding site kassoc diminished by the presence of non-specific sites on DNA. Kns is the equilibrium constant for non-specific binding of TF to DNA, c is the concentration of non-specific DNA, p(r) is the probability distribution that the binding site is free of NAPs and r is the diameter of TF.
| (5) |
where ki is a microscopic (intrinsic) association rate constant (Figure 1B), l is a distance between consecutive base pairs and ζ is a mean distance between neighbouring DNA chains and b is the DNA radius. ζ is calculated as in (8,12) from 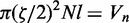 . The probability that a binding site of size r on DNA is free from NAPs is as follows (12,24):
. The probability that a binding site of size r on DNA is free from NAPs is as follows (12,24):
| (6) |
where d is a mean weighted size of NAPs, i.e. 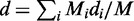 , with M being the total amount of NAPs per nucleoid and
, with M being the total amount of NAPs per nucleoid and 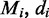 are amount and size of i-th NAP (see Supplementary Table S1), respectively. The fraction of DNA that is free from NAPs is
are amount and size of i-th NAP (see Supplementary Table S1), respectively. The fraction of DNA that is free from NAPs is 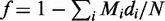 . The sliding enhances the rate constant by effectively increasing the mean target size
. The sliding enhances the rate constant by effectively increasing the mean target size 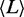 . The size (in bp), under the assumption of semi-stationary positions of NAPs (12), is as follows:
. The size (in bp), under the assumption of semi-stationary positions of NAPs (12), is as follows:
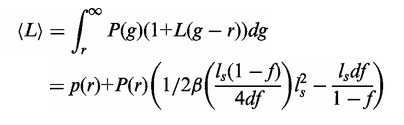 |
(7) |
where P(g) is the probability density function calculated from Equation (6), 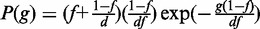 , and
, and 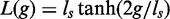 is an effective target size for specific site placed symmetrically between two NAPs (8,12) with
is an effective target size for specific site placed symmetrically between two NAPs (8,12) with 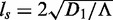 . β is the
. β is the 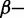 function (34) given by
function (34) given by 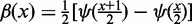 with ψ being an Euler psi function (
with ψ being an Euler psi function (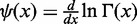 ). The factor 1 denotes that at least one binding site is scanned on TF binding. Λ is the macroscopic dissociation constant (dissociation to the distance
). The factor 1 denotes that at least one binding site is scanned on TF binding. Λ is the macroscopic dissociation constant (dissociation to the distance 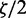 where TF can bind to uncorrelated DNA chain) equal to microscopic dissociation constant λ times the probability of reaching
where TF can bind to uncorrelated DNA chain) equal to microscopic dissociation constant λ times the probability of reaching 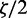 (8), that is
(8), that is
| (8) |
RESULTS
The presence of NAPs highly reduces nucleoid volume
The length of E. coli nucleoid is 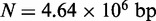 . The Kuhn length of undisturbed DNA is 300 bp,
. The Kuhn length of undisturbed DNA is 300 bp, 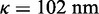 . For the free polymer regime (without NAPs bound to DNA), the nucleoid radius of gyration and volume would be
. For the free polymer regime (without NAPs bound to DNA), the nucleoid radius of gyration and volume would be 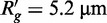 and
and 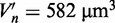 , respectively, that is much larger than E. coli cell size. Abundances of DNA binding proteins [NAPs (35,36), TF (37) and RNA polymerase (RNAP) (38)] for early exponential phase of E. coli growth in rich media are collected in Supplementary Table S1. The calculated fraction of DNA that is free from DNA binding proteins is
, respectively, that is much larger than E. coli cell size. Abundances of DNA binding proteins [NAPs (35,36), TF (37) and RNA polymerase (RNAP) (38)] for early exponential phase of E. coli growth in rich media are collected in Supplementary Table S1. The calculated fraction of DNA that is free from DNA binding proteins is 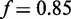 (total level of the proteins is M = 36 600). Number of bending NAPs
(total level of the proteins is M = 36 600). Number of bending NAPs 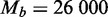 shows that mean distance between these NAPs (178 bp) is below Kuhn length of naked DNA. The random placement of bending protein every 200 bp reduce the persistence length by a factor 2.2 (39). We use the value
shows that mean distance between these NAPs (178 bp) is below Kuhn length of naked DNA. The random placement of bending protein every 200 bp reduce the persistence length by a factor 2.2 (39). We use the value 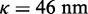 in calculations. Thus, we predict that nucleoid shrinking is due to presence of DNA linking and bending proteins. The observed volume of nucleoid is within range 1–0.1
in calculations. Thus, we predict that nucleoid shrinking is due to presence of DNA linking and bending proteins. The observed volume of nucleoid is within range 1–0.1 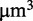 (40–43). In Supplementary Figure S1, we show that the number of cross-links estimated from (44,45) is enough to shrink nucleoid to the observed values. The length of relaxed MukB homodimer is
(40–43). In Supplementary Figure S1, we show that the number of cross-links estimated from (44,45) is enough to shrink nucleoid to the observed values. The length of relaxed MukB homodimer is 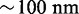 , while on binding to DNA, the protein is compacted to the size below the Kuhn length of DNA (46). Similar case is for SeqA and MatP proteins (47). This is significantly below calculated value of
, while on binding to DNA, the protein is compacted to the size below the Kuhn length of DNA (46). Similar case is for SeqA and MatP proteins (47). This is significantly below calculated value of 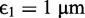 . Therefore, the nucleoid is in the first regime of Equation (3). The mean volume of the nucleoid is between
. Therefore, the nucleoid is in the first regime of Equation (3). The mean volume of the nucleoid is between 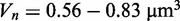 at the early exponential phase of growth (Supplementary text and Figure S1). Two other NAPs are commonly attributed to the group of NAPs forming connections between DNA chains: H-NS/StpA dimers (48,49) and Dps dodecamers through the oligomerization (50). However, presence of divalent cations (Mg2+) precludes formation of bridges between DNA chains by these proteins (50,51).
at the early exponential phase of growth (Supplementary text and Figure S1). Two other NAPs are commonly attributed to the group of NAPs forming connections between DNA chains: H-NS/StpA dimers (48,49) and Dps dodecamers through the oligomerization (50). However, presence of divalent cations (Mg2+) precludes formation of bridges between DNA chains by these proteins (50,51).
Non-specific binding of LacI to DNA is activation-limited
We use the results of single-molecule experimental studies (17,18) of specific-site binding kinetics of LacI dimer (fused with Venus proteins). We obtain parameters for the model in the following fitting procedure: (i) In Ref. (18), the fit giving sliding length 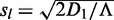 does not include presence of NAPs. We develop a simulation procedure (Supplementary text and Supplementary Figure S2) for this experiment, including NAPs on DNA as crowders. The best fit to the model gives
does not include presence of NAPs. We develop a simulation procedure (Supplementary text and Supplementary Figure S2) for this experiment, including NAPs on DNA as crowders. The best fit to the model gives 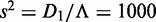 bp2 that corresponds to sliding length equal to
bp2 that corresponds to sliding length equal to 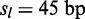 (for the vacancy
(for the vacancy 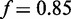 , and mean weighted size of NAPs d = 19 bp). The sliding length is larger in our model since NAPs weaken the spatial correlations between operators. (ii) The 3D diffusion constant of LacI dimer without DNA binding domains equals (17)
, and mean weighted size of NAPs d = 19 bp). The sliding length is larger in our model since NAPs weaken the spatial correlations between operators. (ii) The 3D diffusion constant of LacI dimer without DNA binding domains equals (17) 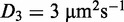 . We use this value for calculation of hydrodynamic radius of LacI dimer fused with Venus proteins from Equation (1). We get
. We use this value for calculation of hydrodynamic radius of LacI dimer fused with Venus proteins from Equation (1). We get 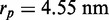 . The calculated 1D diffusion constant for in vivo conditions is
. The calculated 1D diffusion constant for in vivo conditions is 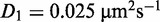 . (iii) From point (i) and (ii), we calculate
. (iii) From point (i) and (ii), we calculate 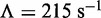 . (iv) The non-specific equilibrium constant Kns is calculated from the equation
. (iv) The non-specific equilibrium constant Kns is calculated from the equation 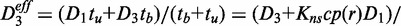
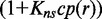 , where tb and tu are the average times spent by the protein diffusing in the cytoplasm (with diffusion constant D3) and spent diffusing on DNA (with diffusion constant D1), respectively. The concentration c of non-specific sites is
, where tb and tu are the average times spent by the protein diffusing in the cytoplasm (with diffusion constant D3) and spent diffusing on DNA (with diffusion constant D1), respectively. The concentration c of non-specific sites is 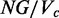 , where G is the amount of DNA in genome equivalents and Vc is the cell volume (Supplementary text). For
, where G is the amount of DNA in genome equivalents and Vc is the cell volume (Supplementary text). For 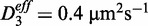 (17), we get the value
(17), we get the value 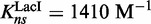 . (v) From Equation (8), we calculate microscopic parameters:
. (v) From Equation (8), we calculate microscopic parameters: 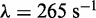 and
and 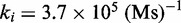 . The
. The 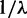 gives the average time spent by the protein on DNA smaller than 5 ms as observed in vivo (17). We get
gives the average time spent by the protein on DNA smaller than 5 ms as observed in vivo (17). We get 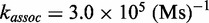 . Such small value of kassoc suggests that the process of binding of the protein to DNA is activation-limited since in the case of the diffusion-limited association this parameter would have the value
. Such small value of kassoc suggests that the process of binding of the protein to DNA is activation-limited since in the case of the diffusion-limited association this parameter would have the value 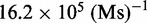 . (vi) Using parameters from points (i-v) we find from Equations (4–8) that searching of the single specific site takes
. (vi) Using parameters from points (i-v) we find from Equations (4–8) that searching of the single specific site takes 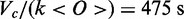 for LacI dimer fused with Venus proteins, where
for LacI dimer fused with Venus proteins, where 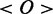 is the average number of operators in the cell volume (Supplementary text). The lower value of s2 in the fit from pt. (i) is
is the average number of operators in the cell volume (Supplementary text). The lower value of s2 in the fit from pt. (i) is 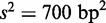 and this gives the time 385 s. The experimentally determined rate is
and this gives the time 385 s. The experimentally determined rate is 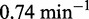 and there are 3–5 LacI dimers (18). Thus, the in vivo binding time is in the range (240–405 s).
and there are 3–5 LacI dimers (18). Thus, the in vivo binding time is in the range (240–405 s).
LacI repressor and RNA polymerase are optimized for fast target location
The average time for target location is a function of microscopic association and dissociation constants and can be reduced by changing these two constants. In the previous paragraph, we have discussed the experiment involving LacI dimer, but LacI protein in the native form attaches to the operator as the tetramer. Molecular weight of LacI tetramer is 154.36 kDa. Its hydrodynamic radius (in nm) is determined from the following equation (52):
| (9) |
The calculated mobility constants (14) 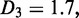
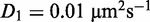 give the overall search time equal to 730 s. Figure 2 depicts the relative average time of location of the specific site as a function of one of the microscopic rate constants (the second is kept constant). We find that the searching time has a minimum as a function of microscopic association rate constant ki. The minimization is an effect of two opposite processes. Increase of microscopic constant enhances the rate of TF binding to non-specific site and increase the overall association rate [Equation (4)]. The increase is limited as kassoc becomes diffusion-limited, i.e. for
give the overall search time equal to 730 s. Figure 2 depicts the relative average time of location of the specific site as a function of one of the microscopic rate constants (the second is kept constant). We find that the searching time has a minimum as a function of microscopic association rate constant ki. The minimization is an effect of two opposite processes. Increase of microscopic constant enhances the rate of TF binding to non-specific site and increase the overall association rate [Equation (4)]. The increase is limited as kassoc becomes diffusion-limited, i.e. for 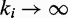 . Additionally it increases non-specific equilibrium constant and decrease the overall rate by tight binding to non-specific sites. For large values of ki, TF is recaptured to the same site after dissociation, and thus the search process is limited by 1D sliding. The association constant for in vivo conditions differ from that in the minimum by a factor 1.002.
. Additionally it increases non-specific equilibrium constant and decrease the overall rate by tight binding to non-specific sites. For large values of ki, TF is recaptured to the same site after dissociation, and thus the search process is limited by 1D sliding. The association constant for in vivo conditions differ from that in the minimum by a factor 1.002.
Figure 2.
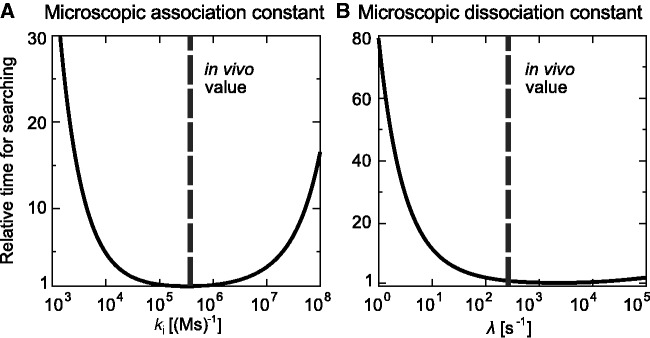
Relative changes of the average time for location of the operator by LacI tetramer. (A) Microscopic association constant is optimized for fast target location. The times are given with respect to the minimal value. (B) The minimum of the time as a function of the microscopic dissociation rate constant is shifted by 1 order of magnitude from determined value. However, the decrease of the time is only by a factor 1.66. The dashed lines point to determined values of microscopic rate constants.
The reduction of association rate due to transient binding to non-specific sites [by a factor 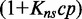 , denominator of Equation (4)] is larger than decrease of the effective target length due to the sliding
, denominator of Equation (4)] is larger than decrease of the effective target length due to the sliding 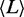 . Therefore, the time for target location reaches a constant value before sliding length is reduced to 1 bp. The minimum is for
. Therefore, the time for target location reaches a constant value before sliding length is reduced to 1 bp. The minimum is for 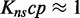 (8). It corresponds to
(8). It corresponds to 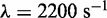 (Figure 2B), i.e. it is 1 order of magnitude larger than calculated for in vivo conditions. However, the minimum is shallow and the calculated difference in search time for in vivo conditions and at the minimum is only 66%. We suspect that high dissociation rate for optimal searching time is not possible for a large macromolecule.
(Figure 2B), i.e. it is 1 order of magnitude larger than calculated for in vivo conditions. However, the minimum is shallow and the calculated difference in search time for in vivo conditions and at the minimum is only 66%. We suspect that high dissociation rate for optimal searching time is not possible for a large macromolecule.
We have done similar analysis for E. coli single promoter localization time during transcription initiation. Calculation of the mobility constants for RNA polymerase holoenzyme complex (with  subunit bound,
subunit bound, 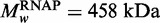 ) gives
) gives 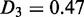 and
and 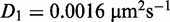 . The microscopic dissociation constant,
. The microscopic dissociation constant, 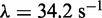 , for RNAP has been recently determined (53) from single molecule measurements. The equilibrium constant for non-specific binding of RNAP to DNA has not been measured so far in vivo, but its estimation for cellular conditions gives a value
, for RNAP has been recently determined (53) from single molecule measurements. The equilibrium constant for non-specific binding of RNAP to DNA has not been measured so far in vivo, but its estimation for cellular conditions gives a value 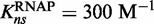 (54) and in vitro measurements for high ionic strength with divalent cations that mimics milieu of E. coli cytoplasm gives
(54) and in vitro measurements for high ionic strength with divalent cations that mimics milieu of E. coli cytoplasm gives 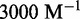 (55). In Supplementary Figure S3, we show that minimum of searching time as a function of microscopic association time is between ki values calculated from the above equilibrium constants. It has been observed recently in independent in vitro experiments that RNA polymerase with
(55). In Supplementary Figure S3, we show that minimum of searching time as a function of microscopic association time is between ki values calculated from the above equilibrium constants. It has been observed recently in independent in vitro experiments that RNA polymerase with 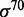 (53) or
(53) or 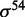 (56) factors does not perform long-range sliding when searching for its promoter. Our calculations also give short-range sliding of the length 28 bp for RNAP-
(56) factors does not perform long-range sliding when searching for its promoter. Our calculations also give short-range sliding of the length 28 bp for RNAP-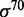 complex in the cell. The measured lifetime of RNAP-
complex in the cell. The measured lifetime of RNAP-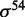 with non-specific DNA is equal to 4.7 s but such long non-specific binding was not observed for
with non-specific DNA is equal to 4.7 s but such long non-specific binding was not observed for 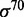 complex (53) pointing that this subunit may interact differently with non-specific DNA.
complex (53) pointing that this subunit may interact differently with non-specific DNA.
Diffusion constants and average time for the location of a single site on DNA for all recognized TFs
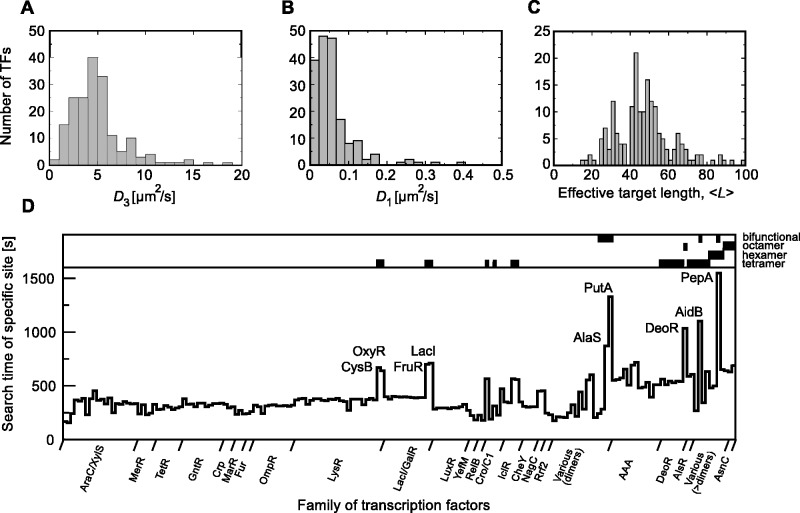
The E. coli genome contains ∼300 genes that encode proteins that regulate promoter activity (57). So far, more than half of them have been verified experimentally. Here we analyse kinetic and dynamic parameters characterizing the process of target location for all 180 verified TFs (regulating ∼1300 operons) for in vivo conditions. We get a list of all TFs and information about their monomer molecular weights (Supplementary Table S2) from E. coli EcoCyc (58) and RegulonDB (59) databases. We extend the EcoCyc database for TF oligomerization levels by mining of this parameter from the literature (References in Supplementary Table S2). We assume oligomerization level that is most common in a given TF family, if this information is unavailable for a given TF from this family. In Figure 3A–B, we show histograms for calculated 3D and 1D diffusion constants for all TFs. The TF mobility on DNA is 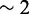 orders of magnitude slower than its cytoplasm mobility. The calculation of single-site localization times on DNA by TFs requires additional assumption about non-specific microscopic rate constants. The unknown rate parameters for all TFs (ki and λ) are assumed to be equal to that for lac repressor. The assumption is justified by the fact that nature of non-specific interactions of TF with DNA is primarily electrostatic (30,60,61) between the positively charged side chains of proteins and the negatively charged backbone phosphate groups of the DNA. Dahirel et al. (62) have shown that DNA–protein non-specific interaction free energy has a minimum at finite separation between interacting interfaces equal to 0.6 nm that is in agreement with the distance observed for the non-specific LacI–DNA complex (30). The minimum is an effect of the concave shape of DNA–proteins and an enhancement of the osmotic pressure at short separation distances. They performed also statistical analysis of 77 DNA binding proteins finding for sequence-specific DNA proteins similar interface area and mean charge density. Thus, it is expected that non-specific interaction should not vary between TFs. Moreover, in vivo experiments give similar values of the fraction of various non-specifically bound TFs on DNA: λ phage regulatory protein CI in λ-infected E. coli cells was measured to be 86% bound to non-specific sites (63) [non-specific equilibrium constants for CI and Cro repressors were identical (63)], 87% of LacI dimers fused with Venus proteins from single-molecule experiments (17) and 90% of LacI tetramers from measurements of the distribution of repressors in minicell-experiment (64).
orders of magnitude slower than its cytoplasm mobility. The calculation of single-site localization times on DNA by TFs requires additional assumption about non-specific microscopic rate constants. The unknown rate parameters for all TFs (ki and λ) are assumed to be equal to that for lac repressor. The assumption is justified by the fact that nature of non-specific interactions of TF with DNA is primarily electrostatic (30,60,61) between the positively charged side chains of proteins and the negatively charged backbone phosphate groups of the DNA. Dahirel et al. (62) have shown that DNA–protein non-specific interaction free energy has a minimum at finite separation between interacting interfaces equal to 0.6 nm that is in agreement with the distance observed for the non-specific LacI–DNA complex (30). The minimum is an effect of the concave shape of DNA–proteins and an enhancement of the osmotic pressure at short separation distances. They performed also statistical analysis of 77 DNA binding proteins finding for sequence-specific DNA proteins similar interface area and mean charge density. Thus, it is expected that non-specific interaction should not vary between TFs. Moreover, in vivo experiments give similar values of the fraction of various non-specifically bound TFs on DNA: λ phage regulatory protein CI in λ-infected E. coli cells was measured to be 86% bound to non-specific sites (63) [non-specific equilibrium constants for CI and Cro repressors were identical (63)], 87% of LacI dimers fused with Venus proteins from single-molecule experiments (17) and 90% of LacI tetramers from measurements of the distribution of repressors in minicell-experiment (64).
Figure 3.
Analysis of TF mobility and kinetic parameters for the set of recognized TFs (Supplementary Table S2) from E. coli EcoCyc (58) and RegulonDB (59) databases. Histograms of TF (A) three-dimensional, D3 [Equation (1)] and (B) one-dimensional diffusion constants, D1 [Equation (2)]. (C) The histogram of the effective target enhancement factors 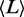 [Equation (7)]. (D) The average time for the location of single specific binding site by TFs [
[Equation (7)]. (D) The average time for the location of single specific binding site by TFs [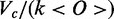 ]. For simplicity, we assume that
]. For simplicity, we assume that 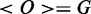 . TFs are grouped into families (the order is the same as in Supplementary Table S2). In the upper belt of the panel (D), TFs that form stable higher oligomers (oligomerization level higher than 2) and/or are bifunctional (TF poses DNA binding domain and various enzymatic activity) are marked with black stripes.
. TFs are grouped into families (the order is the same as in Supplementary Table S2). In the upper belt of the panel (D), TFs that form stable higher oligomers (oligomerization level higher than 2) and/or are bifunctional (TF poses DNA binding domain and various enzymatic activity) are marked with black stripes.
The site-specific location times for higher-order oligomers and bifunctional TFs are substantially decreased. The association rate effective enhancement factor 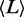 (Figure 3C) due to the sliding changes by 1 order of magnitude (between 10 for TF with large molecular weights/higher level of oligomerization to 100 for TFs that are monomers or small dimers). In Figure 3D, we present the calculated TF searching times for single specific site. The TFs are grouped in the families they belong to (Supplementary Table S2). We mark in the upper belt the TFs that form stable higher-order oligomers in solution (oligomerization level
(Figure 3C) due to the sliding changes by 1 order of magnitude (between 10 for TF with large molecular weights/higher level of oligomerization to 100 for TFs that are monomers or small dimers). In Figure 3D, we present the calculated TF searching times for single specific site. The TFs are grouped in the families they belong to (Supplementary Table S2). We mark in the upper belt the TFs that form stable higher-order oligomers in solution (oligomerization level 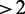 ) or are bifunctional, i.e. contain both DNA binding domain (DBD) and active centres responsible for enzymatic activity (e.g. AlaS - alanyl-tRNA synthetase, PutA - flavoprotein that acts as enzyme catalysing reactions of the proline degradation, AidB - isovaleryl-CoA dehydrogenase activity, PepA - aminopeptidase). Within the same family, the searching times are usually similar (except they belong to one of above groups).
) or are bifunctional, i.e. contain both DNA binding domain (DBD) and active centres responsible for enzymatic activity (e.g. AlaS - alanyl-tRNA synthetase, PutA - flavoprotein that acts as enzyme catalysing reactions of the proline degradation, AidB - isovaleryl-CoA dehydrogenase activity, PepA - aminopeptidase). Within the same family, the searching times are usually similar (except they belong to one of above groups).
TF’s searching strategies (higher oligomers versus dimers)
A TF binding time to the regulatory region of the operon can be reduced by increasing TF concentration. Downside of this is an additional cost of protein production (65,66). The best strategy for fast regulation seems to be reached by small monomers with bipartite helix-turn-helix (HTH) motifs (as for MarA, SoxS) and single binding site in the regulatory region. However, this is not the case observed for E. coli regulatory motifs. Frequently, regulatory regions of the operons contain many binding sites and transcription regulation is a complex function of the number of these sites (67,68). TF dimers are the most frequent oligomeric forms. Stable higher-order oligomers occur for TFs that regulate transcription of the operons involved in transport and catabolism of sugars [e.g. lactose (LacI), glycerol-P (GlpR), glucitol (GutR), l-ascorbate (UlaR), deoxyribonucleoside (DeoR), N-acetylgalactosamine (AgaR), cell wall sugars (MurR), d-allose (AlsR)]. Higher oligomers are capable of forming DNA loops (69) that efficiently stabilize the TF–DNA complex with dissociation rates, e.g. for LacI tetramer as low as 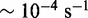 (9,70). DNA looping reduces additionally searching time for the main operator (12) within regulatory region. Many motifs are used to take advantage of DNA looping: presence of auxiliary operators within structural genes (e.g. LacI, GlpR), within regulatory regions (DeoR, AgaR, PepA, IclR), or presence of single regulatory region with multiple operators within divergent operons (AgaR, MurR, AlsR, YqjI). If auxiliary operators are separated by distances higher than sliding length of TF, they reduce searching time by a factor equal to the number of all operators (12,18). On the other side, other repressors, e.g. GalR and GalS, use DNA looping but binds to operators as dimers. Therefore, we pose a question: what is the optimal strategy for fast regulation in case when TFs use DNA looping.
(9,70). DNA looping reduces additionally searching time for the main operator (12) within regulatory region. Many motifs are used to take advantage of DNA looping: presence of auxiliary operators within structural genes (e.g. LacI, GlpR), within regulatory regions (DeoR, AgaR, PepA, IclR), or presence of single regulatory region with multiple operators within divergent operons (AgaR, MurR, AlsR, YqjI). If auxiliary operators are separated by distances higher than sliding length of TF, they reduce searching time by a factor equal to the number of all operators (12,18). On the other side, other repressors, e.g. GalR and GalS, use DNA looping but binds to operators as dimers. Therefore, we pose a question: what is the optimal strategy for fast regulation in case when TFs use DNA looping.
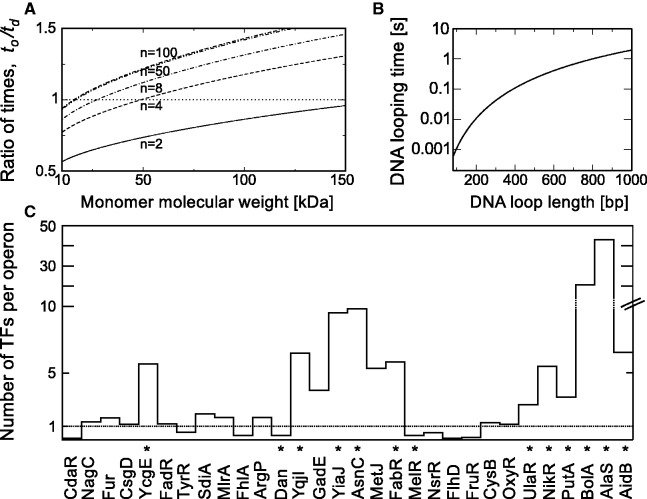
We analyse the simplest searching strategies in which regulatory region is composed of m independent operators and TF is a complex with oligomerization level of dimers equal to o. We consider two scenarios of formation of an oligomeric TF-operators complex: (i) via independent searching by n dimers and oligomerization of dimers when specifically bound via DNA looping (formation time 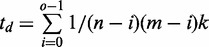 ) or (ii) via searching by n/o oligomers and formation of DNA loops by means of auxiliary operators with time
) or (ii) via searching by n/o oligomers and formation of DNA loops by means of auxiliary operators with time 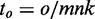 . The optimal strategy depends on the number of searchers (n dimers versus
. The optimal strategy depends on the number of searchers (n dimers versus 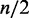 tetramers, case m = o) (Figure 4A) and molecular weight of the TF monomer. Assumption of equal number of TF monomers excludes effect of costs of TF production. Case with one stable tetramer is favoured over independent searching by two dimers. The second scenario is the best strategy for high concentration of TFs. We exclude from calculations the characteristic times of DNA loop formation, since this process for in vivo cellular crowding conditions is expected to take place orders of magnitude faster than specific site searching (Figure 4B).
tetramers, case m = o) (Figure 4A) and molecular weight of the TF monomer. Assumption of equal number of TF monomers excludes effect of costs of TF production. Case with one stable tetramer is favoured over independent searching by two dimers. The second scenario is the best strategy for high concentration of TFs. We exclude from calculations the characteristic times of DNA loop formation, since this process for in vivo cellular crowding conditions is expected to take place orders of magnitude faster than specific site searching (Figure 4B).
Figure 4.
Searching strategy for the formation of TF complex at the operator depends on the number of searchers. (A) Ratio, 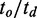 , of the complex formation time via searching by oligomers (abundance n/o) to the time of independent searching by dimers (abundance n). Case with o = 2 (TF complex is a tetramer), and two independent specific sites on DNA within operator region (m = 2). (B) DNA looping times as a function of base pairs that form a loop for in vivo macromolecular crowding. The time calculation is based on DNA relaxation times (71,72) of size L from
, of the complex formation time via searching by oligomers (abundance n/o) to the time of independent searching by dimers (abundance n). Case with o = 2 (TF complex is a tetramer), and two independent specific sites on DNA within operator region (m = 2). (B) DNA looping times as a function of base pairs that form a loop for in vivo macromolecular crowding. The time calculation is based on DNA relaxation times (71,72) of size L from 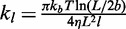 . The hydrodynamic radius of DNA is
. The hydrodynamic radius of DNA is 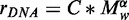 with
with 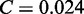 and
and 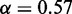 (73) and molecular weight of the 1 bp is 660 Da. The viscosity η experienced by moving DNA is calculated as previously (14). (C) Mean number of oligomeric TFs per regulated operon. Data are taken from single-molecule studies of protein copy number in E. coli cell (37). The dotted line corresponds to the ratio value equal to 1. By means of stars we marked TFs that regulate only one or two operons.
(73) and molecular weight of the 1 bp is 660 Da. The viscosity η experienced by moving DNA is calculated as previously (14). (C) Mean number of oligomeric TFs per regulated operon. Data are taken from single-molecule studies of protein copy number in E. coli cell (37). The dotted line corresponds to the ratio value equal to 1. By means of stars we marked TFs that regulate only one or two operons.
Operons are regulated by low-copy number of TFs. We use data from single-molecule studies of protein copy number in E. coli cell (37) to get abundances of oligomeric TFs. We compare the values with the numbers of regulated operons [taken from RegulonDB (59)]. TFs for which both information are available are shown in Figure 4C. Divergent operons with the same operator are counted as a single unit. The results shed light on the problem of trade-off between TF’s speed of transcription regulation and costs of TF synthesis. TFs that regulate large regulons are present approximately in copies that correspond to single TF per operon. Contrary, regulons that consist of few operons (marked by stars in Figure 4) appeared to be regulated by TF levels higher than one copy per operon. In this group, the most abundant TFs are bifunctional proteins that reduce significantly searching times for specific sites. Possible explanation of these results arises from the fact that the average binding time of TF to the operator of the specific operon in a regulon decreases with the regulon size (Supplementary Figure S4). We consider the simplified problem in which the number of TFs equals the number of regulated operons and every operon contains single operator. Even regulon that consists of four operons and four TFs reduces, on average, the binding time to the particular operator by 50%. When the number of operons increases to 40, the reduction is 90%. The same effect can be achieved when TF regulates single operon by increasing the number of TFs 2- and 10-fold, respectively.
DISCUSSION
We present a first comprehensive study of the impact of macromolecular crowding on the gene regulation kinetics in E. coli. Our approach combines the theory of scale-dependent diffusion (13,26), nucleoid shrinking by NAPs (31) and model of facilitated target location (8,12). This methodology is crucial for predictions of 3D and 1D diffusion constants and specific-site location times for all currently known TFs. From the parameterization of the model, we find that binding of LacI repressor to non-specific DNA is activation-limited reaction. Moreover, the microscopic association rate is optimized for fast target location for in vivo conditions.
From the seminal work of Riggs et al. (74) on association kinetics of LacI repressor to the operator site, there is broad interest in processes involved in a target location by TF (75). Recent measurements of TF searching (17,18) point that TF make use of the processes of facilitated diffusion to locate its specific site in vivo. Here, our analysis shows that presence of 1D diffusion speeds up the specific-site location for all TFs by 1–2 orders of magnitude as compared with the case where the TF binds with the same affinity to non-specific sites but does not slide.
The previous models concerning in vivo facilitated diffusion of LacI repressor (76,77) pointed that the repressor search can be close to the optimal conditions for target location. Koslover et al. (76) used value of LacI dissociation constant equal to 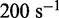 that comes from the observation that the upper value of LacI unbinding time is 5 ms (17). Both models pointed that the DNA configuration has insignificant impact on the search time. In our work, we calculated all the parameters for LacI repressor on the basis of in vivo data. Specifically, taking the results of experiments by Hammar et al. (18) and the macromolecular crowding present in the cell, we calculated the value of microscopic parameters proving that LacI optimizes the target location with respect to the binding rate constant. Thus the parameters include implicitly (78) macromolecular crowding present in the cell.
that comes from the observation that the upper value of LacI unbinding time is 5 ms (17). Both models pointed that the DNA configuration has insignificant impact on the search time. In our work, we calculated all the parameters for LacI repressor on the basis of in vivo data. Specifically, taking the results of experiments by Hammar et al. (18) and the macromolecular crowding present in the cell, we calculated the value of microscopic parameters proving that LacI optimizes the target location with respect to the binding rate constant. Thus the parameters include implicitly (78) macromolecular crowding present in the cell.
Measured 1D diffusion constant in vitro is 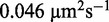 (17) in a reaction buffer characterized by water viscosity. The value is less than predicted from Equation (2). The difference is explained by presence of small energy barrier in helical sliding of TF along DNA during transition between consecutive base pairs (16). This effect was incorporated by additional factor
(17) in a reaction buffer characterized by water viscosity. The value is less than predicted from Equation (2). The difference is explained by presence of small energy barrier in helical sliding of TF along DNA during transition between consecutive base pairs (16). This effect was incorporated by additional factor 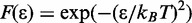 (79) multiplying right-hand side of Equation (2), where ε is root-mean-squared roughness of the interaction potential. The value of ε for LacI was determined to be
(79) multiplying right-hand side of Equation (2), where ε is root-mean-squared roughness of the interaction potential. The value of ε for LacI was determined to be 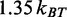 (16) for experiment carried out without Mg2+ in the reaction buffer. One-dimensional diffusion on DNA is much faster in the presence of Mg2+ (70,80). It has been shown (70) that magnesium cations change the diffusion constant of LacI repressor from
(16) for experiment carried out without Mg2+ in the reaction buffer. One-dimensional diffusion on DNA is much faster in the presence of Mg2+ (70,80). It has been shown (70) that magnesium cations change the diffusion constant of LacI repressor from 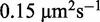 in the presence of 10 mM Mg2+ to 0.045 µm2s−2 for buffers without Mg2+ [the divalent cations stabilize the double helix of DNA (81) and concentration of Mg2+
in vivo is evaluated to be 5–10 mM (61) or even 100 mM (82)]. More studies are required to quantify this effect as a function of Mg2+ concentration. We simply assume that protein 1D diffusion constant is decreased only by cytoplasm viscosity. Including energy barrier
in the presence of 10 mM Mg2+ to 0.045 µm2s−2 for buffers without Mg2+ [the divalent cations stabilize the double helix of DNA (81) and concentration of Mg2+
in vivo is evaluated to be 5–10 mM (61) or even 100 mM (82)]. More studies are required to quantify this effect as a function of Mg2+ concentration. We simply assume that protein 1D diffusion constant is decreased only by cytoplasm viscosity. Including energy barrier 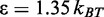 into the model gives the following values: 1D diffusion constant
into the model gives the following values: 1D diffusion constant 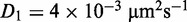 , microscopic dissociation constant
, microscopic dissociation constant 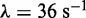 and target location time 3000 s. The values disagree with the experimental observation for which
and target location time 3000 s. The values disagree with the experimental observation for which 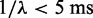 and searching times are 1 order of magnitude smaller.
and searching times are 1 order of magnitude smaller.
TF must scan DNA base pairs (18) to recognize specific site (23,83,84). We neglected scanning in the model at non-specific sites because the scanning rates are unknown. It has been shown by Sheinman et al. (84) that model in which the scanning energy barrier varies between non-specific sites gives appreciable increase of searching times. The free-energy calculations between DNA and protein give argument for the existence of such barriers (62). The model allows also for optimal search that is equivalent to the classical model of facilitated target location. Since the calculated value of searching time for LacI dimer is similar to the experimentally measured one, we suspect that the TF trapping at non-specific sites does not affect significantly binding times.
We assume that motion of all TFs and RNAP is diffusive. This is confirmed by many experimental results for proteins of large range of sizes [(14,22,25) and references therein]. Genomic loci (85) and large RNA–protein complexes (86) subdiffuse in the E. coli cytoplasm. The reason of subdifussive motion is confined diffusion in a polymer chain and activation processes (87).
We assume in our model that TFs are initially uniformly distributed within a cell. Therefore, our calculations concern a case in which initial position of TF is not correlated with the region of its synthesis (88–90). The assumption is justified by the observation that for fast-growth conditions, considered here, DNA is less densely packed and distribution of LacI does not depend on the spatial location of its encoding gene (89). Also repression strength as a function of intergenic distance is well reproduced by model assuming spatial homogeneity of the repressor (89). Intersegmental jumps that speed up the target location in coiled DNA (91–93) are implicitly embedded in classical model of facilitated diffusion. Since the values of the microscopic and macroscopic dissociation constants are comparable, most of the unbinding events lead to the association to uncorrelated DNA sites.
Our findings also shed light on trade-off between facilitated target location by TFs due to DNA looping, increase of TF concentration and costs of TF production. The increase of TF concentration weaken also the fluctuations in the gene products (94–97). The precise regulation of transcription initiation is easier for dimers with regulation at the dissociation step and direct interactions between dimers. Here, more parameters can be adjusted to reach a gene transfer function that effectively couples the expression of genes to environmental signals. Searching by dimers (98) with further oligomerization at the operator is also preferred in AAA family of TFs (ATPase associated with diverse cellular activities) (Supplementary Table S3). These TFs regulate gene expression in  -RNAP–dependent promoters. AAA TFs when bound to upstream activating sequences (UAS) function by coupling the energy yielded from ATP hydrolysis to the isomerization of RNAP from closed to open complex. TF oligomerization is required for the formation of functional ATPase and it is promoted by the binding to UAS (99). Therefore, searching by high-oligomeric TF is disadvantageous owing to consumption of ATP when not bound with UAS.
-RNAP–dependent promoters. AAA TFs when bound to upstream activating sequences (UAS) function by coupling the energy yielded from ATP hydrolysis to the isomerization of RNAP from closed to open complex. TF oligomerization is required for the formation of functional ATPase and it is promoted by the binding to UAS (99). Therefore, searching by high-oligomeric TF is disadvantageous owing to consumption of ATP when not bound with UAS.
System biology studies of genetic networks require enormous amount of data. Most of them are still unavailable from experiments. Database built here on TF mobilities and searching kinetics can now be used in studies of functioning of these networks with proper temporal resolution to give accurate predictions of gene products fluctuations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science of Poland for the Iuventus-Plus program [IP2010 052570 (2011) to M.T.]; National Science Center [DEC1-2011/01/N/ST3/00865 to T.K.]; Foundation for Polish Science for START scholarship [to T.K.]; National Science Center for MAESTRO grant [DEC-2011/02/A/ST3/00143 to R.H.]. Funding for open access charge: MAESTRO grant from National Science Center of Poland [DEC-2011/02/A/ST3/00143].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
M.T. and R.H. designed research; M.T. and R.H. performed research; M.T., R.H. analysed data with input from T.K.; and M.T. and R.H. wrote the article. We thank N. Bobrovska for help with preparation of figures.
REFERENCES
- 1.Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Bioph. Biomol. Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 2.Ellis R. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 3.Ellis R. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 4.Zhou H, Rivas G, Minton A. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Ann. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richter PH, Eigen M. Diffusion controlled reaction rates in spheroidal geometry. Application to repressor–operator association and membrane bound enzymes. Biophys. Chem. 1974;2:255–263. doi: 10.1016/0301-4622(74)80050-5. [DOI] [PubMed] [Google Scholar]
- 6.Berg OG, Blomberg C. Association kinetics with coupled diffusional flows. Special application to the lac repressor–operator system. Biophys. Chem. 1976;4:367–381. doi: 10.1016/0301-4622(76)80017-8. [DOI] [PubMed] [Google Scholar]
- 7.Berg OG, Blomberg C. Association kinetics with coupled diffusion. An extension to coiled-chain macromolecules applied to the lac repressor-operator system. Biophys. Chem. 1977;7:33–39. doi: 10.1016/0301-4622(77)87012-9. [DOI] [PubMed] [Google Scholar]
- 8.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 9.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 10.von Hippel PH, Berg OG. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 11.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li GW, Berg OG, Elf J. Effects of macromolecular crowding and DNA looping on gene regulation kinetics. Nat. Phys. 2009;5:294–297. [Google Scholar]
- 13.Kalwarczyk T, Ziebacz N, Bielejewska A, Zaboklicka E, Koynov K, Szymański J, Wilk A, Patkowski A, Gapiński J, Butt HJ, et al. Comparative analysis of viscosity of complex liquids and cytoplasm of mammalian cells at the nanoscale. Nano Lett. 2011;11:2157–2163. doi: 10.1021/nl2008218. [DOI] [PubMed] [Google Scholar]
- 14.Kalwarczyk T, Tabaka M, Hołyst R. Biologistics–diffusion coefficients for complete proteome of Escherichia coli. Bioinformatics. 2012;28:2971–2978. doi: 10.1093/bioinformatics/bts537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336:1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 19.Bagchi B, Blainey PC, Xie XS. Diffusion constant of a nonspecifically bound protein undergoing curvilinear motion along DNA. J. Phys. Chem. B. 2008;112:6282–6284. doi: 10.1021/jp077568f. [DOI] [PubMed] [Google Scholar]
- 20.Fulton A. How crowded is the cytoplasm? Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 21.McGuffee S, Elcock A. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comp. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mika JT, Poolman B. Macromolecule diffusion and confinement in prokaryotic cells. Curr. Opin. Biotechnol. 2011;22:117–126. doi: 10.1016/j.copbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys. J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flyvbjerg H, Keatch S, Dryden D. Strong physical constraints on sequence-specific target location by proteins on DNA molecules. Nucleic Acids Res. 2006;34:2550–2557. doi: 10.1093/nar/gkl271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elowitz MB, Surette MG, Wolf P-E, Stock JB, Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 1999;181:197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hołyst R, Bielejewska A, Szymański J, Wilk A, Patkowski A, Gapiński J, Zywociński A, Kalwarczyk T, Kalwarczyk E, Tabaka M, et al. Scaling form of viscosity at all length-scales in poly(ethylene glycol) solutions studied by fluorescence correlation spectroscopy and capillary electrophoresis. Phys. Chem. Chem. Phys. 2009;11:9025–9032. doi: 10.1039/b908386c. [DOI] [PubMed] [Google Scholar]
- 27.Szymanski J, Patkowski A, Wilk A, Garstecki P, Holyst R. Diffusion and viscosity in a crowded environment: from nano-to macroscale. J. Phys. Chem. B. 2006;110:25593–25597. doi: 10.1021/jp0666784. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 29.Tabaka M, Sun L, Kalwarczyk T, Hołyst R. Implications of macromolecular crowding for protein–protein association kinetics in the cytoplasm of living cells. Soft Matter. 2013;9:4386–4389. [Google Scholar]
- 30.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 31.Vilgis T, Solf M. Cross-linked polymer chains: scaling and exact results. In: Rubi M, Perez-Vicente C, editors. Complex Behaviour of Glassy Systems. Berlin Heidelberg: Springer; 1997. pp. 403–413. [Google Scholar]
- 32.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nir G, Lindner M, Dietrich HR, Girshevitz O, Vorgias CE, Garini Y. HU protein induces incoherent DNA persistence length. Biophys. J. 2011;100:784–790. doi: 10.1016/j.bpj.2010.12.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gradshteuin IS, Ryzhik IM. In: Table of Integrals, Series, and Products. Jeffrey A, Zwillinger D, editors. Burlington: Academic Press; 2000. [Google Scholar]
- 35.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrushenko ZM, Lai CH, Rybenkov VV. Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB. J. Biol. Chem. 2006;281:34208–34217. doi: 10.1074/jbc.M606723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bremer H, Dennis P, Ehrenberg M. Free RNA polymerase and modeling global transcription in Escherichia coli. Biochimie. 2003;85:597–609. doi: 10.1016/s0300-9084(03)00105-6. [DOI] [PubMed] [Google Scholar]
- 39.Czapla L, Swigon D, Olson WK. Effects of the nucleoid protein HU on the structure, flexibility, and ring-closure properties of DNA deduced from Monte Carlo simulations. J. Mol. Biol. 2008;382:353–370. doi: 10.1016/j.jmb.2008.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hecht R, Taggart R, Pettijohn D. Size and DNA content of purified E. coli nucleoids observed by fluorescence microscopy. Nature. 1975;253:60–62. doi: 10.1038/253060a0. [DOI] [PubMed] [Google Scholar]
- 41.Van Ness J, Pettijohn DE. A simple autoradiographic method for investigating long range chromosome substructure: size and number of DNA molecules in isolated nucleoids of Escherichia coli. J. Mol. Biol. 1979;129:501–508. doi: 10.1016/0022-2836(79)90509-6. [DOI] [PubMed] [Google Scholar]
- 42.Valkenburg JA, Woldringh CL. Phase separation between nucleoid and cytoplasm in Escherichia coli as defined by immersive refractometry. J. Bacteriol. 1984;160:1151–1157. doi: 10.1128/jb.160.3.1151-1157.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science. 2012;338:528–531. doi: 10.1126/science.1227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cagliero C, Grand RS, Jones MB, Jin DJ, O’Sullivan JM. Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic Acids Res. 2013;41:6058–6071. doi: 10.1093/nar/gkt325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matoba K, Yamazoe M, Mayanagi K, Morikawa K, Hiraga S. Comparison of MukB homodimer versus MukBEF complex molecular architectures by electron microscopy reveals a higher-order multimerization. Biochem. Biophys. Res. Commun. 2005;333:694–702. doi: 10.1016/j.bbrc.2005.05.163. [DOI] [PubMed] [Google Scholar]
- 47.Dupaigne P, Tonthat NK, Espéli O, Whitfill T, Boccard F, Schumacher MA. Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome. Mol. Cell. 2012;48:560–571. doi: 10.1016/j.molcel.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 2004;32:5935–5944. doi: 10.1093/nar/gkh915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dill KA, Ghosh K, Schmit JD. Physical limits of cells and proteomes. Proc. Natl Acad. Sci. USA. 2011;108:17876–17882. doi: 10.1073/pnas.1114477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang F, Redding S, Finkelstein IJ, Gorman J, Reichman DR, Greene EC. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat. Struct. Mol. Biol. 2013;20:174–181. doi: 10.1038/nsmb.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klumpp S, Hwa T. Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc. Natl Acad. Sci. USA. 2008;105:20245–20250. doi: 10.1073/pnas.0804953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeHaseth P, Lohman T, Burgess R, Record M., Jr Nonspecific interactions of Escherichia coli RNA polymerase with native and denatured DNA: differences in the binding behavior of core and holoenzyme. Biochemistry. 1978;17:1612–1622. doi: 10.1021/bi00602a006. [DOI] [PubMed] [Google Scholar]
- 56.Friedman LJ, Mumm JP, Gelles J. RNA polymerase approaches its promoter without long-range sliding along DNA. Proc. Natl Acad. Sci. USA. 2013;110:9740–9745. doi: 10.1073/pnas.1300221110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pérez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:583–590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muñiz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, García-Sotelo JS, López-Fuentes A, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:98–105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Record MT, deHaseth PL, Lohman TM. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977;16:4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- 61.von Hippel PH. From”simple” DNA-protein interactions to the macromolecular machines of gene expression. Annu. Rev. Biophys. Biomol. Struct. 2007;36:79–105. doi: 10.1146/annurev.biophys.34.040204.144521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dahirel V, Paillusson F, Jardat M, Barbi M, Victor JM. Nonspecific DNA-protein interaction: why proteins can diffuse along DNA. Phys. Rev. Lett. 2009;102:228101–228101. doi: 10.1103/PhysRevLett.102.228101. [DOI] [PubMed] [Google Scholar]
- 63.Bakk A, Metzler R. In vivo non-specific binding of lambda CI and Cro repressors is significant. FEBS Lett. 2004;563:66–68. doi: 10.1016/S0014-5793(04)00249-2. [DOI] [PubMed] [Google Scholar]
- 64.Kao-Huang Y, Revzin A, Butler AP, O’Conner P, Noble DW, Von Hippel PH. Nonspecific DNA binding of genome-regulating proteins as a biological control mechanism: measurement of DNA-bound Escherichia coli lac repressor in vivo. Proc. Natl Acad. Sci. USA. 1977;74:4228–4232. doi: 10.1073/pnas.74.10.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 66.Tănase-Nicola S, ten Wolde PR. Regulatory control and the costs and benefits of biochemical noise. PLoS Comput. Biol. 2008;4:e1000125. doi: 10.1371/journal.pcbi.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R. Transcriptional regulation by the numbers: applications. Curr. Opin. Genet. Dev. 2005;15:125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matthews K. DNA looping. Microbiol. Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barkley MD. Salt dependence of the kinetics of the lac repressor-operator interaction: role of nonoperator deoxyribonucleic acid (DNA) in the association reaction. Biochemistry. 1981;20:3833–3842. doi: 10.1021/bi00516a026. [DOI] [PubMed] [Google Scholar]
- 71.Meiners JC, Quake SR. Femtonewton force spectroscopy of single extended DNA molecules. Phys. Rev. Lett. 2000;84:5014–5017. doi: 10.1103/PhysRevLett.84.5014. [DOI] [PubMed] [Google Scholar]
- 72.Morelli MJ, Ten Wolde PR, Allen RJ. DNA looping provides stability and robustness to the bacteriophage λ switch. Proc. Natl Acad. Sci. USA. 2009;106:8101–8106. doi: 10.1073/pnas.0810399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robertson RM, Laib S, Smith DE. Diffusion of isolated DNA molecules: dependence on length and topology. Proc. Natl Acad. Sci. USA. 2006;103:7310–7314. doi: 10.1073/pnas.0601903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction: III. Kinetic studies. J. Mol. Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 75.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 76.Koslover EF, Díaz de la Rosa MA, Spakowitz AJ. Theoretical and computational modeling of target-site search kinetics in vitro and in vivo. Biophys. J. 2011;101:856–865. doi: 10.1016/j.bpj.2011.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bauer M, Metzler R. In vivo facilitated diffusion model. PLoS One. 2013;8:e53956. doi: 10.1371/journal.pone.0053956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morelli MJ, Allen RJ, Wolde PR. Effects of macromolecular crowding on genetic networks. Biophys. J. 2011;101:2882–2891. doi: 10.1016/j.bpj.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zwanzig R. Diffusion in a rough potential. Proc. Natl Acad. Sci. USA. 1988;85:2029–2030. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berg OG, Blomberg C. Association kinetics with coupled diffusion: III. Ionic-strength dependence of the lac repressor-operator association. Biophys. Chem. 1978;8:271–280. doi: 10.1016/0301-4622(78)80010-6. [DOI] [PubMed] [Google Scholar]
- 81.Eichhorn GL. The effect of metal ions on the structure and function of nucleic acids. In: Eichhorn GL, Marzilli LG, editors. Metal Ions in Genetic Information Transfer. New York: Elsevier; 1981. pp. 1–46. [Google Scholar]
- 82.Moncany M, Kellenberger E. High magnesium content of Escherichia coli B. Experientia. 1981;37:846–847. doi: 10.1007/BF01985672. [DOI] [PubMed] [Google Scholar]
- 83.Hu L, Grosberg AY, Bruinsma R. Are DNA transcription factor proteins maxwellian demons? Biophys. J. 2008;95:1151–1156. doi: 10.1529/biophysj.108.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheinman M, Bénichou O, Kafri Y, Voituriez R. Classes of fast and specific search mechanisms for proteins on DNA. Rep. Prog. Phys. 2012;75:026601–026601. doi: 10.1088/0034-4885/75/2/026601. [DOI] [PubMed] [Google Scholar]
- 85.Weber SC, Spakowitz AJ, Theriot JA. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys. Rev. Lett. 2010;104:238102–238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Golding I, Cox EC. Physical nature of bacterial cytoplasm. Phys. Rev. Lett. 2006;96:098102–098102. doi: 10.1103/PhysRevLett.96.098102. [DOI] [PubMed] [Google Scholar]
- 87.Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc. Natl Acad. Sci. USA. 2012;109:7338–7343. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wunderlich Z, Mirny LA. Spatial effects on the speed and reliability of protein-DNA search. Nucleic Acids Res. 2008;36:3570–3578. doi: 10.1093/nar/gkn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuhlman TE, Cox EC. Gene location and DNA density determine transcription factor distributions in Escherichia coli. Mol. Syst. Biol. 2012;8:610. doi: 10.1038/msb.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pulkkinen O, Metzler R. Distance matters: the impact of gene proximity in bacterial gene regulation. Phys. Rev. Lett. 2013;110:198101–198101. doi: 10.1103/PhysRevLett.110.198101. [DOI] [PubMed] [Google Scholar]
- 91.Gowers DM, Halford SE. Protein motion from non-specific to specific DNA by three-dimensional routes aided by supercoiling. EMBO J. 2003;22:1410–1418. doi: 10.1093/emboj/cdg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van den Broek B, Lomholt MA, Kalisch SM, Metzler R, Wuite GJ. How DNA coiling enhances target localization by proteins. Proc. Natl Acad. Sci. USA. 2008;105:15738–15742. doi: 10.1073/pnas.0804248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lomholt MA, van den Broek B, Kalisch SM, Wuite GJ, Metzler R. Facilitated diffusion with DNA coiling. Proc. Natl Acad. Sci. USA. 2009;106:8204–8208. doi: 10.1073/pnas.0903293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 95.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc. Natl Acad. Sci. USA. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shahrezaei V, Swain PS. Analytical distributions for stochastic gene expression. Proc. Natl Acad. Sci. USA. 2008;105:17256–17261. doi: 10.1073/pnas.0803850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ochab-Marcinek A, Tabaka M. Bimodal gene expression in noncooperative regulatory systems. Proc. Natl Acad. Sci. USA. 2010;107:22096–22101. doi: 10.1073/pnas.1008965107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tucker NP, Ghosh T, Bush M, Zhang X, Dixon R. Essential roles of three enhancer sites in sigma54-dependent transcription by the nitric oxide sensing regulatory protein NorR. Nucleic Acids Res. 2010;38:1182–1194. doi: 10.1093/nar/gkp1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.