Abstract
R2Bm is a non-long-terminal-repeat (non-LTR) retrotransposon that was identified at a specific target site in the 28S rRNA genes of the silkworm, Bombyx mori. Although in vitro analysis has revealed that the 3′ end of R2Bm is integrated into the target site by means of target-primed reverse transcription (TPRT), the mechanism of the 5′ end integration is not well understood. We established a novel in vivo system to assay the insertion mechanism of R2Bm using a cultured cell line, C65, and a baculovirus, AcNPV, as host and vector, respectively. The 3′ end of R2Bm integrated at the target site in the rRNA genes of C65 cells when an AcNPV containing both the full-length 3′ UTR and the entire open reading frame (ORF) of R2Bm was introduced while the 5′ end integration was incorrect. The 5′ end of R2Bm was integrated, however, when the 28S gene sequence upstream of the R2Bm target site was added to the R2Bm sequence. Thus, in our assay, homologous sequences were likely essential for the successful integration of the entire R2Bm into the host cell genome. We also demonstrated that the failure to integrate caused by a frame-shifted ORF was rescued by co-infection with a helper virus that contained only the R2Bm ORF. This indicates that R2 retrotransposition can be complemented in trans. These findings suggest that the host’s mechanism for DNA repair may be necessary for the integration of the 5′ end of R2Bm and that R2Bm protein may only have the ability to integrate the 3′ end of the element by TPRT.
INTRODUCTION
The analysis of the integration mechanism of non-long-terminal-repeat (non-LTR) retrotransposons into a host cell genome has been carried out by the in vitro analysis of R2Bm. R2Bm was identified at a specific site in the 28S rRNA genes of the silkworm, Bombyx mori (1,2). Detailed studies of the integration mechanism of R2Bm in vitro revealed that the protein, which is encoded by a single open reading frame (ORF) within R2Bm (3), has both reverse transcriptase (RT) and endonuclease (EN) activities, and this protein catalyzes RNA-mediated retrotransposition (4–6). First, the RT domain of the R2Bm protein binds to the 3′ UTR (∼250 bp) of the R2Bm RNA (7–9). Secondly, a RNP complex recognizes and binds to the target site in the 28S rRNA genes. Thirdly, one of the DNA strands is nicked by the EN activity, the reverse transcription of the R2Bm RNA is primed by the hydroxyl at the 3′ end exposed by this nick, and the other strand is cleaved two bases upstream of the first nicked site (5). This process for the integration mechanism into a host genome is called target-primed reverse transcription (TPRT) (5,10), and it is believed that other non-LTR retrotransposons as well as the mobile group II introns may have a similar mechanism of integration into host cell genomes (11–14). Although the 3′ integration is definitely promoted by TPRT, the integration mechanism of the 5′ end of R2Bm is still uncertain. It is possible that additional factors may be required to accomplish the integration of R2Bm into the target site of the host cell genome.
To look for these factors, we constructed a novel in vivo system for analyzing the integration mechanism of R2Bm into a host cell genome. For the effective study of retrotransposition mechanism, it is necessary that the host cell genome contain no retrotransposons of the same type because the endogenous activity of these elements may supply factors not generated by the added element. From this standpoint, the cultured cell line, C65, is useful as a host for the introduction of R2Bm. The C65 cell line was established from embryos of Bombyx mori, and its genome contains no detectable insertion sequences for R2Bm or for R1Bm, which also inserts at a specific site in the 28S rRNA genes (Hashido et al., unpublished data). Thus, it should be possible to detect the insertion of introduced sequences by a simple PCR assay. The baculovirus, AcNPV (Autographa californica Nuclear Polyhedrosis Virus), was used as a vector for expressing the R2Bm protein in the C65 cells. AcNPV has the ability to infect a B.mori cell line, BmN (15), although it is speculated that this baculovirus cannot replicate in BmN because of differences in host range (16). Therefore, AcNPV may be a suitable vector to introduce R2Bm into C65 cells. AcNPV has also been reported to be utilized for the analysis of the integration mechanism of SART1, which is a non-LTR retrotransposon identified at a specific site in the telomeric repeats of the B.mori genome (17).
We demonstrate in this study that it is possible to integrate successfully the entire R2Bm retrotransposon into the host cell genome using this in vivo assay system. Because this system has the advantages of rapidity and facility to manipulate and to determine the integration mechanism, we expect that it can be applied to analyze the insertion of other retrotransposons.
MATERIALS AND METHODS
Animals and cells
R2Bm was amplified using PCR from genomic DNA extracted from B.mori strain, N4, which was a gift from Nagoya University and has been maintained in our laboratory.
The cultured cell line, C65, was used as a host for the introduction of R2Bm. The C65 cell line was derived from a single cell of the subculture line, l 30T (18), of NEN-Bm-l 30 (19), which was established from 10-day embryos of B.mori strain, w30.
Cloning of R2Bm
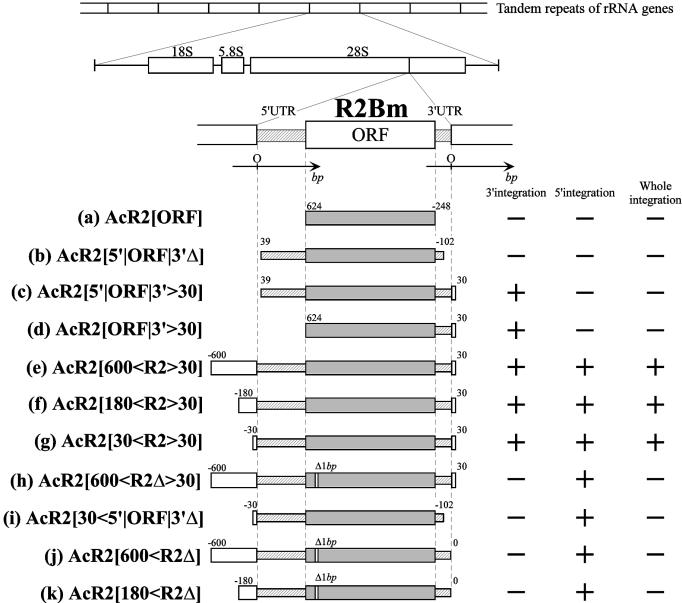
For cloning the R2Bm sequence, genomic DNA was prepared from posterior silk glands of fifth instar larvae (20). Regions that contained a portion of R2Bm were amplified using primers designed inside and outside of R2Bm, and PCR products were cloned into the plasmid, pAcYM1, which is a transfer vector for making recombinant AcNPV (21). Oligonucleotides used in this study as PCR primers are listed in the Supplementary Material, and regions introduced into recombinant AcNPV are shown in Figure 1. Each region is flanked by a set of primers: (a) AcR2[ORF]; ORF-s and ORF-as, (b) AcR2[5′|ORF|3′Δ]; A and B, (c) AcR2[5′|ORF|3′>30]; A and 06, (d) AcR2[ORF|3′>30]; ORF-s and 06, (e) AcR2[600<R2>30]; 011 and 06, (f) AcR2[180<R2>30]; 013 and 06, (g) AcR2[30<R2>30]; 03 and 06, (h) AcR2[600<R2Δ>30], 011 and 06, (i) AcR2[30<5′|ORF|3′Δ]; 03 and B, (j) AcR2[600<R2Δ]; 011 and 210, (k) AcR2[180<R2Δ]; 013 and 210. It was difficult to amplify the entire sequence of R2Bm using sequences flanking the target site (ex. 011 and 06) because the fragments lacking R2Bm were differentially amplified relative to fragments containing R2Bm (see Fig. 5A). To overcome this problem, we anchored the amplification reaction by using two primers both from within R2Bm and a sequence flanking the target site (ORF-s and 015, 017 and 027), and then nested PCR was performed using 011 and 06 as primers and a mixture of both purified products of the first PCR reaction as a template. The final PCR product was cloned into pAcYM1.
Figure 1.
Regions of R2Bm introduced into recombinant AcNPV and the integration of 3′ end, 5′ end and whole integration of R2Bm into the host cell genome. Letters indicating each construct [from (a) to (k)] correspond with those in subsequent figures. Numbers on the left/right of each region shown by open, striped and shaded bars indicate the position of the beginning/ending of the sequence in bases measured from the 5′ and 3′ end of R2Bm with the direction from 5′ to 3′ indicated as a plus. ‘Δ1bp’ denotes a 1 bp deletion inside the R2Bm ORF [(h), (j) and (k)]. In this study, these mutations, which could not produce a functional R2Bm protein because of a frame-shift, were used as controls for confirming the function of the R2Bm protein, although it was found that these constructs had other mutations (e.g. Fig. 6B; the region for 212Wt and 212Mu contains three replacements). Whether the integration of 3′ end, 5′ end and entire R2Bm was detected within cell genomes infected by each recombinant AcNPV was illustrated by ‘+’ (detected) and ‘–’ (not detected), respectively, on the right part of figure. The integration of 3′ and 5′ end was tested by PCR assay (see Fig. 2) and the whole integration was tested by the Southern hybridization performed by same means shown in Figure 5.
Figure 5.
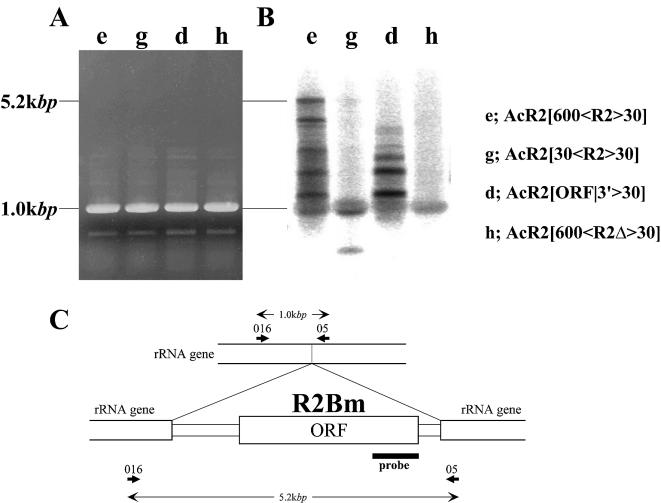
Detection of the integration of the complete sequence of R2Bm by Southern hybridization. (A) Image of an agarose gel of PCR performed using the cell genomes infected by variable recombinant AcNPVs as templates and the set of primers designed on the 28S rRNA genes that are diagrammed in (C). (B) The 5′ region of the R2Bm ORF that is diagrammed in (C) was labeled by 32P and hybridized with the PCR products shown in (A). (C) Locations of primers for PCR and the region of the probe used for the Southern hybridization. Numbers in the diagram indicate predicted sizes for successful integration of the entire R2Bm into the host cell genome (5.2 kb) or no integration (1.0 kb).
Sequences of all clones were determined using an ABI PRISM 310 Genetic Analyzer (PERKIN ELMER) (accession no. AB076841). The total length of R2Bm was 4185 bp, which included 596 bp of the 5′ UTR, 3345 bp of the single ORF and 245 bp of the 3′ UTR. A mutation that has a 1 bp deletion at the 200 bp from the beginning of the ORF was also found (see Fig. 6B). In this study, these mutations, which could not produce a functional R2Bm protein because of a frame-shift, were used as controls for confirming the function of the R2Bm protein.
Figure 6.
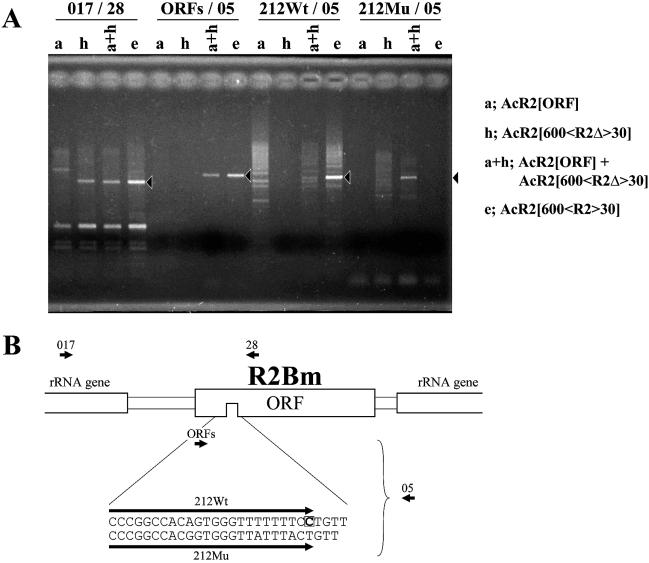
Rescue of the failure of frame-shifted R2Bm ORF to integrate by co-infection with the helper virus, AcR2[ORF]. (A) An image of an agarose gel of the PCR amplification products for the 5′ junction (017/28) and the 3′ junction (ORFs/05) between R2Bm and the 28S rRNA genes is shown. Genomes of C65 cells infected by AcR2[ORF] (a), AcR2[600<R2Δ>30] (h), AcR2[600<R2>30] (e) and co-infected by AcR2[ORF] and AcR2[600<R2Δ>30] (a + h) were used as templates for PCR. To check whether sequences were integrated into the host cell genome, primers were designed to include a deletion site located at the 3′ end of each construct and used for PCR (212Wt/05, 212Mu/05). Solid arrowheads indicate predicted sizes in each reaction. (B) Diagram of the location of each primer. Below the diagram, sequences of primers designed to include the mutation site are indicated. AcR2[600<R2Δ>30] has a 1 bp deletion indicated by an open boxed letter in 212Wt.
Construction of recombinant AcNPV and infection of C65 cultured cells
Recombinant AcNPVs were made inside sf9 cells in which virus DNAs and transfer vector were co-transfected. Sf9 cells at 5 × 105 cells/dish were cultured on 35 mm dish with 1 ml of the serum-free medium, Sf-900II (GIBCO-BRL). Both 4 µg of transfer vector, pAcYM1, that including R2Bm sequences and 1 µg of BD BaculoGold™ Baculovirus DNA (PharMingen) were mixed and added into sf9 cell culture with Lipofectin Reagent (Invitrogen). After incubation for 1 day at room temperature, TC-100 medium supplemented with 10% fetal bovine serum (filtron) was added. After 4 days the supernatant was collected and used for plaque purification and propagation of recombinant virus [details are described in (22)]. The virus solution of 100 µl (1 × 109 p.f.u./ml) was added to the C65 cell culture that was adjusted to 1 × 106 cells/dish. Under these conditions, the multiplicity of infection (m.o.i.) was estimated at 100. Templates for PCR were prepared from cultured cells 4 days after the infection according to the method of Funatsuki et al. (23). A solution of 3.0 µl including infected cells was used for the PCR.
Detection of the 3′ and 5′ integration of R2Bm
The 3′ and 5′ end integration were detected by PCR performed with genomic DNA extracted from C65 cells infected by recombinant AcNPV as a template. Primers used for the detection were 26 and 05 for the 3′ end integration and 016 and 27 for the 5′ end integration. Reactions were conducted under the following conditions: [PTC-200 Pelititer Thermal Cycler (MJ Reserch), GeneAmp PCR System 2400 (PERKIN ELMER)]; 94°C, 1 min denaturation, 60°C, 1 min annealing, 72°C, 3 min extension, for 30 cycles. Products were loaded onto 1% agarose gels and stained with ethidium bromide. When the 3′/5′ end of R2Bm is successfully integrated into the host cell genome, products of 1.2/2.4 kb should be observed, respectively. DNA fragments observed at the predicted sizes were excised from an agarose gel and purified using a QIAspin gel extraction kit (QIAGEN). Purified DNA fragments were cloned into a pGEM-T TA cloning vector (Promega) and sequenced using T7 and sp6 universal sequencing primers.
Southern hybridization
The integration of the entire sequence of R2Bm was detected by Southern hybridization. The PCR products, which were amplified using primers 016 and 05 with genomic DNA extracted from C65 cells infected by recombinant AcNPV as template were loaded onto a 1% agarose gel. After staining with ethidium bromide and photographing, the gel was soaked in denaturing solution (0.5M NaOH, 1.5M NaCl), and DNA was transferred onto a Hybond-N+ membrane (Amersham Pharmacia Biotech). After washing in distilled water, the membrane was soaked in hybridization buffer [5× SSPE (43.3 mM NaH2PO4, 750 mM NaCl, 6.3 mM EDTA), 5× Denhardt solution (0.1% Ficoll400, 0.1% polyvinylpyrrolidone, 0.1% BSA (bovine serum albumin)], 0.5% SDS (sodium dodecyl sulfate) and 5 mg salmon sperm DNA (Sigma) and incubated at 65°C for 2 h. The 5′ end of the R2Bm ORF (850 bp) was labeled by [α-32P]dCTP using a Random Primer DNA Labeling Kit Ver.2 (TaKaRa) and used for the probe (see Fig. 5C). The hybridization was carried out at 65°C for 12 h. The membrane was incubated in successive wash buffers [0.1× SSC (16.7 mM sodium citrate, 16.7 mM NaCl), 0.1% SDS and 2× SSC, 0.1% SDS]. For signal detection, the membrane was put in contact with an imaging plate for 2 h, and signals were visualized using an imaging plate reader [BAS2000 (Fuji film)].
RESULTS
The integration of R2Bm into the host cell genome by TPRT
The sequences introduced into recombinant AcNPV in this study are shown in Figure 1. After the infection of C65 cells by a recombinant virus, the genomic DNA of infected cells was extracted and employed as a template for PCR to amplify regions that included the 28S rRNA gene and the 3′ or 5′ end of R2Bm (Fig. 2).
Figure 2.
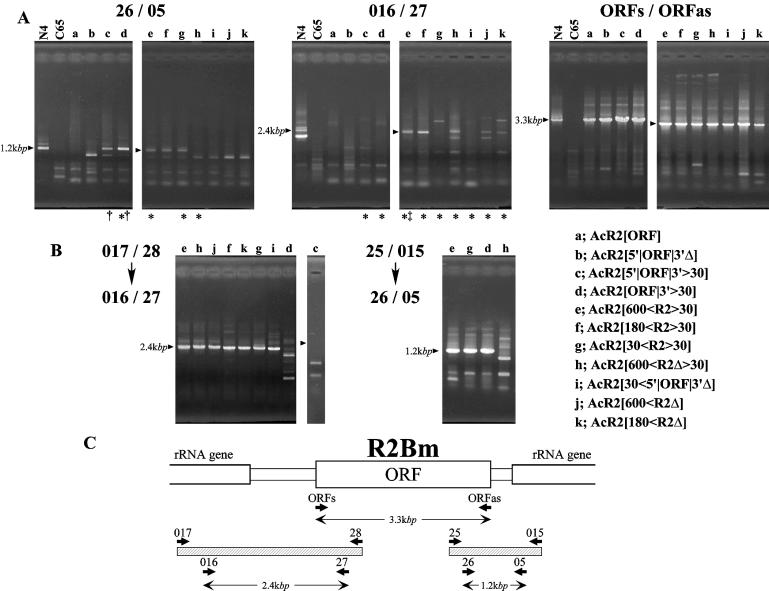
PCR and nested PCR assay of the 3′ and 5′ end integration of R2Bm into genomes of infected cells by various recombinant AcNPVs. (A) Images of ethidium bromide-stained agarose gels of each reaction. Each lane indicates the template for PCR; the genome of B.mori strain, N4, as a positive control, C65 cultured cells as a negative control and C65 cells that were infected by each recombinant AcNPV. The left and center sections in (A) show the products obtained with the combination of primers used to amplify the region including the junctions between the 3′ and 5′ ends of R2Bm and rRNA genes (26/05 and 016/27, respectively). The right section is the result of amplifying with primers inside the R2Bm ORF. The presence of PCR products means that infection by each recombinant AcNPV was successful. Lanes marked with asterisks indicate that the template of each reaction was used for nested PCR, shown in (B). PCR products of lanes marked with daggers and a double dagger were cloned and sequenced (see Figs 3 and 4, respectively). (B) The 5′ and 3′ end integration of R2Bm into rRNA genes were retested by nested PCR. In each reaction, the first PCR was performed using a combination of primers (017/28 and 25/015) and then the second PCR was carried out using a set of primers (16/27 and 26/05) to detect the 5′ and 3′ end integration, respectively. Note that PCR products were sometimes observed even if few products amplified by single PCR (compare A, 016/27, lanes g and i with B, 017/28 → 016/27, lanes g and i). (C) Location of primers and predicted sizes of each PCR product.
As the first step, we tested whether R2Bm could insert into the host cell genome by itself. PCR products with the predicted size of 1.2 kb (primer pair 26 and 05, Fig. 2C) were observed when the introduced sequence contained the entire ORF and 3′ UTR (Fig. 2A, lanes c–g). When sequences contained a frame-shifted ORF (Fig. 2A, lanes h, j and k), PCR products with the size of 1.2 kb were not observed, suggesting that a functional R2Bm protein might not be translated. Similarly, no PCR products were observed when an introduced sequence contained only a part of the 3′ UTR (Fig. 2A, lanes b and i) or did not contain the 3′ UTR (Fig. 2A, lane a). PCR products purified from the amplification using genomic DNA from cells infected by AcR2[5′|ORF|3′>30] and AcR2[ORF|3′>30] (Fig. 1) were cloned and sequenced (Fig. 3). All of the investigated clones had a consecutive sequence carrying both the region introduced into the recombinant AcNPV and the downstream region of the R2Bm target site in the 28S rRNA gene, showing that the 3′ end of R2Bm inserted into the host cell genome successfully. Thus, an intact ORF and the full-length of the 3′ UTR are required for the 3′ end integration of R2Bm. These findings were consistent with the results of the in vitro analysis reported previously (5). One of 12 clones had four extra bases (AAGG) at the junction between R2Bm and the rRNA gene (Fig. 3). Such additional sequences have also been reported previously (5,10,24).
Figure 3.
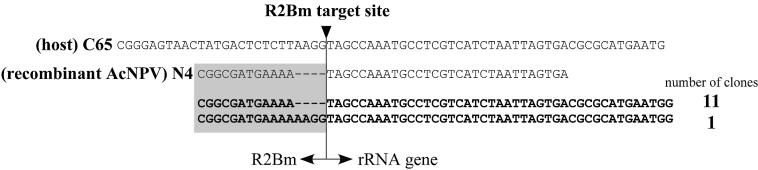
Confirmation of the 3′ end integration by sequencing the region of the 3′ junction between R2Bm and the host cell genome. The sequence around the boundary of the C65 28S rRNA gene is shown on the top of the diagram [(host) C65]. It was confirmed that this region is completely conserved in the N4 genome. The second sequence indicates the common region between sequences introduced into recombinant AcNPVs derived from the N4 strain [(rcAcNPV) N4] and the sequence of the 28S rRNA genes that was derived from C65 cells. Sequences below the diagram are those determined from C65 cell genomes infected by AcR2[5′|ORF|3′>30] and AcR2[ORF|3′>30]. Sequences derived from R2Bm were shown in the shaded box. One of 12 clones had four extra bases at the junction between R2Bm and the rRNA gene.
In contrast, no PCR products with the predicted size of 2.4 kb using primers, 016 and 27 (Fig. 2C), corresponding to the 5′ end of the insertion, were observed when the introduced sequence contained only R2Bm sequences (Fig. 2A, lanes a and b, primers 016 and 27) or R2Bm with 30 bp of the 28S rRNA gene downstream of the R2Bm target site (Fig. 2A, lanes c and d on 016/27). These results suggested that R2Bm does not have the ability to integrate its 5′ end in this assay.
An incorrect insertion mechanism of R2Bm is supplied by the repair system of host cells
According to a report of Eickbush et al., when the R2Bm protein and an RNA containing 28S rRNA sequences upstream of the R2Bm target site in addition to both the 5′ UTR and 3′ UTR of R2Bm were injected into embryos of Drosophila melanogaster, cDNA reverse transcribed from the injected RNA can be integrated into the target site in the host genome using the repair system of the host cells (24). This suggested that it might be possible for the entire R2Bm to insert successfully into the silkworm genome in our assay if we included upstream 28S rRNA sequences.
To make the 5′ end of R2Bm integrate into host cell genome, sequences homologous to the 28S rRNA genes upstream of the R2Bm target site were added upstream of the introduced sequences (Fig. 1, from e to k). PCR products with the size (2.4 kb) that was predicted for successful integration of the 5′ end of R2Bm into the host cell genome were observed when the introduced sequences contained 600 or 180 bp upstream 28S rRNA gene sequences (Fig. 2A, lanes e, f, h, j and k of the middle panel).
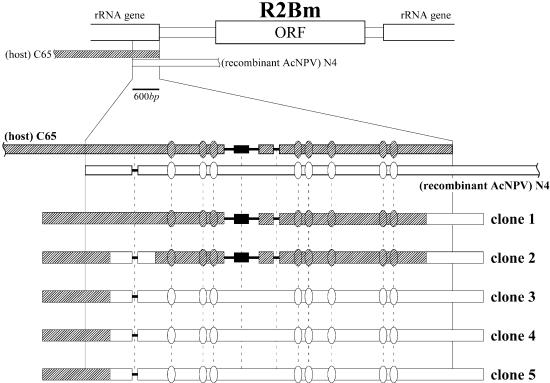
PCR products purified with the construct, AcR2[600<R2>30], were cloned and sequenced (Fig. 4). All of the sequenced clones had a consecutive sequence with both the region introduced into the recombinant AcNPV and the 28S rRNA genes upstream of the introduced sequence, indicating that the 5′ end of R2Bm integrated successfully into the host cell genome. There were three gaps and eight replacements in the overlapping region (600 bp) between the sequence introduced into recombinant AcNPV that was cloned from the genome of the N4 strain and the sequence of the C65 host cells. Three of five clones had N4 type sequences in this region (Fig. 4, clones 3–5); clone 1 had a C65 type sequence. In the case of clone 2, the position of the first gap was an N4 type, while the other gaps and replacements were C65 type. The variation of positions in which the two sequences exchanged with one another suggested that the 5′ end insertion of R2Bm is carried out by homologous recombination between the C65 genome and the R2Bm cDNA. Such homologous recombination was also observed in cells of D.melanogaster (24).
Figure 4.
5′ End integration after infection with a construct containing a sequence homologous to the 28S rRNA gene upstream of the R2Bm target site. PCR products amplified from the C65 cell genome infected by AcR2[600<R2>30], which contains 600-bp of a sequence homologous to the 28S rRNA gene upstream of the R2Bm target site (clones 1–5) are shown. Three gaps and eight replacements were found in the overlapping region between the genome of C65 cells [(Host) C65] and the genome of recombinant AcNPV that was derived from the N4 strain [(rcAcNPV) N4]; gaps and replacements are illustrated with solid lines and ovals, respectively. The fragment inserted into the second gap is indicated with a solid box. The sequence presumably derived from the C65 cell genome is diagrammed with a diagonally shaded box and the sequence presumably derived from the sequence introduced into AcR2[600<R2>30] is diagrammed with an open box. Crossing-over is presumed to have occurred within regions where sequences derived from host cells and AcR2[600<R2>30] exchanged with each other.
Products that had the same size observed when the 5′ end of R2Bm integrated into the host cell genome were sometimes detected at low levels when we repeated the PCR amplification using genomic DNA extracted from cells infected by AcR2[30<R2>30] and AcR2[30<5′|ORF|3′Δ]. In the sequence introduced into these recombinant AcNPVs, 30 bp of the 28S rRNA genes upstream of the R2Bm target site were added to the R2Bm sequences. It seems that the 5′ end of R2Bm may insert into the host genome at a low frequency even if only 30-bp of the upstream region of the R2Bm target site are added. The result of nested PCR supports this possibility (Fig. 2B, left). When genomic DNA extracted from cells infected with AcR2[600<R2>30], AcR2[180<R2>30], AcR2[30<R2>30] and AcR2[30<5′|ORF|3′Δ], which contained variable length regions of the host genome, were employed as templates, products with the size (2.4 kbp) predicted for successful integration of the 5′ end of R2Bm were amplified (Fig. 2B, left; lanes e–g and i), whereas, products with such a predicted size were not amplified when genomic DNA extracted from cells infected by AcR2[ORF|3′>30] and AcR2[5′|ORF|3′>30], which did not contain a homologous sequence, was employed as a template (Fig. 2B, left; lanes d and c). These results suggested that homologous recombination occurred between the C65 genome and a cDNA of R2Bm even if only 30 bp of the upstream region of the R2Bm target site are added to the R2Bm sequence.
These findings raised the question of whether integration of the 3′ end of R2Bm into the host cell genome was also brought about by recombination, since some constructs contained 30 bp of downstream 28S gene sequences. We tested this possibility using nested PCR (Fig. 2B, right). When genomic DNA extracted from cells infected by AcR2[600<R2Δ>30], which contained 30 bp of the downstream region of the R2Bm target site and a frame-shifted ORF, were employed as templates, products with the size (1.2 kb) predicted for the integration of the 3′ end of R2Bm were not detected (Fig. 2B, right; lane h); whereas, products with the predicted size were observed when genomic DNA extracted from cells infected by AcR2[600<R2>30], AcR2[30<R2>30] and AcR2[ORF|3′> 30], which contained both 30 bp of the downstream regions of the R2Bm target site and the whole R2Bm ORF, were employed as templates (Fig. 2B, right; lanes e, g and d). These results suggest that, in this study, homologous recombination would occur only when the 5′ end of R2Bm integrates into the host cell genome and that the 3′ end integration of R2Bm could be caused by TPRT mechanism.
It should be noted that DNA extracted from cells infected by AcR2[600<R2Δ>30], AcR2[600<R2Δ] or AcR2[180<R2Δ], which contained 180 or 600 bp upstream of 28S sequence and did not have complete ORFs, did give rise to the amplification of products with predicted sizes for the 5′ ends (Fig. 2B, left; lanes h, j and k). This finding suggests that the 5′ integration into host cell genome may occur when constructs that could not produce a functional R2Bm protein because of a frame-shift inside R2Bm ORF were introduced. In these cases, it is supposed that the introduced sequence into recombinant virus would be directly recombined with the host cell genome, because the baculovirus has been reported possible to utilize for the gene targeting by homologous recombination (25).
The fact that the integration of the 5′ end of R2Bm had occurred when the integration of the 3′ end of R2Bm was not observed (compare lane h in each panel) indicates that the two events occurred independently. Therefore, detecting the integration of either the 3′ end or the 5′ end of R2Bm independently would not confirm the complete integration of an intact R2Bm.
The detection of the complete integration of R2Bm
It is difficult to detect the complete integration of R2Bm by PCR using primers designed outside R2Bm (016 and 05 in Fig. 5) because 5.2 kb products are out competed by the 1.0 kb products generated from unoccupied sites. To demonstrate that the entire R2Bm integrated into the host cell genome in this in vivo system, the 3′ region of the R2Bm ORF was used as a probe to hybridize the PCR products using as templates genomic DNA extracted from cells infected by AcR2[600<R2>30], AcR2[30<R2>30], AcR2[ORF|3′>30] and AcR2[600<R2Δ>30] (Fig. 5). When an introduced sequence contained the entire R2Bm with the 600 bp of sequence homologous to the 28S rRNA genes upstream of the R2Bm target site, a DNA band of 5.2 kb predicted for integration of the entire R2Bm into the host cell genome was detected (Fig. 5B, lane e). A weak signal of this size was also detected when the length of the upstream 28S sequences was 30 bp (Fig. 5B, lane g). On the other hand, no signals were observed at the same size when an introduced sequence lacked the upstream region of the R2Bm target site (Fig. 5B, lane d) or contained a frame-shifted ORF (Fig. 5B, lane h). Thus, an entire R2Bm was inserted into the host cell genome only when the entire R2Bm element together with the sequence homologous to the 28S rRNA genes upstream of the R2Bm target site was introduced into the cultured cells.
Many extra signals were detected in the size range between 5.2 and 1.0 kb (Fig. 5B, lanes e and d). This finding suggested that the insertion mechanism of R2Bm is not perfect, and that truncation at the 5′ end has often occurred. Moreover, the fact that these signals were not smeared but laddered indicated that there are several hot spots where truncation tended to occur. It appeared that not only the 5′ end of R2Bm but also the 28S rRNA gene upstream of the R2Bm target site could be truncated because extra signals were also present below 1.0 kb, although no R2Bm could be contained in this region.
Rescue of the frame-shifted mutation by co-infection with a helper virus
Direct evidence that the R2Bm protein catalyzes the integration of R2Bm into the host cell genome would be obtained if the frame-shifted mutation were rescued by co-infection with the helper virus, AcR2[ORF], which contained only the R2Bm ORF. Subsequently, AcR2[600<R2Δ>30] (Fig. 1, h), which contains 600 bp of the sequence homologous with the 28S rRNA genes upstream of the R2Bm target site and the complete R2Bm sequence with a frame-shifted ORF was co-infected into C65 cells with AcR2[ORF] (Figs 1, a and 6). The 3′ end of R2Bm could not integrate into the host cell genome when both recombinant viruses were used to infect host cells independently (Fig. 6A, lanes a and h under ORFs/05). On the other hand, both the 5′ end and the 3′ end of R2Bm were integrated into the R2Bm target site when the two recombinant viruses were co-infected (Fig. 6A, lanes a + h under 017/28 and ORFs/05). Complete integration of R2Bm was confirmed using the Southern hybridization performed by the same means shown in Figure 5 (data not shown).
These results suggested that the R2Bm protein that can be translated from AcR2[ORF] may act in trans on the R2Bm RNA transcribed from AcR2[600<R2Δ>30]. If this interpretation is correct, then the integrated sequence should have a 1 bp deletion within the coding region of R2Bm. To test this prediction, PCR was performed using forward primers whose 3′ ends were designed at the deletion site (Fig. 6A, lanes 212Wt/05 and 212Mu/05). Compared with the situation when the template DNA was extracted from cells infected by AcR2[600<R2>30] (Fig. 6A, lane e in each column), PCR products that were predicted for integration of the sequence with a 1 bp deletion were observed when both recombinant viruses were used to co-infect (Fig. 6A, lanes a + h on the column 212Wt/05 and 212Mu/05). A product with the same size was not observed when these recombinant viruses were used to infect host cells independently (Fig. 6A, lanes a and h under 212Wt/05 and 212Mu/05). This suggested that the sequence with a 1 bp deletion in the coding region of the R2Bm protein was integrated into the R2Bm target site only when both recombinant viruses were used to co-infect. Therefore, we concluded that the R2Bm protein catalyzes the 3′ end integration of R2Bm into the host cell genome in this in vivo system.
DISCUSSION
Whether the 3′ end, 5′ end and entire sequence of R2Bm were integrated correctly into C65 cell genome in our assay was summarized in Figure 1. Both the full-length 3′ UTR and the complete ORF of R2Bm are necessary for the 3′ end integration into the R2Bm target site in 28S rRNA genes (Fig. 1, c–g), suggesting that the 3′ end of R2Bm integrates by TPRT catalyzed by the R2Bm protein and the R2Bm RNA. However, in this assay, R2Bm does not have the ability to integrate its 5′ end correctly and adding the 28S gene sequences upstream of the R2Bm target site to the construct enables the 5′ end of R2Bm to be integrated into host genome (Fig. 1, e–k). In such cases, the 5′ end integration into the host cell genome seems to be a result of homologous recombination using the function of the DNA repair system of the host cells. And only when all conditions described above, i.e. full-length R2Bm with homologous sequence, are cleared, whole sequence of R2Bm are integrated into host cell genome (Fig. 1, e–g).
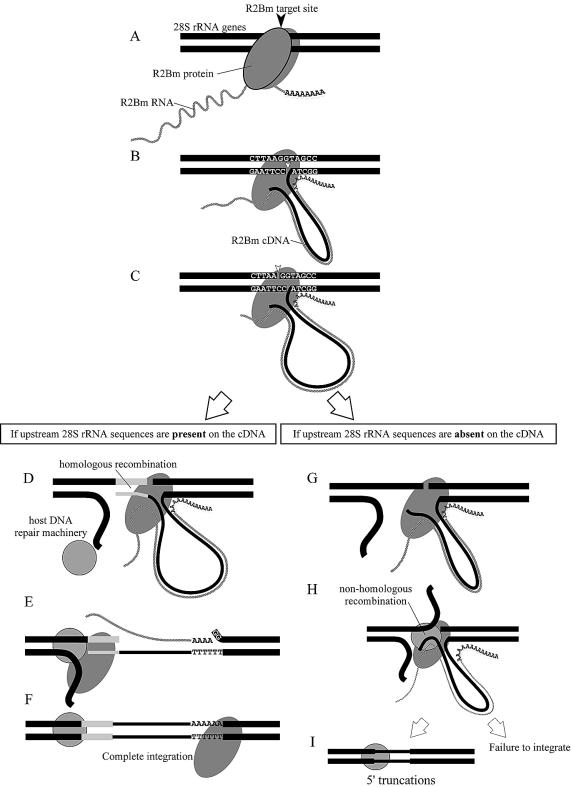
The insertion mechanism of R2Bm into a host cell genome deduced from the result of our assay is modeled in Figure 7. After the R2Bm cDNA is synthesized by TPRT (Fig. 7A–C), the 5′ end of R2Bm may integrate by homologous recombination if the R2Bm cDNA has a sequence homologous to the 28S rRNA genes upstream of the R2Bm target site (Fig. 7D–F). On the other hand, if the R2Bm cDNA does not have such a sequence homologous to the 28S rRNA genes, a non-homologous recombination occurs between the R2Bm cDNA and the upstream region of the R2Bm target site (Fig. 7G–I) with truncations both of the 5′ end of R2Bm and the 28S rRNA genes upstream of the R2Bm target site, or there may be a case that its 5′ end cannot be integrated into a host cell genome. Truncations on the 5′ end of R2Bm are observed for R2 elements in the genus Drosophila (26,27) and this model of the 5′ integration mechanism almost corresponds with one proposed in the previous report using a Drosophila assay system (24). We believe that the model deduced with an alternative approach would strongly support results of those serial previous works.
Figure 7.
Model for the integration mechanism of R2Bm into the host cell genome. The 3′ end integration of R2Bm into rRNA genes is accomplished by an interaction between the host cell genome and a complex of the R2Bm RNA and the R2Bm protein by means of TPRT (A–C). In contrast, the 5′ end integration of R2Bm may be carried out by homologous recombination when the R2Bm cDNA contains a sequence homologous with the 28S rRNA genes upstream of the R2Bm target site (D–F). When the R2Bm cDNA does not have such a homologous sequence with 28S rRNA genes, a non-homologous recombination must occur between the R2Bm cDNA and the upstream region of the R2Bm target site (G–I), or there may be a case that its 5′ end cannot be integrated into a host cell genome. In a case that a non-homologous recombination has occurred, not only the 5′ end of R2Bm but also the 28S rRNA genes upstream of the R2Bm target site could be truncated.
In this study, the homologous recombination at the upstream region of the R2Bm target site occurs whether or not the introduced sequences carried a complete ORF (Fig. 2B, left). This suggests that the recombination occurred between the genome of the cultured cell and the viral DNA when an introduced sequence contained a frame-shifted ORF. However, it is difficult to explain why homologous recombination did not occur in the region downstream of the R2Bm target site, but did occur at the upstream of the R2Bm target site even when both homologous sequences had the same lengths (30 bp) (compare Fig. 2B, left; lane g with right, lane h).
How does the endogenous R2Bm carry out retrotransposition in the cells of the host organisms? In this study, we demonstrated one of the possibilities to enable R2Bm to integrate successfully into a host cell genome. If the endogenous R2Bm has the ability to transpose by means of the model shown in Figure 7, RNAs containing both the entire R2Bm flanking rRNA sequences are necessary for successful retrotransposition. Such RNAs may be transcribed only when a 28S region of rRNA genes is read through with an R2Bm sequence. The lack of a promoter in the 5′ UTR of Drosophila R2 elements supports this hypothesis (28). But thus far, no such RNAs containing an R2Bm sequence have been found in the EST database of the silkworm constructed from cDNA libraries for several strains, tissues and developmental stages (details described in Silkbase; http://samia.ab.a.u-tokyo.ac.jp/silkbase/). Moreover, RNA transcribed from an endogenous R2Bm has not been found by northern blot analysis (24). These results suggest that the frequency of transcription might be at a very low level, or that the period when R2Bm is transcribed might be different from the stages from which the cDNA libraries were constructed.
R2 elements are widespread among the genomes of arthropods (29), and the domain structure and the copy number in the rRNA gene cluster is highly conserved in the genome of each strain (30–32). Thus, we suppose that it would be difficult to find some direct evidences of the retrotransposition of the R2 element. Recently, we succeeded in visualizing the approximate location of R2Bm inside the rRNA gene cluster using fluorescence in situ hybridization (Ejiri et al., in preparation). We suppose it might be possible to find direct evidence of the retrotransposition in the cell genome by tracing the location of R2Bm sequences in the rRNA gene cluster for several generations using the FISH analysis after introducing R2Bm into the C65 cell genome, which is a virgin from R2Bm, using the in vivo system demonstrated in this study.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We greatly thank Dr M. R. Goldsmith for critical reading and thoughtful advice. We also truly thank Dr T. H. Eickbush for useful comments. This research was partly supported by Nuclear Energy Infrastructure Crossover Research Project and grants of Ministry of Education, Culture, Sports, Science and Technology.
DDBJ/EMBL/GenBank accession no. AB076841
REFERENCES
- 1.Eickbush T.H. and Robins,B. (1985) Bombyx mori 28S ribosomal genes contain insertion elements similar to the Type I and II elements of Drosophila melanogaster. EMBO J., 4, 2281–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujiwara H., Ogura,T., Takada,N., Miyajima,N., Ishikawa,H. and Maekawa,H. (1984) Introns and their flanking sequence of Bombyx mori rDNA. Nucleic Acids Res., 12, 6861–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke W.D., Calalang,C.C. and Eickbush,T.H. (1987) The site-specific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol. Cell. Biol., 7, 2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Y.E. and Eickbush,T.H. (1988) Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell, 55, 235–246. [DOI] [PubMed] [Google Scholar]
- 5.Luan D.D., Korman,M.H., Jakubczak,J.L. and Eickbush,T.H. (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell, 72, 595–605. [DOI] [PubMed] [Google Scholar]
- 6.Yang J., Malik,H.S. and Eickbush,T.H. (1999) Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl Acad. Sci. USA, 96, 7847–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathews D.H., Banerjee,A.R., Luan,D.D., Eickbush,T.H. and Turner,D.H. (1997) Secondary structure model of the RNA recognized by the reverse transcriptase from the R2 retrotransposable element. RNA, 3, 1–16. [PMC free article] [PubMed] [Google Scholar]
- 8.Luan D.D. and Eickbush,T.H. (1996) Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol. Cell. Biol., 16, 4726–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J. and Eickbush,T.H. (1998) RNA-induced changes in the activity of the endonuclease encoded by the R2 retrotransposable element. Mol. Cell. Biol., 18, 3455–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luan D.D. and Eickbush,T.H. (1995) RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol., 15, 3882–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anzai T., Takahashi,H. and Fujiwara,H. (2001) Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)(n) by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol. Cell. Biol., 21, 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen S., Pont-Kingdon,G. and Carroll,D. (2000) Target specificity of the endonuclease from the Xenopus laevis non-long terminal repeat retrotransposon, Tx1L. Mol. Cell. Biol., 20, 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prak E.T. and Kazazian,H.H.,Jr (2000) Mobile elements and the human genome. Nat. Rev. Genet., 1, 134–144. [DOI] [PubMed] [Google Scholar]
- 14.Zimmerly S., Guo,H., Perlman,P.S. and Lambowitz,A.M. (1995) Group II intron mobility occurs by target DNA-primed reverse transcription. Cell, 82, 545–554. [DOI] [PubMed] [Google Scholar]
- 15.Volkman L.E. and Goldsmith,P.A. (1982) Generalized immunoassay for Autographa californica nuclear polyhedrosis virus infectivity in vitro. Appl. Environ. Microbiol., 44, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo A. and Maeda,S. (1991) Host range expansion by recombination of the baculoviruses Bombyx mori nuclear polyhedrosis virus and Autographa californica nuclear polyhedrosis virus. J. Virol., 65, 3625–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi H. and Fujiwara,H. (2002) Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J., 21, 408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okano K., Miyajima,N., Takada,N., Kobayashi,M. and Maekawa,H. (1992) Basic conditions for the drug selection and transient gene expression in the cultured cell line of Bombyx mori. In Vitro Cell. Dev. Biol., 28A, 779–781. [DOI] [PubMed] [Google Scholar]
- 19.Ninaki O., Takada,N., Fujiwara,H., Ogura,T., Miyajima,N., Kiyota,A. and Maekawa,H. (1989) Gene analysis by blot hybridization on the silkworm, Bombyx mori, cell lines. J. Inverteb. Cell Sys. Appl., 1, 143–149. [Google Scholar]
- 20.Suzuki Y., Gage,L.P. and Brown,D.D. (1972) The genes for silk fibroin in Bombyx mori.J. Mol. Biol., 70, 637–656. [DOI] [PubMed] [Google Scholar]
- 21.Matsuura Y., Possee,R.D., Overton,H.A. and Bishop,D.H. (1987) Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J. Gen. Virol., 68, 1233–1250. [DOI] [PubMed] [Google Scholar]
- 22.Christopher D.R. (ed.) (1995) Methods in Molecular Biology Volume 39: Baculovirus Expression Protocols. Humana Press Inc., Totowa, NJ. [Google Scholar]
- 23.Funatsuki K., Hashido,K., Matsunami,M., Kameoka,Y., Iwabuchi,K., Tsukeda,H., Tsuchida,K., Takada,N., Nakajima,Y. and Maekawa,H. (2001) Rapid indentification of Bombyx mori cells using PCR amplification following a direct procedure for genomic DNA preparation. J. Insect Biotech. Seric., 70, 129–136. [Google Scholar]
- 24.Eickbush D.G., Luan,D.D. and Eickbush,T.H. (2000) Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster. Mol. Cell. Biol., 20, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamao M., Katayama,N., Nakazawa,H., Yamakawa,M., Hayashi,Y., Hara,S., Kamei,K. and Mori,H. (1999) Gene targeting in the silkworm by use of a baculovirus. Genes Dev., 13, 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George J.A., Burke,W.D. and Eickbush,T.H. (1996) Analysis of the 5′ junctions of R2 insertions with the 28S gene: implications for non-LTR retrotransposition. Genetics, 142, 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Gonzalez C.E. and Eickbush,T.H. (2001) Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics, 158, 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George J.A. and Eickbush,T.H. (1999) Conserved features at the 5 end of Drosophila R2 retrotransposable elements: implications for transcription and translation. Insect Mol. Biol., 8, 3–10. [DOI] [PubMed] [Google Scholar]
- 29.Jakubczak J.L., Burke,W.D. and Eickbush,T.H. (1991) Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc. Natl Acad. Sci. USA, 88, 3295–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burke W.D., Malik,H.S., Jones,J.P. and Eickbush,T.H. (1999) The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol., 16, 502–511. [DOI] [PubMed] [Google Scholar]
- 31.Jakubczak J.L., Xiong,Y. and Eickbush,T.H. (1990) Type I (R1) and type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J. Mol. Biol., 212, 37–52. [DOI] [PubMed] [Google Scholar]
- 32.Maekawa H., Takada,N., Mikitani,K., Ogura,T., Miyajima,N., Fujiwara,H., Kobayashi,M. and Ninaki,O. (1988) Nucleolus organizers in the silkworm, Bombyx mandarina and the domesticated silkworm B. mori. Chromosoma, 96, 263–269. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.