Abstract
CTCF (CCCTC-binding factor) is a highly conserved multifunctional DNA-binding protein with thousands of binding sites genome-wide. Our previous work suggested that differences in CTCF’s binding site sequence may affect the regulation of CTCF recruitment and its function. To investigate this possibility, we characterized changes in genome-wide CTCF binding and gene expression during differentiation of mouse embryonic stem cells. After separating CTCF sites into three classes (LowOc, MedOc and HighOc) based on similarity to the consensus motif, we found that developmentally regulated CTCF binding occurs preferentially at LowOc sites, which have lower similarity to the consensus. By measuring the affinity of CTCF for selected sites, we show that sites lost during differentiation are enriched in motifs associated with weaker CTCF binding in vitro. Specifically, enrichment for T at the 18th position of the CTCF binding site is associated with regulated binding in the LowOc class and can predictably reduce CTCF affinity for binding sites. Finally, by comparing changes in CTCF binding with changes in gene expression during differentiation, we show that LowOc and HighOc sites are associated with distinct regulatory functions. Our results suggest that the regulatory control of CTCF is dependent in part on specific motifs within its binding site.
INTRODUCTION
CCCTC-binding factor (CTCF) is an essential zinc-finger transcription factor (TF) that shows high conservation from flies to mammals and exhibits nearly ubiquitous expression in all tissue types (1). Having tens of thousands of binding sites in these genomes, CTCF exhibits a wide and variable effect on gene expression. When bound proximal to promoters, CTCF has been shown to be associated with activating or repressing activity on various genes, including Myc and App (2,3). CTCF can also act as an enhancer-blocker, having the ability to impede downstream enhancers at the H19/Igf2 and Hbb loci (4,5). Similarly, CTCF has been implicated as a chromatin barrier, with CTCF binding being significantly enriched at boundaries between repressive and active chromatin domains (6,7). Furthermore, the formation of CTCF-dependent chromatin loops is mechanistically tied to and likely required for CTCF to exert its transcriptional effect at many of its binding sites (8–11).
The varied regulatory activities of CTCF underlie its crucial role in development. CTCF is required during oocyte and preimplantation embryo maturation. CTCF knockdown at these early developmental stages results in mis-regulation of imprinted gene expression, mitotic defects and ultimately wide-spread apoptosis (12,13). Recent work has also shown that heterozygous CTCF mutations are associated with intellectual disability, microcephaly and growth retardation (14).
Accordingly, CTCF can also play an important role in cell-type–specific gene expression. While many CTCF binding sites (CBSs) are thought to be invariantly bound across cell types, ∼20–50% of these sites show some cell-type–specific binding (15–18). Regulated CTCF binding has been shown to be important for cell-type–specific chromatin loops at the Hbb locus and within the protocadherin gene cluster (19,20). CTCF also directly recruits TAF3, a critical developmental regulator and core promoter factor, resulting in developmentally regulated chromatin loops (21). Additionally, CTCF is implicated in the control of differentiation through its regulation of a variety of lineage-specific genes including Myc, Pax6 and Myod (22–24). Mis-expression of CTCF in progenitors leads to changes in expression of these key cell-fate determinants, resulting in improper transcriptional programming and incomplete differentiation.
The mechanisms by which CTCF carries out its multiple regulatory functions are largely unknown. It is hypothesized that differential recruitment of CTCF’s 11 zinc fingers may allow CTCF to adopt several distinct conformations and ultimately carry out distinct regulatory activities (25). Individual CTCF zinc fingers display unique preferences in binding to various CBS sequences both in vitro and in vivo, suggesting that characteristics of CBSs play an important role in CTCF regulation (26,27). Variation in binding site sequence has been shown to affect the function and recruitment of other TFs, including Glucocorticoid receptor, the NF-κB complex and Pit-1 (28–30). Such differences in activity can stem from changes in protein conformation and cofactor recruitment linked to single nucleotide differences in the TF’s binding site (30,31). Preferential binding of CTCF’s zinc fingers to specific sequences could similarly affect CTCF, either through direct conformational changes or by altering the recruitment of CTCF’s numerous cofactors.
To explore how binding site sequence affects the characteristics of CTCF binding genome-wide, we previously separated human and mouse CBSs into three classes (Low, Medium and High Occupancy) based on their sequence similarity to the published consensus (15). Using published ChIP-Seq and microarray data from multiple cell-types in mouse and human, we reported that these classes of sites were associated with distinct transcriptional functions and varying levels of cell-type–specific binding (32). Additionally, we found that these classes showed differences in CTCF occupancy as measured via ChIP-Seq. As their names suggest, Low Occupancy (LowOc) and High Occupancy (HighOc) sites showed lower and higher ChIP-Seq tag counts, respectively. These observations suggested that CBS sequence may control the transcriptional effect of CTCF and the developmental regulation of its binding through differences in binding affinity.
To examine these trends further, it is critical to analyze CTCF binding dynamics and expression differences during a developmental process where genetic and technical variations between data sets are minimized. To this end, we have measured CTCF binding and global gene expression during induced mouse embryonic stem (ES) cell differentiation. We found that developmentally regulated CTCF binding occurs preferentially at LowOc sites, and that binding is more often maintained during differentiation at HighOc sites. Furthermore, sites where binding was lost during differentiation are enriched in motifs associated with weaker in vitro affinity for CTCF. Conversely, sites where binding was maintained are enriched in motifs that can confer stronger affinity binding. These results suggest that high affinity binding of CTCF may act as a barrier to the regulation of CTCF recruitment, and that certain positions in the binding site may play a more important role in this mode of regulation. Specifically, the 18th position of the CBS is differentially enriched for T and C among regulated and constitutive sites, respectively, and the identity of this position can predictably affect CTCF affinity in vitro. Finally, by correlating CTCF binding and expression changes during differentiation, we show that developmentally regulated LowOc and HighOc sites are associated with distinct transcriptional functions. Taken together, these results suggest that the regulation of CTCF binding and function is dependent in part on specific motifs within its binding site.
MATERIALS AND METHODS
ES cell culture and differentiation
E14 mouse ES cells were grown in Dulbecco’s modified Eagle’s medium, high glucose (DMEM, GIBCO® #11965-084), 15% fetal bovine serum (FBS, HyClone #SH30071.03), 2 mM l-glutamine (GIBCO® #25030-081), 0.1% 2-mercaptoethanol (GIBCO® #21985-023) and 1000 u/ml ESGRO® supplement containing Leukemia Inhibitory Factor (Chemicon/Millipore #ESG1107), on mitomycin C-treated mouse embryonic fibroblasts (MEF). Before differentiation and collection for RNA or chromatin preparation, ES cells were dissociated into single cell suspension using 0.25% Trypsin-EDTA (GIBCO® #25200056), and adsorbed twice, for 45 min to remove MEF. For ES cell differentiation, ES cells were plated at 1E4 cells/cm2 on gelatinized cell culture plates and grown in DMEM, 10% FBS, 2 mM glutamine, 0.1% 2-mercaptoethanol and 0.1 µM all-trans-retinoic acid (Sigma #R2625). After 4.5 days, cells were dissociated using 0.25% trypsin-EDTA and collected for chromatin or RNA preparation.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as previously described (33) after cross-linking cells for 10 min with 1% formaldehyde. Sonication was performed using a Diagenode Bioruptor to obtain fragments ranging mostly between 100 and 250 bp. Chromatin was immunoprecipitated with 2.5 µg of CTCF antibody (Millipore) or IgG (SantaCruz) for 1E6 cells, and immune complexes were collected using A-sepharose beads (Millipore).
For ChIP-sequencing, cells were cross-linked for 15 min with 1% formaldehyde and chromatin was sonicated with a Diagenode Bioruptor. Chromatin was immunoprecipitated using 10 µg of CTCF antibody (Millipore 07–729, lot DAM1682158) or 10 µg of IgG (Santa Cruz) for 50E6 cells. In each case, for both undifferentiated and differentiated cells, ∼15 ng of CTCF or IgG immunoprecipitated DNA were recovered after combining two technical replicates from the same biological sample. DNA fragments of ∼150–300 bp range were isolated by agarose gel purification, ligated to primers and then subject to Solexa sequencing using manufacturers recommendations (Illumina, Inc.). Analysis of ChIP-Seq data is described in Supplementary Methods.
Defining a CBS as regulated or constitutive
To define a CTCF site as regulated or constitutive, we scanned the 200-bp genomic regions flanking the ChIP-Seq peak positions for the best-scoring CTCF site using the published CTCF motif in the form of a positional weight matrix (PWM) (15) and our PWM scanning tool (34). Given the position of the CTCF site at one of the time points (day 0 or day 4.5), if no CTCF site was detected within 200 bp in the other time point, we deemed the site ‘regulated’ and otherwise ‘constitutive’. Regulated site could be ‘gained’ (absent in day 0) or lost (absent in day 4.5).
RNA extraction and reverse transcription
RNA was extracted and purified using Trizol Reagent (Ambion®, #15596-026), according to manufacturer's instructions. For reverse transcription (RT)-qPCR analysis, RNA was reverse-transcribed using the Superscript III Reverse Transcriptase (Invitrogen #18080-051) with random hexamer primers, according to manufacturer's instructions.
RNA-sequencing
The quality of total RNA was verified on a Bioanalyzer 2100 using the RNA 6000 Nano Total RNA kit (Agilent #5067–1511). Starting from 5 µg of total RNA, samples were prepared according to the Illumina mRNA sequencing sample preparation protocol (# RS-930-1001), with purification of ∼250 bp cDNA templates from 2% agarose gel run at 100 V for 1 h. Two lanes were sequenced for each biological sample, with 36 bp single-end reads, on an Illumina Genome Analyzer IIx using Cluster Generation kits (v4) and Sequencing kits (v4). Analysis of RNA-sequencing data is described in the Supplementary Methods.
Measurement of binding affinity using fluorescence polarization
Fluorescence polarization (FP) experiments were performed using conditions and methods as previously described (35). Briefly, 2 nM of FAM-6–labeled 36-bp dsDNA probe with a known dissociation constant of 17 nM for CTCF11ZF (CTCF site HighOc1, Supplementary Table S1) was added to increasing concentrations of unlabelled 36 bp dsDNA probe (0–5 μM). CTCF11ZF (17 nM) was then added to a final volume of 30 μl for each well and incubated for 60 min at 4°C. Experimental data were analyzed using the Prism 3.0 software (GraphPad) and the inhibition constants were determined by nonlinear regression.
Selection of CBSs for affinity measurements
A total of four comparisons were made: (i) regulated LowOc versus regulated HighOc, (ii) constitutive LowOc versus constitutive HighOc, (iii) regulated LowOc versus constitutive LowOc and (iv) regulated HighOc versus constitutive HighOc. Here, by ‘regulated’ we refer to sites that were occupied at day 0 and lost at day 4.5. Each of the four comparisons, say, between group-A and group-B sites were performed identically as follows:
Separately for group-A and group-B sites, we constructed a 4-mer position weight array (PWA) (36). PWA is a generalized PWM. A 4-mer PWA is a matrix with 256 rows (corresponding to all possible 4-bp oligonucleotides) and 17 columns (corresponding to the 17 4-mers in a 20-bp CTCF site. The entry corresponding to row-i and column-j in the PWA contains the normalized frequency of the ith 4-mer at the jth position (a small pseudocount of 1 was used to ensure that no entry was equal to zero). We thus constructed PWAA and PWAB for the two groups of sites. For each of the 17 columns, say j, we computed the relative entropy (RE) of column-j in PWAA versus column-j in PWAB (37), yielding REA. Similarly, we computed REB. The entries in the RE vectors indicate how different the 4-mer distributions are between the two groups, higher the RE, the greater the difference. Given PWAA, PWAB, REA, REB and given a 20-bp CTCF site X belonging to one of the groups, say group-A, we computed Score (X) as follows:
 |
(1) |
where ij refers to the index of the jth 4-mer of X. Scoring a site in group-B is done analogously. The scores calculated for each CBS and each 4-mer in the different comparisons are indicated in Supplementary Table S3 and represented graphically on the heatmaps in Supplementary Figure S8. A CBS's score captures how much it is ‘similar’ to sites in a group and ‘dissimilar’ to sites in the other group. It sums overall the intergroup differences in frequency of all 4-mers, weighted by the ‘importance’ (measured by RE) of each of the 17 positions.
Definition of the ‘differential expression’ insulator function
To characterize the differential expression (DE) insulator function, we considered only sites that are flanked by divergent promoters, consistent with previous studies (32,38). For sites uniquely bound in undifferentiated cells, we developed a score that captures the fact that, in undifferentiated cells, exactly one of the promoters is expressed and, in differentiated cells, both genes are expressed (i.e. loss of insulation with the binding loss, leading to the co-expression of the flanking promoters; this scenario is referred to as DE1). The score varies between 0 and 1 (1 being the ideal case scenario), with a probabilistic interpretation, and measures the decrease in differential between the expression of the two flanking promoters; the higher the score, the greater the decrease in differential. Formally, the score is calculated as follows.
Denoting the two flanking promoters by x and y, let x0, x4, y0, y4 be the expression values from these promoters in undifferentiated (day 0) and differentiated (day 4.5) cells. We normalize the four values into a percentile value such that the expression level indicates the fraction of all genes whose expression is below the given value, i.e. the transformed expression is the probability that the given gene has expression greater than a randomly selected gene. To capture the fact that, in undifferentiated cells, exactly one of the genes is expressed and, in differentiated cells, both genes are expressed, the score is defined as max(x0(1-y0)x4y4, (1-x0)y0x4y4).
We divided the lost sites in two groups, one with a score among the top 25% (Decrease in DE) and the other in the bottom 50% (No Decrease). We then use Fisher's exact test to determine whether one occupancy class of CTCF sites is relatively enriched in one of the groups.
We also calculated the score, which captures the fact that, in undifferentiated cells, exactly one of the promoters is expressed and, in differentiated cells, neither promoter is expressed (i.e. the insulation is lost with the binding loss, leading to co-repression of the flanking promoters), referred to as DE0. The score and the analysis are analogous to DE1. Similar calculations were also made in cases where CTCF binding was gained, as opposed to lost.
Definition of the ‘correlated expression’ insulator function
To characterize the correlated expression (CE) function of a pair of CTCF sites, we defined transcript blocks as genomic intervals with lengths between 50 kb and 1 Mb flanked by CBS on either side. We only consider the blocks where both flanking CBS are occupied in undifferentiated cells and at least one of them is not occupied in differentiated cells. Within block variance (V), the transcript expression level is computed for each block. Normalized increase in variance from undifferentiated (V0) to differentiated cell (V4) is calculated for each block as dV = (V4 −V0)/(V4 + V0). All blocks are classified into two groups based on whether dV is among the top 20% of dV for all blocks, or among the bottom 80%. The blocks are labeled as LowOc-LowOc, MedOc-MedOc and HighOc-HighOc, if both flanking CBS are LowOc, MedOc or HighOc, respectively. We compare the relative proportions of LowOc-LowOc, MedOc-MedOc, HighOc-HighOc blocks between the two classes based on dV, using a Fisher's exact test.
RESULTS
LowOc sites are associated with regulated binding during ES cell differentiation
Using ChIP-Seq data from human cell lines, we previously established that LowOc-binding sites tend to be cell-type specific, whereas HighOc sites tend to be bound in multiple cell types by CTCF (32). These findings suggested that LowOc sites are more prone to developmental regulation than HighOc sites. To test this hypothesis directly, we used in vitro differentiation of mouse ES cells as a developmental model. E14 mouse ES cells were differentiated for 4.5 days in the presence of retinoic acid, and genome-wide CTCF binding was measured by ChIP-Seq before and after differentiation. At 4.5 days of treatment, the expression of the pluripotency factors Nanog and Oct4 was lost, confirming that cells were fully differentiated (Supplementary Figure S1D). Overall, 15 330 and 9016 CTCF peaks were identified before and after differentiation, respectively. The 20-bp motif with the highest similarity to the previously published CTCF binding consensus was determined for each peak using the PWM_SCAN tool (15,34). These CBSs were then separated into the LowOc, MedOc or HighOc class based on their low, medium or high similarity to the CTCF consensus motif as previously described [(32); Supplementary Table S2].
Sites were defined as lost if no CBS was observed within 200 bp after differentiation (8263 sites). CBSs in undifferentiated cells were considered ‘constitutive’ if a CBS was detected within 200 bp in differentiated cells (7067 sites). Similarly, binding sites in differentiated cells were considered either constitutive or gained depending on whether they could be matched to a site in undifferentiated cells (7102 and 1914 sites, respectively). Examples of sites where CTCF binding is lost, gained or constitutive, some of which have been previously characterized, are shown in Figure 1A–C (39,40). As expected, LowOc sites comprised a larger proportion of CBSs where binding was lost or gained as compared with sites where binding was constitutive (Figure 1D). These enrichments for LowOc sites were significant when compared with that of the HighOc class, which comprised a larger proportion of CBSs, where binding was maintained (Fisher’s exact test P = E-81 and P = E-18, respectively). These trends hold when comparing another subsequently generated undifferentiated ES cell data set (26 614 sites) and our differentiated ChIP-Seq data set (Fisher’s exact test P = 3.2 E-144 and P = 1.6 E-36 lost and gained, respectively). Additionally, when comparing mouse ENCODE CTCF ChIP-Seq data sets generated from two mouse ES cell lines and seven distinct adult mouse tissues, we also observed a significant enrichment of LowOc sites among ES cell-specific sites compared with ubiquitously bound sites. This provides an additional independent confirmation of our observed trends (Supplementary Figure S2).
Figure 1.
CTCF binding during ES cell differentiation. Representative CTCF ChIP-Seq peaks where binding is lost (A), maintained (B) and gained (C) during differentiation are shown. Y-axis shows relative enrichment above IgG. The relative proportion of LowOc, MedOc and HighOc sites among sites that are lost, gained or maintained during ES cell differentiation are also shown (D). The proportion of sites from each class was compared between the different groups via Fisher's exact test. LowOc sites are significantly enriched among sites where CTCF binding is lost (P = E-81) and gained (P = E-18) as compared with those that maintain binding.
Because our analysis in differentiating ES cells relies on comparing changes in CTCF binding between two states, it is important that the specificity and sensitivity of binding detection is similar for the two ChIP-Seq data sets. Strong differences in detection specificity are unlikely, as in both undifferentiated and differentiated cells, 10 randomly chosen CTCF-bound sites from our ChIP-Seq data sets were confirmed via ChIP-qPCR in two biological replicates (Supplementary Figure S3A and B, Supplementary Table S4). To test for possible differences in sensitivity, we first randomly selected 10 sites whose binding is observed in publically available ChIP-Seq CTCF data sets generated from seven adult mouse tissues (see Supplementary Methods). We then confirmed binding of CTCF at these sites in undifferentiated and differentiated cells via RT-qPCR. Finally, we determined if these sites were also detected in our ChIP-Seq data sets. If our data sets identified drastically different numbers of these commonly bound CTCF sites before and after differentiation, this would suggest a difference in detection sensitivity. For undifferentiated and differentiated cells, 3 of 10 and 4 of 10 sites were detected, respectively, indicating that differences in detection sensitivity are likely limited (Supplementary Figure S3C and D, Supplementary Table S4).
To further ensure that sites were not mistakenly characterized as regulated due to discrepancy in detection sensitivity between the two differentiation states, we compared the tag count of CTCF sites between undifferentiated and differentiated cells. We first verified that, for constitutive sites, tag counts showed correlation between the undifferentiated and differentiated states (Pearson = 0.596; Supplementary Figure S4C and D). We then reasoned that, if sites had mistakenly been considered as regulated, their tag count should be correlated between the two differentiation states, as for the constitutive sites. Conversely, if these sites are truly regulated, there should be little correlation because in the nonbound state, tag-count would only reflect detection background. The correlation coefficients for sites where binding is lost (Pearson r = 0.326; Supplementary Figure S4A) or gained (Pearson r = 0.172; Supplementary Figure S4B) are indeed much lower than that for constitutive sites, suggesting that a majority of these sites are truly regulated.
Because LowOc sites have a lower average ChIP-Seq tag count than HighOc sites, it is also possible that the greater variability of binding we observe at LowOc sites only reflects a lower chance of detection of CTCF binding. To exclude this possibility, we repeated our analysis after correcting for tag count, using a binning and sampling technique (see Supplementary Methods). LowOc sites still appeared significantly more enriched than MedOc and HighOc sites among CBSs for which CTCF binding was lost (Fisher’s exact P = 3.9 E-23 and P = 2.7 E-57, respectively) or gained (Fisher’s exact P = 5.7 E-5 and P = 2.9 E-10, respectively), as compared with sites whose binding was maintained. This finding confirms that the greater variability of CTCF binding at LowOc sites cannot be explained entirely by their lower tag count and supports that CTCF recruitment is more developmentally regulated at LowOc sites than at HighOc sites.
As shown above and in our previous work, MedOc sites generally appear to have properties intermediate between those of the LowOc and HighOc sites. Therefore, we focused our subsequent analysis on comparing the properties of the LowOc and HighOc class to characterize more efficiently how binding site sequence modulates CTCF's ability to be developmentally regulated.
Different classes of binding sites have distinct in vitro affinity for CTCF
We previously observed that HighOc sites have a higher in vivo occupancy than LowOc sites as approximated by tag counts (32). This trend is also observed in our ChIP-Seq data sets for both undifferentiated and differentiated ES cells (Wilcoxon P = 6.04 E-11 and P = 2.44 E-4, respectively; Supplementary Figure S5). The higher occupancy of HighOc sites could reflect a higher binding affinity, which may explain why CTCF binding at these sites is constitutive. Conversely, LowOc sites may have a lower binding affinity, making their recruitment of CTCF more susceptible to the effect of development cues.
Because the partition between the LowOc and HighOc class is based on binding site sequence, we expect differences in binding affinity to arise from the presence of different sequence motifs characteristic of each class. Sequence within the CBS core motifs (nucleotides 4–8 and 10–18) has previously been shown to be the most critical determinant for CTCF binding in vitro, making it a good candidate to explain possible affinity differences between the LowOc and HighOc class (26). Thus, as a first approach, we measured binding affinity for two LowOc and two HighOc sites selected to have core motifs that are unique to the LowOc and HighOc class, respectively (see Supplementary Methods). Binding affinity was measured by electrophoretic mobility shift assay and a high-throughput FP-based method (see ‘Materials and Methods’ section). A construct consisting of CTCF’s DNA-binding 11 zinc-finger domain (CTCF11ZF) was used for these experiments because full-length CTCF tends to self-associate through N and C termini that flank the 11 zinc-finger domain (41,42). As expected, the tested LowOc sites showed a markedly (∼2.5-fold) lower binding affinity than the tested HighOc sites (Supplementary Figure S6). We also measured the affinity of two CBSs from two extensively characterized loci: a LowOc site in the H19/Igf2 imprinting control region, and a HighOc site in the Hbb locus control region. These sites showed a similar difference in affinity for CTCF11ZF (Supplementary Figure S6). These results suggest that differences in occupancy between the LowOc and HighOc classes may arise from differences in binding affinity.
We then refined our method of comparing the LowOc and HighOc classes to address the link between low binding affinity and developmental regulation. We considered two alternate possibilities to explain the difference in occupancy between LowOc and HighOc sites: (i) Motifs that cause low and high binding affinity are enriched in the LowOc and HighOc class. (ii) Motifs that cause low and high binding affinity are found among regulated and constitutive sites, which are enriched in the LowOc and HighOc class, respectively.
Thus, to determine if occupancy class or developmental regulation of binding is more predictive of CTCF binding affinity, we performed two sets of comparisons: between LowOc and HighOc sites (regulated LowOc versus regulated HighOc and constitutive LowOc versus constitutive HighOc), and between regulated and constitutive sites (regulated LowOc versus constitutive LowOc and regulated HighOc versus constitutive HighOc). To select sites to be tested for each comparison, we further hypothesized that differences in sequence-encoded binding affinity would arise from changes in the interaction of individual zinc fingers with DNA. Out of CTCF's 11 zinc fingers, 10 are C2H2 zinc fingers (43). Zinc fingers of this family have been shown to interact with 4-bp motifs (44). We thus identified 4-bp motifs differentially enriched between each group being compared. CBSs were then scored based on the presence of differentially enriched 4-bp motifs at each position (see Methods, Supplementary Figure S8A and Supplementary Table S3). The binding affinity of the six or seven CBSs with the highest score for each comparison was measured using FP.
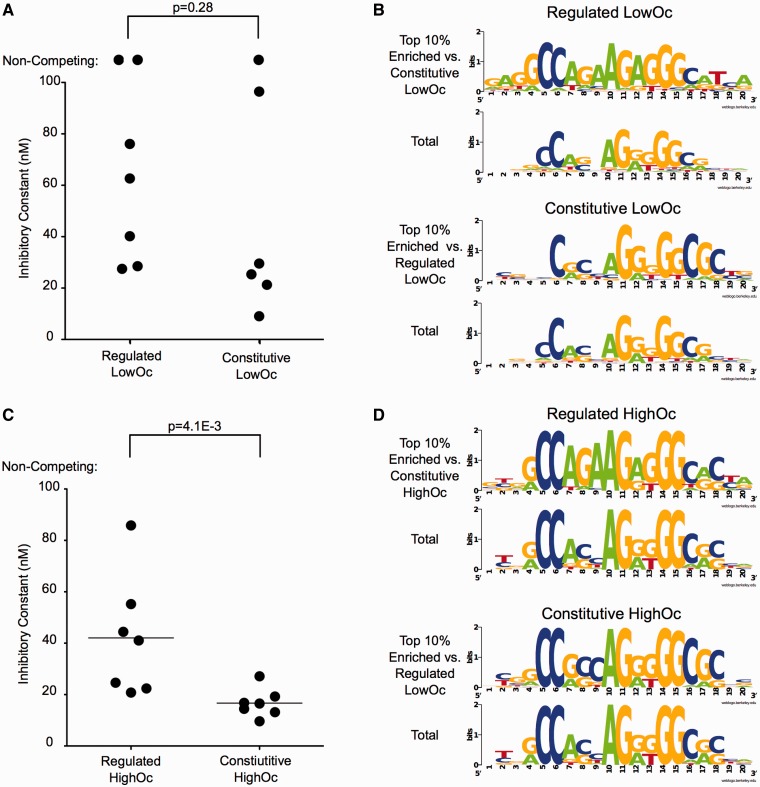
Strikingly, when comparing regulated LowOc and regulated HighOc sites, five of the six tested regulated LowOc sites had no measurable in vitro binding to CTCF11ZF (noncompeting, Figure 2A). These sites had comparable binding characteristics to a purposefully mutated CTCF site (Supplementary Figure S7). All tested regulated HighOc sites, however, bound in vitro. In contrast, when comparing constitutive LowOc and constitutive HighOc sites, four of seven constitutive LowOc sites had a measurable affinity for CTCF, as did five of six constitutive HighOc sites (Figure 2C). In both comparisons, LowOc sites tend to have a lower affinity than HighOc sites, but this trend is significant only for regulated sites (Wilcoxon P = 0.002).
Figure 2.
Comparison CTCF’s in vitro affinity for selected LowOc and HighOc sites. The in vitro affinities of CTCF11ZF for sites from the LowOc and HighOc class were compared by measuring inhibitory constants for selected binding sites via FP. Low inhibitory constants reflect high binding affinity and vice versa. Two comparisons were made, first among sites whose binding was regulated during differentiation (A) and then among sites whose binding was constitutive (C). For each comparison, sites were selected based on the enrichment of 4-bp motifs in one group of sites as compared with the other (see Supplementary Figure S8 and ‘Materials and Methods’ section). Shown are the motif logos generated from sites with the highest differential enrichment of motifs for each comparison (B, D- ‘Top 10% Enriched’), from which sites were selected for testing. Also shown are the motif logos generated from all sites in a corresponding group for comparison (B, D- ‘Total’).
Interestingly, 4-bp sequence motifs that are the most differentially enriched between the LowOc and HighOc class are located at the same positions (nucleotides 2–7 and 15–18) and overlap with CBS core motifs (nucleotides 4–8 and 10–18). Furthermore, differentially enriched motifs at these positions are observed for a majority of sites for both comparisons (Supplementary Figure S8B). Thus, motifs enriched within the core regions may explain why LowOc sites as a whole tend to have a lower affinity than HighOc sites. However, they do not explain why this difference in binding affinity is more pronounced for regulated than for constitutive sites. On closer examination, we found that top-scoring constitutive LowOc sites showed an enrichment for C or G at the 18th position. This enrichment is also found in HighOc sites, but not in regulated LowOc sites (Figure 2B and D). Strikingly, most tested LowOc sites that effectively bound CTCF11ZF had a C or G at the 18th position (five of six). The majority (four of six) of tested LowOc sites that did not bind CTCF had an A or T at this position (Supplementary Table S1). These results suggest that the presence of C or G at the 18th position is critical to stabilize CTCF binding, which may explain why differences in binding affinity between LowOc and HighOc sites are more pronounced for regulated sites. In sum, these results suggest that LowOc sites tend to have a lower affinity than HighOc sites due to motifs characteristic of each class as a whole. Additionally, distinct sequence motif characteristic of either regulated or constitutive sites may modulate this binding affinity.
To directly characterize sequence motifs associated with the developmental regulation of CTCF binding, we performed a second set of comparisons: regulated versus constitutive LowOc sites, and regulated versus constitutive HighOc sites. The binding affinity was lower at regulated sites than at constitutive sites within the HighOc class, but surprisingly, no significant difference was observed within the LowOc class (Figure 3A and C; Wilcoxon P = 0.004 and P = 0.528, respectively). Thus, developmental regulation of HighOc sites may be facilitated by sequence motifs that reduce binding affinity.
Figure 3.
Comparison of CTCF’s in vitro affinity for selected regulated and constitutive sites. The in vitro affinities of CTCF11ZF for sites that show regulated and constitutive binding were compared by measuring inhibitory constants via FP. Low inhibitory constants reflect high binding affinity and vice versa. Two comparisons were made, first among LowOc sites (A) and then among HighOc sites (C), as a complement to the analysis shown in Figure 2. For each comparison, sites were selected based on the enrichment of 4-bp motifs in one group of sites as compared with the other (see Supplementary Figure S8 and ‘Materials and Methods’ section). Shown are the motif logos generated from sites with the highest differential enrichment of motifs for each comparison (B, D- ‘Top 10% Enriched’), from which sites were selected for testing. Also shown are the motif logos generated from all sites in a corresponding group for comparison (B, D- ‘Total’).
Unlike our previous set of comparisons, a strong differential enrichment of 4-bp motifs at specific positions was only observed in high-scoring sites (Supplementary Figure S8C). Because of the small proportion of sites containing such motifs, it is possible that their association with regulated or constitutive binding is particular to our experimental system. To exclude this possibility, we verified that similar motif enrichments were found when comparing ES cell-specific and ubiquitously bound CTCF sites identified using public ChIP-Seq data sets from two mouse ES cell lines and seven distinct mouse adult tissues. Specifically, we tested whether a motif-based model trained on our data can distinguish the corresponding classes in independent public data sets. In all cases, we found this to be true (P ∼ 0 for regulated LowOc versus constitutive LowOc, P = E-227 for constitutive LowOc versus regulated LowOc, P ∼ 0 for regulated HighOc versus constitutive HighOc, P = E-039 for constitutive HighOc versus regulated HighOc; see Supplementary Methods).
Differences in sequence between high-scoring regulated and high-scoring constitutive sites are likely responsible for the observed affinity differences within the HighOc class. Specifically, high-scoring regulated sites showed an enrichment for A at the 7th and 9th position as compared with their high-scoring constitutive counterparts (Figure 3D). This is reflected in the sequence of our tested sites, as all regulated HighOc sites examined had an A at their 7th and 9th position (Supplementary Table S1), and no constitutive HighOc sites examined had an A at either position. A similar enrichment of A at the 7th and 9th position coupled with an enrichment of T at the 18th position was observed for high-scoring regulated LowOc sites compared with their constitutive counterparts (Figure 3B). However, this did not lead to a strong reduction in binding affinity. Therefore, nucleotide preferences at the 7th, 9th and 18th positions are associated with the regulation of CTCF binding, but their observable effect on in vitro binding affinity likely varies depending on other positions within the CBS.
Identity of position 18 in the binding core motif has a predictable effect on in vitro CTCF affinity
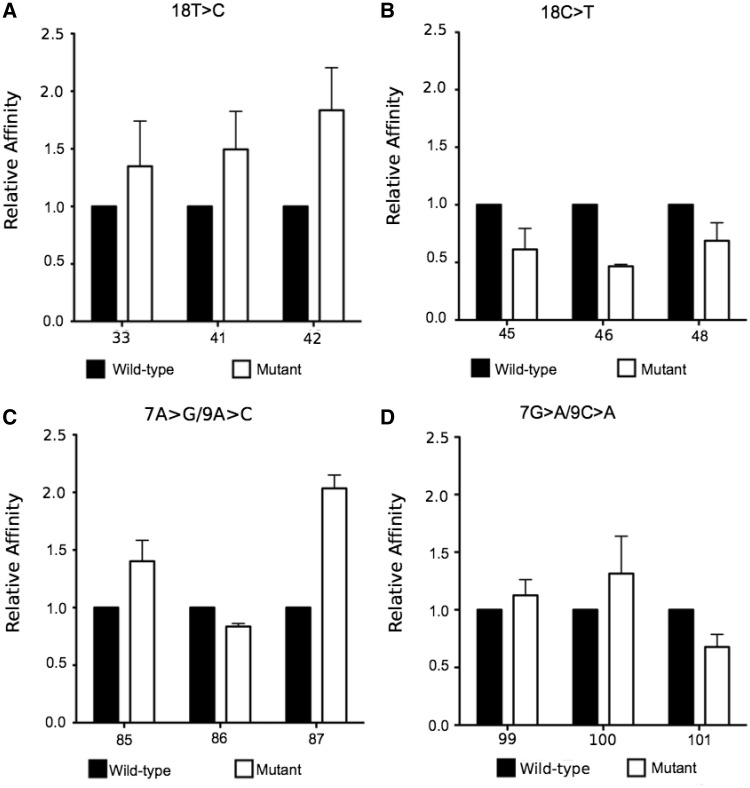
To directly test the effect of nucleotide identity at the 7th, 9th and 18th position on CTCF affinity, we mutated several previously characterized binding sites at these positions and measured the relative change in affinity as compared with wild type. LowOc sites with nonmeasurable binding affinity often have an A or a T at position 18. Thus, we predicted that an 18 T > C mutation in a binding site would increase the affinity for CTCF, while an 18 C > T mutation would conversely decrease affinity. Our results follow this trend, with an average 45% increase in affinity resulting from an 18 T > C mutation for three regulated LowOc sites, and an average 55% decrease in affinity resulting from a 18 C > T mutation for three constitutive LowOc sites (Figure 4A and B). These results suggest that the nucleotide identity of position 18 in a CBS modulates CTCF’s affinity for that sequence in a predictable manner.
Figure 4.
Changes in CTCF binding affinity following mutation of specific nucleotides in CBSs. The relative change in in vitro affinity for CTCF was measured via FP following 18 T > C (A), 18 C > T (B), 7 A > G/9 A > C (C) or 7 G > A/9 C > A (D) mutations. For each mutation, three wild-type and three mutated probes were tested. Probe names are listed below each measure of relative affinity.
We similarly tested the effect of nucleotide identity at the 7th and 9th position in CBSs. We observed an enrichment for G at the 7th position and C at the 9th position in constitutive HighOc sites associated with increased affinity. Thus, we predicted that a 7 A > G/9 A > C double mutation would result in an increase in CTCF affinity and a 7 G > A/9 C > A double mutation would result in a decrease in CTCF affinity. Only two out of the three regulated HighOc probes with 7 A > G/9 A > C mutations showed an increase in affinity, and only one out of three constitutive HighOc probes with 7 G > A/9 C > A mutations showed a decrease in affinity (Figure 4C and D). These results suggest that, unlike at the 18th position, the nucleotides at the 7th and 9th positions do not modulate CTCF’s affinity for a given sequence in a predictable manner.
Distinct TF motifs are differentially enriched within CTCF site classes
Although the sequence and affinity of CBSs likely contribute to the regulation of binding, additional regulatory signals must influence actual changes in CTCF recruitment. CTCF binding is thus likely affected by the regulatory context surrounding its binding site. This context is defined by a multitude of criteria, including the capacity for proximal binding of other TFs. To assess possible differences in the regulatory context at CTCF sites, we conducted a differential motif enrichment analysis based on vertebrate TF motifs from the TRANSFAC database (45) following the same comparison scheme used for the binding affinity experiments (regulated LowOc versus regulated HighOc, constitutive LowOc versus constitutive HighOc, regulated LowOc versus constitutive LowOc, regulated HighOc versus constitutive HighOc).
Across the four comparisons, motifs corresponding to 80 unique TFs were enriched within 100 bp of CBSs (false discovery rate ≤ 10%; Supplementary Table S5). Interestingly, only a small number of motifs were differentially enriched in the LowOc versus HighOc comparison, and a majority of those were enriched near HighOc sites, especially among constitutively bound sites. However, numerous motifs were identified in the regulated–constitutive comparison. Many of these motifs were enriched in regulated or constitutive sites irrespective of being of the LowOc or HighOc class. This is particularly the case of certain TF motifs (AP-2, Elk-1, E2F-1, HIC1, ZF5) previously reported to be enriched near constitutive and syntenic CTCF sites (46). Overall, the TRANSFAC analysis suggests that the capacity for TF recruitment may be different at regulated and constitutive CTCF sites, but less variable between the LowOc and HighOc class.
Regulated LowOc and HighOc sites are associated with distinct gene expression patterns
CTCF binding at different loci has been shown to be associated with various transcriptional activities (16). To address the question of whether the binding site sequence plays a role in determining CTCF activity, we previously inferred CTCF function by correlating CTCF occupancy and gene expression from published data sets (32). We found that LowOc sites were associated with transcriptional activation and with DE of flanking divergent promoters, a hallmark of insulators. HighOc sites were associated with transcriptional repression and were more often located at the boundary of co-regulated gene domains, indicating a different type of insulator activity. However, a more accurate genome-wide determination of CTCF function can be achieved by correlating changes in CTCF binding with changes in gene expression during a dynamic biological process. For this reason, we measured genome-wide expression by RNA-Seq, from the same pool of undifferentiated and differentiated cells characterized by CTCF ChIP-Seq. The accuracy of the RNA-Seq was confirmed by comparing the reads per kilobase per million (RPKM) values for 10 genes with expression levels measured via RT-qPCR (Supplementary Figure S1A–C). Global quantification of gene expression allowed us to investigate more accurately the contribution of the LowOc, MedOc and HighOc classes to the transcriptional activation, transcriptional repression and insulation functions of CTCF at developmentally regulated binding sites.
A CTCF site was defined as a transcriptional activator if during differentiation, expression from the nearest promoter decreased below a certain threshold when CTCF binding was lost, or increased above this threshold when CTCF binding was gained (Figure 5A). Conversely, a CTCF site was defined as a transcriptional repressor if gene expression increased above that same threshold when CTCF binding was lost, or decreased below this threshold when binding was gained. To discriminate between low-level and actively regulated transcription, we used a gene-expression threshold of 10 RPKM, above which ∼10% of genes are expressed in either state. Consistent with our previous findings, we found that the LowOc class made up a larger proportion of sites where CTCF was likely exerting an activator activity as compared with MedOc sites (Fisher’s exact P = 0.014), or MedOc and HighOc sites combined (Fisher’s exact P = 0.029; Figure 5B). Contrary to our previous finding, HighOc sites were not preferentially associated with transcriptional repression. This could be explained by the restriction of the analysis to developmentally regulated sites, among which HighOc sites are underrepresented, thereby reducing the statistical power.
Figure 5.
Association of CTCF site classes with distinct regulatory functions. The expression of genes associated with regulated LowOc, MedOc and HighOc sites was analyzed to determine possible trends in activation, repression or two different measures of insulation activity. The expression patterns used as proxies for these activities are depicted (A). The relative proportion of LowOc, MedOc and HighOc sites associated with particular transcriptional functions are shown (B–D). The proportion of classes was compared between each functional group and significant Fisher's exact test P-values are indicated.
To characterize where CTCF has discernible insulator activity, we used two distinct definitions of insulation. First, we considered CBSs flanked within 50 kb by divergent promoters. Such CBSs were considered to be DE insulators if only one of the flanking promoters showed strong gene expression when CTCF is bound and either both (DE1) or neither (DE0) promoters showed strong expression when CTCF is not bound (Figure 5A; see ‘Materials and Methods’ section). In this instance, we hypothesized that CTCF may be preventing regulatory elements on one flank from acting on the opposing flank, allowing for greater DE of the two genes. Secondly, we considered CTCF a CE insulator if, for 50 kb–1 Mb genomic regions flanked by CBSs, the variance in transcript expression within the domain is lower when CTCF is bound than when at least one CTCF site is unoccupied (Figure 5A; see ‘Materials and Methods’ section). In this instance, genes within the CTCF-defined domain are hypothesized to be only affected by regulatory elements within this domain, resulting in greater co-regulation.
When CTCF is bound in undifferentiated ES cells and its binding is lost during differentiation, we found that LowOc sites are significantly enriched among the DE1 insulators as compared with MedOc sites (Fisher’s exact P = 0.028), or MedOc and HighOc sites combined (Fisher’s exact P = 0.036; Figure 5C). Interestingly, we did not observe such trends for DE0 insulators, suggesting that LowOc sites may preferentially protect flanking promoters from being co-expressed and not from being co-repressed. Conversely, HighOc sites are significantly enriched among CE insulators as compared with MedOc sites (Fisher’s exact P = 0.005) or LowOc and MedOc sites combined (Fisher’s exact P = 0.011). This preferential association of LowOc sites with DE insulators and of HighOc sites with CE insulators is consistent with our previous observations in human cells. It supports the notion that different types of insulator activity are associated with different CTCF sites, and that the site sequence is important to determine this activity.
To further confirm our analysis of CTCF function, we performed stable knockdown of CTCF in mouse ES cells and quantified changes in expression at a number of putative CTCF target genes. If CTCF knockdown resulted in a similar expression change as observed when CTCF was lost during differentiation, this suggests that the function inferred for the corresponding CBS does not depend on differentiation and relies mostly on this single site. We carried out two knockdown experiments, in which we confirmed CTCF depletion by western blot (98 and 95% knockdown from wild type, data not shown). We additionally confirmed this knockdown at individual sites via ChIP and measured changes in expression of the relevant target gene by RT-qPCR for four putative activator sites and four putative DE1 insulator sites. For both tested functions, we observed changes in gene expression concordant with the expectation for two of four tested loci (Supplementary Figures S9 and S10). This suggests that, at least for these sites, our approach of monitoring changes during differentiation was successful at predicting CTCF function. It should be noted, however, that definite proof could only be obtained by targeted mutagenesis, for which the characterized loci would be good candidates.
DISCUSSION
In this study, we aimed to ascertain how binding site sequence plays a role in the regulation and function of CTCF binding. In the context of a controlled developmental system, we observed that sites with a lower similarity to the CBS consensus (LowOc sites) are more likely to show changes in binding during differentiation. We also observed that the regulation of CTCF binding is often associated with specific DNA motifs within CBSs, leading to a lower in vitro affinity for CTCF. This suggests the possibility of a mechanism regulating CTCF recruitment dependent in part on sequence-based affinity. Accordingly, we show that certain nucleotide preferences within particular classes of binding sites can contribute predictably to CTCF affinity. We also show that binding site sequence differences are associated with distinct transcriptional functions genome-wide. Our results suggest that small changes to the CBS sequence likely play a contributing role to CTCF’s recruitment and its effect on transcription.
Comparison of LowOc and HighOc sites
Our analysis reveals that CTCF binding at LowOc sites is more likely to be regulated during ES cell differentiation. We also show that LowOc sites are more cell-type specific within mouse ENCODE data sets, suggesting that low similarity to the consensus motif may be a fundamental characteristic underlying cell-type–specific CTCF recruitment. Additionally, our findings are among recent works that highlight the functional importance of cell-type–specific CTCF binding genome-wide (11,47). While these studies highlight key characteristics associated with differential CTCF binding across species and cell types, our results are the first to identify a class of CTCF sites more prone to such differential occupancy during differentiation.
We demonstrated that LowOc and HighOc sites are enriched in specific motifs that are associated with lower and higher binding affinity for CTCF, respectively. This suggests that differences in binding occupancy observed between the LowOc and HighOc classes are, at least in part, caused by differences in binding affinity of CTCF to the binding site sequence. Importantly, this relationship between occupancy and affinity is generally assumed but, to our knowledge, has never been tested in the case of CTCF. Because the sequence differences between LowOc and HighOc sites occur primarily within the two core motifs (nucleotides 4–8 and 10–18) of the 20 bp consensus, these regions likely define any sequence-based affinity differences observed between these two classes.
It is notable that the majority of regulated LowOc sites selected to have distinct motifs from regulated HighOc sites showed no binding to CTCF11ZF in vitro. These binding sites likely require additional cues for effective in vivo recruitment, which could be mediated by CTCF’s N/C termini, cofactor recruitment or posttranslational modifications, all of which have been previously implicated as important for CTCF function (38,48,49). It is also possible that these sites require additional sequence outside of the core motif. For example, recent work has identified motifs upstream of the 20 bp core motif that confer stronger in vivo recruitment of CTCF (27). It is possible that these sites may require such extra sequence elements for effective binding in vitro and in vivo. In contrast, constitutively bound LowOc sites, selected to have distinct motifs from constitutive HighOc sites, showed robust in vitro binding in our assays. Their binding to CTCF could thus rely principally on the interaction of CTCF 11 zinc-finger domain with nucleotides within the core motif, and be less dependent on other factors.
It is also important to note that while we focus our analysis on the HighOc and LowOc class, many CBSs fall within the MedOc class. Our results suggest that the properties of each class reflect a difference in enrichment of certain motifs rather than the existence of motifs unique to each class. Thus, as in our previous study, it is unsurprising we observe the MedOc class of CBS displays intermediate characteristics as compared with the LowOc and HighOc classes.
Comparison of regulated and constitutive sites
The direct comparison of regulated and constitutive sites within either the LowOc or the HighOc class further elucidated the relationship between CTCF affinity and dynamic binding. In contrast to the comparison of LowOc and HighOc sites, motifs enriched between regulated and constitutive sites were concentrated in only a small subset of all regulated and constitutive sites, respectively. This suggests that our definitions of LowOc and HighOc may effectively capture a majority of sites associated with regulated and constitutive binding.
We observed that regulated HighOc sites selected to have distinct motifs from constitutive HighOc sites showed significantly lower affinity for CTCF than their constitutive HighOc counterparts. Conversely, regulated LowOc sites selected to have distinct motifs from constitutive LowOc sites showed no significant affinity differences from their constitutive LowOc counterparts. In both groups of LowOc sites, however, binding affinity was lower than for the tested constitutive HighOc sites. Together, these results indicate that high binding affinity is an obstacle to dynamic regulation of CTCF recruitment, although low affinity binding is unlikely to be sufficient to prompt such regulation.
Importance of specific positions within the CBS
We observed that at least one specific position of CBS can predictably modulate affinity for CTCF. HighOc sites and sites that are constitutively bound are enriched for C or G at position 18, suggesting a role of these nucleotides in the stabilization of CTCF binding. Accordingly, we show that 18 T > C mutations result in an increase in CTCF affinity, whereas 18 C > T mutations result in a decrease. It is worth noting that the majority of CTCF sites whose functions have been experimentally characterized have a C or G at the 18th position (Supplementary Table S6), likely because these studies examined only CBS that exhibited strong binding in vitro. Thus, it is possible that our current understanding of CTCF function may apply only to a subset of sites with strong binding.
Similar enrichments for G and C at the 7th and 9th positions, respectively, were observed within constitutively bound sites. Unlike mutations at the 18th position, however, 7 A > G/9 A > C double mutations inconsistently affected CTCF affinity. This is surprising, as these changes represent a more extensive mutation of the binding site and are located within a region critical for CTCF recruitment (26). A possible explanation is that the modulation of CTCF affinity by positions 7 and 9 depends on the sequence at other positions within the CBS. While our PWM model accounts for interdependency between noncontiguous positions only to a limited extent, this has been shown to be an important contributor to the affinity of other TFs (50). Recent study of polymorphic CBSs has also shown that the effect of a single nucleotide change on ChIP-Seq occupancy is highly dependent on local context, further supporting this possibility (51).
Such differences at specific positions in the core motif are likely to affect interactions with specific zinc fingers within CTCF’s DNA binding domain. A recent study mutating individual CTCF zinc fingers has assessed the contribution of each zinc finger to CTCF’s recruitment in vivo (27). Its findings suggest that individual zinc fingers are critical for the recruitment to unique subsets of binding sites, with zinc fingers that bind specifically to the core motif being more important for general recruitment. Further study of the interplay between individual zinc fingers and specific base pair positions within the core motif is required to further illuminate mechanisms controlling CTCF recruitment.
Possible mechanisms for changes in CTCF binding
Our results support a model of CTCF regulation where generally weaker LowOc sites are more amenable to developmental cues that affect CTCF recruitment, which necessarily involve stabilization or destabilization by cofactors and epigenetic modifications (16). Conversely, the generally stronger HighOc-binding sites would be more resistant, but not completely impervious, to such cues. A similar mode of regulation has been suggested to control the recruitment of OCT-1 and OCT-2, where weaker binding sites initiate cell-type–specific expression at immunoglobulin promoters, but stronger binding sites direct ubiquitous expression (52). Additionally, another zinc-finger TF, NRSF-REST, has been observed to have more cell-type–specific binding at binding sites that are a weaker match to its consensus (53).
Specifically, a critical developmental cue that could affect CTCF recruitment is CpG DNA-methylation, which is highly dynamic during differentiation and inhibits CTCF binding (5,54). Importantly, CTCF binding actively prevents methylation of DNA, possibly by hindering the recruitment of DNA methyltransferases (55). It is thus possible that, as a consequence of the weaker binding of CTCF at LowOc sites, CTCF inhibits DNA methylation at these sites in a weaker fashion, making them more permissive to regulation by DNA methylation. Specific methylated CpG positions within binding sites have been shown to be more important for the inhibition of CTCF binding (51), which provides an additional level of regulation by which CTCF activity is impacted by CBS sequence. Therefore, a detailed examination of the interplay of CBS sequence, CTCF binding affinity and DNA methylation during development would be of particular interest.
Regulatory context and transcriptional regulatory function of CTCF sites
We show that several TF binding motifs are differentially enriched in the vicinity of CBSs when comparing LowOc and HighOc sites and when comparing constitutive and regulated sites. Interestingly, the comparison between constitutive and regulated sites shows a wider variety of motif enrichment than the comparison between the LowOc and the HighOc class. This is consistent with the idea that, while CBS sequence modulates the ability for CTCF to be regulated, actual regulation or maintenance of binding relies on the activity of cofactors. It is important to note, however, that the vast majority of known CTCF cofactors do not have a known consensus motif or DNA-binding domain. Whether CBS sequence affects CTCF conformation in a way that regulates its interaction with cofactors, possibly leading to the coevolution of CBS and surrounding sequences, as well as to differentiated activities of CTCF, will be interesting to explore.
A first indication that CBS sequence affects CTCF activity was our observation that our binding site classes (LowOc, HighOc) correlated with distinct patterns of gene expression, as established in our previous study (32). While we did confirm our previous finding that regulated LowOc sites are more likely to act like transcriptional activators, we were not able to confirm our previous finding that HighOc sites are more likely to be repressors. This is likely because our experimental design limits our study to sites that show regulated binding during differentiation, among which there are significantly fewer HighOc sites. We did confirm, however, that LowOc and HighOc sites are associated with two distinct measures of insulator activity, which suggests distinct mechanisms for transcriptional insulation associated with differences in binding site sequence.
Because sites exhibiting activator, repressor and both types of insulator activity are present in each CBS class, the full relationship between CTCF function and binding site sequence remains unclear. Further work examining motifs associated with particular transcriptional outputs could lend great insight into the regulation CTCF function.
ACCESSION NUMBERS
The ChIP-Seq and RNA-Seq data generated for the study have been submitted to the NCBI Gene Expression Omnibus (GEO) under accession number GSE39523.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [56–64].
ACKNOWLEDGEMENT
We acknowledge the support of the Wistar Institute Genomics and Bioinformatics Cores.
FUNDING
NIH [R01HD042026 to M.S.B., R01CA140652 to P.M.L., R01GM085226 to S.H., R01-GM052880 to R.M., K99AI099153 to I.T.]; the Wistar Cancer Center core grant [P30 CA10815]; the Commonwealth Universal Research Enhancement Program, PA Department of Health; a postdoctoral fellowship from the American Heart Association (to S.V.); [T32GM008216 to R.P.]. Funding for open access charge: NIH
Conflict of interest statement. None declared.
REFERENCES
- 1.Filippova G, Fagerlie S, Klenova E, Myers C, Dehner Y, Goodwin G, Neiman P, Collins S, Lebenankov V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian cmyc oncogenes. Mol. Cell. Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- 3.Burton T, Liang B, Dibrov A, Amara F. Transforming growth factor-beta-induced transcription of the Alzheimer beta-amyloid precursor protein gene involves interaction between the CTCF-complex and Smads. Biochem. Biophys. Res. Commun. 2002;295:713–723. doi: 10.1016/s0006-291x(02)00725-8. [DOI] [PubMed] [Google Scholar]
- 4.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc. Natl Acad. Sci. USA. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 6.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2008;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CWH, Ye C, Ping JLH, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Publishing Group. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engel N, Raval AK, Thorvaldsen JL, Bartolomei SM. Three-dimensional conformation at the H19/Igf2 locus supports a model of enhancer tracking. Hum. Mol. Genet. 2008;17:3021–3029. doi: 10.1093/hmg/ddn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribeiro de Almeida C, Stadhouders R, Thongjuea S, Soler E, Hendriks RW. DNA-binding factor CTCF and long-range gene interactions in V(D)J recombination and oncogene activation. Blood. 2012;119:6209–6218. doi: 10.1182/blood-2012-03-402586. [DOI] [PubMed] [Google Scholar]
- 10.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl Acad. Sci. USA. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan LB, Pan H, Hannenhalli S, Cheng Y, Ma J, Fedoriw A, Lobanenkov V, Latham KE, Schultz RM, Bartolomei MS. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development. 2008;135:2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 14.Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S, et al. De novo mutations in the genome organizer CTCF cause intellectual disability. Am. J. Hum. Genet. 2013;93:124–131. doi: 10.1016/j.ajhg.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim TH, Abdullaev Z, Smith A, Ching K, Loukinov D, Green R, Zhang M, Lobanenkov V, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Splinter E. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl Acad. Sci. USA. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torrano V, Chernukhin I, Docquier F, D'Arcy V, León J, Klenova E, Delgado MD. CTCF regulates growth and erythroid differentiation of human myeloid leukemia cells. J. Biol. Chem. 2005;280:28152–28161. doi: 10.1074/jbc.M501481200. [DOI] [PubMed] [Google Scholar]
- 23.Wu D, Li T, Lu Z, Dai W, Xu M, Lu L. Effect of CTCF-binding motif on regulation of PAX6 transcription. Invest. Ophthalmol. Vis. Sci. 2006;47:2422–2429. doi: 10.1167/iovs.05-0536. [DOI] [PubMed] [Google Scholar]
- 24.Delgado-Olguin P, Brand-Arzamendi K, Scott IC, Jungblut B, Stainier DY, Bruneau BG, Recillas-Targa F. CTCF promotes muscle differentiation by modulating the activity of myogenic regulatory factors. J. Biol. Chem. 2011;286:12483–12494. doi: 10.1074/jbc.M110.164574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 26.Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV. Critical DNA binding interactions of the insulator protein CTCF. J. Biol. Chem. 2007;282:33336–33345. doi: 10.1074/jbc.M706213200. [DOI] [PubMed] [Google Scholar]
- 27.Nakahashi H, Kwon KRK, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morin B, Nichols LA, Holland LJ. Flanking sequence composition differentially affects the binding and functional characteristics of glucocorticoid receptor homo- and heterodimers. Biochemistry. 2006;45:7299–7306. doi: 10.1021/bi060314k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung TH, Hoffmann A, Baltimore D. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Shewchuk BM, Ho Y, Liebhaber SA, Cooke NE. A single base difference between Pit-1 binding sites at the hGH promoter and locus control region specifies distinct Pit-1 conformations and functions. Mol. Cell. Biol. 2006;26:6535–6546. doi: 10.1128/MCB.00267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009;10:R131. doi: 10.1186/gb-2009-10-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Z, Lezina L, Chen C-J, Shtivelband S, So W, Lieberman PM. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell. 2002;9:493–503. doi: 10.1016/s1097-2765(02)00476-8. [DOI] [PubMed] [Google Scholar]
- 34.Levy S, Hannenhalli S. Identification of transcription factor binding sites in the human genome sequence. Mamm. Genome. 2002;13:510–514. doi: 10.1007/s00335-002-2175-6. [DOI] [PubMed] [Google Scholar]
- 35.Deng Z, Wang Z, Stong N, Plasschaert R, Moczan A, Chen H-S, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei MS, et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012;31:4165–4178. doi: 10.1038/emboj.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 37.Hannenhalli S. Eukaryotic transcription factor binding sites—modeling and integrative search methods. Bioinformatics. 2008;24:1325–1331. doi: 10.1093/bioinformatics/btn198. [DOI] [PubMed] [Google Scholar]
- 38.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl Acad. Sci. USA. 2007;104:7145. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes Dev. 2008;22:575–580. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc. Natl Acad. Sci. USA. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 42.Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of De Novo methylation upon maternal inheritance. Mol. Cell. Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filippova GN, Qi CF, Ulmer JE, Moore JM, Ward MD, Hu YJ, Loukinov DI, Pugacheva EM, Klenova EM, Grundy PE, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 44.Persikov AV, Singh M. An expanded binding model for Cys 2His 2zinc finger protein–DNA interfaces. Phys. Biol. 2011;8:035010. doi: 10.1088/1478-3975/8/3/035010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin D, Pantoja C, Miñán AF, Valdes-Quezada C, Moltó E, Matesanz F, Bogdanović O, de la Calle-Mustienes E, Domínguez O, Taher L, et al. Genome-wide CTCF distribution in vertebrates defines equivalent sites that aid the identification of disease-associated genes. Nat. Struct. Mol. Biol. 2011;18:708–714. doi: 10.1038/nsmb.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves Â, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol. Cell. Biol. 2011;31:2174–2183. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 50.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurano MT, Wang H, Kutyavin T, Stamatoyannopoulos JA. Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet. 2012;8:e1002599. doi: 10.1371/journal.pgen.1002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kemler I, Bucher E, Seipel K, Müller-lmmerglück MM, Schaffner W. Promoters with the octamer DNA motif (ATGCAAAT) can be ubiquitous or cell type-specific depending on binding affinity of the octamer site and Oct-factor concentration. Nucleic Acids Res. 1991;19:237–242. doi: 10.1093/nar/19.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruce AW, Lopez-Contreras AJ, Flicek P, Down TA, Dhami P, Dillon SC, Koch CM, Langford CF, Dunham I, Andrews RM, et al. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res. 2009;19:994–1005. doi: 10.1101/gr.089086.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 55.Engel N, Thorvaldsen JL, Bartolomei MS. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 2006;15:2945–2954. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 56.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Meth. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 57.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, Davuluri RV. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 2011;21:1260–1272. doi: 10.1101/gr.120535.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12.1–S14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim H, Bi Y, Pal S, Gupta R, Davuluri RV. IsoformEx: isoform level gene expression estimation using weighted non-negative least squares from mRNA-Seq data. BMC Bioinformatics. 2011;12:305. doi: 10.1186/1471-2105-12-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rasmussen LM, Hansen PR, Nabipour MT, Olesen P, Kristiansen MT, Ledet T. Diverse effects of inhibition of 3-hydroxy-3-methylglutaryl-CoA reductase on the expression of VCAM-1 and E-selectin in endothelial cells. Biochem. J. 2001;360:363. doi: 10.1042/0264-6021:3600363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 64.Lin S, Ferguson-Smith AC, Schultz RM, Bartolomei MS. Nonallelic transcriptional roles of CTCF and cohesins at imprinted loci. Mol. Cell. Biol. 2011;15:3094–3104. doi: 10.1128/MCB.01449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.