Abstract
The Hop2–Mnd1 complex functions with the DMC1 recombinase in meiotic recombination. Hop2–Mnd1 stabilizes the DMC1-single-stranded DNA (ssDNA) filament and promotes the capture of the double-stranded DNA partner by the recombinase filament to assemble the synaptic complex. Herein, we define the action mechanism of Hop2–Mnd1 in DMC1-mediated recombination. Small angle X-ray scattering analysis and electron microscopy reveal that the heterodimeric Hop2–Mnd1 is a V-shaped molecule. We show that the protein complex harbors three distinct DNA binding sites, and determine their functional relevance. Specifically, the N-terminal double-stranded DNA binding functions of Hop2 and Mnd1 co-operate to mediate synaptic complex assembly, whereas ssDNA binding by the Hop2 C-terminus helps stabilize the DMC1-ssDNA filament. A model of the Hop2-Mnd1-DMC1-ssDNA ensemble is proposed to explain how it mediates homologous DNA pairing in meiotic recombination.
INTRODUCTION
During meiosis, homologous chromosomes undergo recombination that is critical for their proper segregation in the first cell division (1–3). Besides its meiotic role, homologous recombination (HR) serves as a major tool for chromosome damage repair and is also relevant for telomere maintenance (4,5). The vast majority of eukaryotes, including humans, possess two recombinases, namely, RAD51 and DMC1. These recombinases co-operate with their cohort of accessory factors to mediate the HR reaction (5,6). Although RAD51 functions in both mitotic and meiotic cells, DMC1 is expressed only during meiosis (3,7).
HR is often initiated by DNA double-strand breaks (DSBs), either in a programmed manner involving the meiosis-specific Spo11 protein complex (8) or in an unscheduled manner, mounted in response to the exposure of cells to DSB causative agents, such as ionizing radiation (5,6). For HR to occur, the 5' ends of the DSBs must first be resected to yield 3' single-stranded DNA (ssDNA) tails (9–12). The RAD51 or DMC1 recombinase then polymerizes on these ssDNA tails, in an ATP-dependent manner, to form a highly ordered helical polymer known as the presynaptic filament (5,6). The presynaptic filament engages and searches for homology in dsDNA, leading to the assembly of an ensemble termed the synaptic complex, in which the recombining DNA molecules are aligned in homologous registry (5,13). The assembly of the synaptic complex thus represents an obligatory step during HR (5).
In Saccharomyces cerevisiae, the HOP2 and MND1 genes are indispensable for meiotic HR. In hop2 and mnd1 mutants, meiosis arrests at prophase, DSBs are not repaired and chromosomes are more frequently synapsed with a non-homologous counterpart (14–19). Similarly, ablation of the Hop2 gene in the mouse and of either the Hop2 or Mnd1 gene in Arabidopsis thaliana renders meiotic DSB repair defective (20–24). The Hop2 and Mnd1 proteins form a complex (15,17,19,25–28) that binds ssDNA and dsDNA (28–30). Importantly, the mammalian Hop2–Mnd1 complex interacts with RAD51 and DMC1 and greatly enhances their ability to mediate homologous pairing between the recombining DNA molecules to form the D-loop (26,27,29). However, Hop2–Mnd1 from S. cerevisiae and Saccharomyces pombe appears to functionally interact with DMC1 only (25,29). Hop2–Mnd1 acts at two steps during the D-loop reaction, by stabilizing the recombinase presynaptic filament and synergizing with the presynaptic filament to assemble the synaptic complex (30,31). However, the detailed molecular mechanism of Hop2–Mnd1 in synaptic complex formation remains unknown. Herein, we describe biochemical, biophysical, mutagenic and structural analyses that allow us to delineate the action mechanism of the Hop2–Mnd1 in DMC1-mediated homologous DNA pairing during meiosis.
MATERIALS AND METHODS
Protein expression plasmids
The bacterial expression plasmids pRSF-(His)6-Hop2 and pRSF-(His)6-Hop2144−217 were constructed by introducing the relevant Hop2 cDNAs harboring an N-terminal (His)6-tag into the pRSF-duet vector. Plasmids pRSF-(His)6-Hop21−84 and pRSF-(His)6-Hop21−125 were generated by inserting a stop codon into the Hop2 coding region of pRSF-(His)6-Hop2. pRSF-(His)6-Hop2-Mnd1 and pRSF-(His)6-Hop2126−217-Mnd1 were made by inserting the Mnd1 cDNA into pRSF-(His)6-Hop2 and pRSF-(His)6-Hop2126−217, respectively. pRSF-(His)6-Hop2-Mnd1100−205 was constructed by introducing the cDNA coding for Mnd1100−205 into pRSF-(His)6-Hop2. Maltose Binding Protein (MBP) and Flag tags were introduced into pRSF-(His)6-Hop2-Mnd1 to yield pRSF-MBP-(His)6-Hop2-Mnd1-Flag [encoding N-terminally MBP-(His)6-tagged Hop2 and C-terminally Flag-tagged Mnd1], pRSF-(His)6-Hop2-MBP-Mnd1 [encoding N-terminally (His)6-tagged Hop2 and N-terminally MBP-tagged Mnd1] or pRSF-MBP-(His)6-Hop2-MBP-Mnd1-Flag [encoding N-terminally MBP-(His)6-tagged Hop2 and N-terminally MBP-tagged and C-terminally Flag-tagged Mnd1]. The protein expression plasmids pET15b-Hop21−143-Mnd1 and pET15b-Hop2-Mnd11−113 have been described (28). Construction of mutants was done using the Quickchange mutagenesis kit (Stratagene).
Protein purification procedures
Mouse Hop2, Hop2–Mnd1 and their mutant derivatives were expressed in Escherichia coli and purified as described (28,30). The fragments of Hop2 were expressed in E. coli and purified using the same chromatographic steps as for full-length Hop2. Hop21−125 and Hop2144−217 elute from SP Sepharose at 150–250 and 75–150 mM KCl, respectively, from hydroxyapatite at 180–230 and 100–130 mM KH2PO4, respectively, and from Mono S at 200–230 and 100–120 mM KCl, respectively. Hop21−84, purified without the hydroxyapatite step, elutes from SP Sepharose at 100–200 mM KCl and from Mono S at 120–160 mM KCl. Singly and doubly MBP-tagged Hop2–Mnd1 species were expressed in E. coli and purified as for Hop2–Mnd1 but with an additional affinity step using Amylose resin [New England Biolabs; for Hop2-(MBP–Mnd1)] or anti-Flag resin [Sigma-Aldrich; for (MBP–Hop2)-Mnd1 and (MBP–Hop2)-(MBP–Mnd1)] following the protocols provided by the manufacturers. Human DMC1 was purified from insect cells infected with a recombinant DMC1 baculovirus, as described (32).
Light scattering analysis of Hop2–Mnd1
Light scattering studies were performed with a micro volume size-exclusion chromatographic system (Ettan LC, GE Healthcare) connected inline to an 18-angle light scattering and refractive index detector (Wyatt Technology, Santa Barbara, CA, USA), where detector 12 was modified for quasi-elastic light scattering detection. The sample (in 50 µl) was purified using a 2.4 ml Superdex 75 PC 3.2 column eluted at 0.04 ml/min in buffer A (40 mM Na2HPO4, pH 7.5, 10% glycerol, 150 mM NaCl and 1 mM 2-mercaptoethanol). Band broadening and detector calibration were performed with 10 mg/ml xylanase (Hampton Research) using a refractive index increment of 0.178. The light scattering experiments were performed in buffer A.
SAXS data collection and evaluation
Small-angle X-ray scattering (SAXS) data were collected at the ALS beam line 12.3.1 (SIBYLS) at the Lawrence Berkeley National Laboratory (33,34). The wavelength λ = 1.0 Å and sample-to-detector distances were set to 1.5 m resulting in scattering vectors, q, ranging from 0.01 Å−1 to 0.32 Å−1. The scattering vector is defined as q = 4π sinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20°C, and data were processed as described (33). Briefly, the data acquired at short and long time exposure (0.5 s, 5 s) were merged for calculations using the entire scattering spectrum. The experimental SAXS data for different protein concentrations were judged to be free of aggregation by using Guinier plots (35). The radius of gyration, Rg, was derived by the Guinier approximation I (q) = I (0) exp (−q2Rg2/3) with the limits qRg < 1.3. The program GNOM (36) was used to compute the P(r) function, which is the pair-distance distribution function that is used in SAXS to describe the paired-set of distances between all of the electrons within a macromolecular structure. This approach also provided the maximum dimension of the macromolecule, Dmax. The overall shapes for Hop2–Mnd1 and Hop21−84 were restored using the program DAMMIF (37). The program MONSA (38) was used to locate MBP tags relatively to the Hop2–Mnd1 complex. MONSA is multiphase bead modeling that allows the simultaneous fitting of multiple SAXS curves. The program represents the particle as a collection of densely packed beads inside a sphere with the diameter Dmax. To describe the overall and internal structure of the complex particle, each bead can be assigned to the phases, either to the Hop2–Mnd1 or to one of the MBP tags. The singly MBP-tagged constructs are represented by two phases, and the doubly MBP-tagged construct is represented by three phases (Hop2–Mnd1 for phase 1, MBP for phase 2 and second MBP for phase 3). The volumes of the Hop2–Mnd1 V = 115 000 Å3 and MBP V = 65 000 Å3 have been determined from SAXS curves by the Porod–Debey law as described by Rambo and Tainer (39) and used to restrain the modeling. Simulated annealing is used to search, starting from a random phase distribution, which simultaneously fits the multiple SAXS curves from untagged, singly MBP-tagged and doubly MBP-tagged Hop2–Mnd1 species, to minimize overall discrepancy. The Hop2–Mnd1 shapes obtained by this strategy displayed the relative position of MBP and allowed us to determine the position of the Hop2 and Mnd1 N-termini. In the shapes, one can clearly distinguish the V-liked structure of the Hop2–Mnd1 complex (yellow) and the position of the N-terminal MBP tag (red).
DNA binding assay
DNA binding was done with 80-mer ssDNA and dsDNA as substrates, as described (30). The 32P-labeled ssDNA (2.4 µM nucleotides) and dsDNA (2.4 µM bp) were mixed with the indicated amount of the Hop2–Mnd1 and Hop2 species at 37°C in 10 µl of buffer B [25 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM dithiothreitol and 100 µg/ml bovine serum albumin (BSA)] for 10 min. After the addition of 2 µl of gel loading buffer [50% glycerol, 20 mM Tris-HCl, pH 7.4, 0.5 mM ethylenediaminetetraacetic acid (EDTA) and 0.05% orange G], nucleoprotein complexes were resolved by 8% native polyacrylamide gel electrophoresis in TBE buffer (90 mM Tris-borate, pH 8.3 and 2 mM EDTA) at 4°C. The gels were dried and analyzed in the Personal Molecular Imager FX (Bio-Rad) using the Quantity One software.
Southwestern analysis
The procedure followed that of Chi et al. (2007) with slight modifications. Hop2–Mnd1 and its mutant variants were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), electro-transferred onto Immobilon-P PVDF Membrane (Millipore) at 4°C in transfer buffer (25 mM Tris, 192 mM glycine, pH 8.3, 20% methanol). After being soaked in buffer C (25 mM Tris-HCl, pH 7.5, 2 mg/ml BSA, 1 mM DTT, 0.1% Triton X-100, 10% glycerol and 100 mM KCl) for 20 h at 4°C, the membrane was rinsed twice with buffer D (25 mM Tris-HCl, pH 7.5, 200 µg/ml BSA, 50 mM KCl, 4 mM MgCl2 and 1 mM DTT) before being incubated in 10 ml of buffer D containing 32P-labeled 80-bp duplex DNA (200 nM bp) for 1 h at 25°C. After washing the membrane four times with 10 ml buffer D, radioactive nucleoprotein species were visualized by phosphorimaging analysis.
Affinity pulldown assay
To prepare DMC1 affinity resin to test for Hop2–Mnd1 interaction, 100 µg DMC1 or BSA was coupled to 100 µl Affi-Gel 15 resin (Bio-Rad) following the instructions of the manufacturer. The DMC1-Affi or BSA-Affi resin (10 µl) was incubated with 5 µM of Hop2–Mnd1 or mutants at 4°C for 60 min in 30 µl buffer E [25 mM Tris-HCl, pH 7.5, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal CA-630 (Sigma), 1 mM 2-mercaptoethanol and 50 mM KCl] with gentle mixing. For affinity pull-down using amylose resin (New England Biolabs), DMC1 (5 µM), together with 5 µM of Hop2-(MBP–Mnd1) or Hop2-(MBP–Mnd11−185), was mixed gently in 30 µl buffer E with 12 µl of the resin (which binds the MBP tag on Mnd1) at 4°C for 60 min. In each pull-down reaction, after washing the resin three times with 100 µl buffer E, bound proteins were eluted with 20 µl 2% SDS. The supernatant (S), last wash (W) and SDS eluate (E) fractions, 8 µl each, were analyzed by SDS-PAGE.
D-loop reaction
For the D-loop reaction (12.5 µl, final volume), 5′-32P–labeled 90-mer oligonucletide (2.4 µM nt, derived from pBluescript SK II sequence 1932–2021) was preincubated with DMC1 (1.0 µM) for 5 min at 37°C in buffer F (25 mM Tris-HCl, pH 7.5, 60 mM KCl, 1 mM Dithiothreitol (DTT), 5 mM MgCl2, 2 mM ATP or Adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP) and 100 µg/ml BSA). This was followed by the incorporation of Hop2–Mnd1 or mutant in 1 µl and a 5-min incubation at 37°C. The D-loop reaction was initiated by adding pBluescript SK replicative form I DNA (37 µM base pairs) in 1 µl. The completed reaction was incubated at 37°C for 7 min, deproteinized by treatment with SDS (0.5%) and proteinase K (0.5 mg/ml) and then subjected to electrophoresis in 1% agarose gel in TAE buffer (40 mM Tris-acetate, pH 8.0, and 1 mM EDTA). The gels were dried and subjected to phosphorimaging analysis to quantify the level of D-loop product (40)
Exonuclease I protection assay
Protection of ssDNA against the action of E. coli Exo I by DMC1 with and without Hop2–Mnd1 was conducted as described (31). Presynaptic filaments were assembled with DMC1 (0.3 µM) and 5' 32P-labeled 100-mer ssDNA (3 µM nucleotides) and then incubated with ExoI (2–6 U, as indicated in the figures; New England Biolabs).
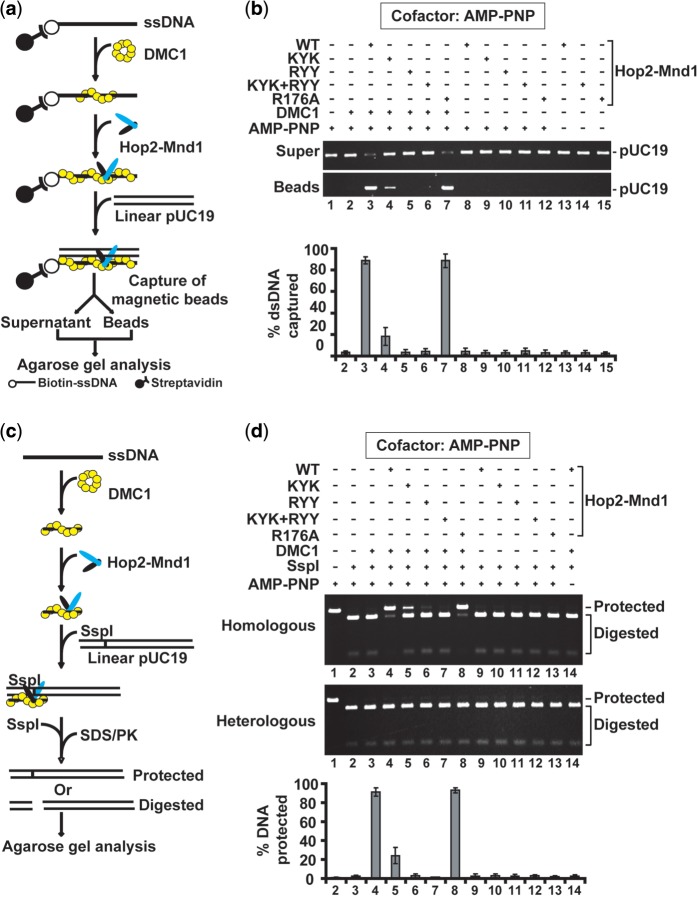
Duplex capture
Duplex capture by the Hop2-Mnd1-DMC1-ssDNA ensemble was examined as described (31). Presynaptic filaments were assembled with DMC1 (2.5 µM) and biotinylated 83-mer oligo dT (8 µM nt).
Synaptic complex formation
Synaptic complex assembly was examined as described (41). Presynaptic filaments were assembled with DMC1 (4 µM) and 60-mer ssDNA (12 µM nt) that are homologous or heterologous to the Ssp I restriction site in the target pUC19 dsDNA.
RESULTS
Biophysical and structural analyses of the Hop2–Mnd1 heterodimer
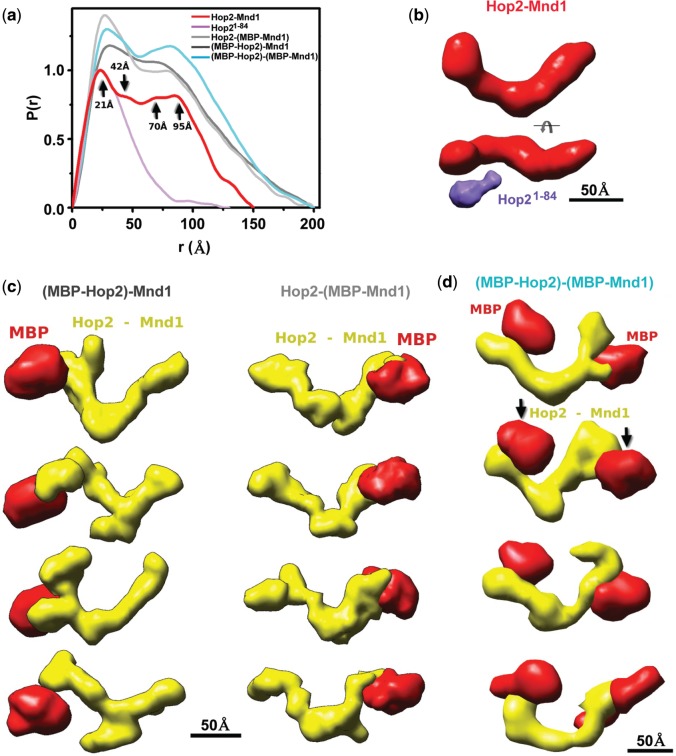
We verified the heterodimeric nature of the complex harboring mouse Hop2 (217 residues) and Mnd1 (205 residues) (27,28) by multi-angle and quasi-elastic light scattering analyses (Supplementary Figure S1a). To further define the architecture and solution assembly (42), we generated SAXS profiles of the Hop2–Mnd1 heterodimer (Supplementary Figure S1b and c), which showed a typical pair distribution function P(r) for multi-modular assembly with a length of ∼150 Å (Figure 1a). The distinct peaks observed in the P(r) at 21 Å and 42 Å indicate a large content of coiled-coil interfaces. Repeated spacing at r ∼70 Å and ∼95 Å indicates the symmetrical arrangement of the molecule. The SAXS envelopes reveal a V-shaped molecule (Figure 1b). We analyzed a different Hop2–Mnd1 preparation and obtained the same P(r) distribution and shape for the complex.
Figure 1.
SAXS and EM analysis reveals a V-shaped structure of Hop2-Mnd1. (a) P(r) of the Hop2-Mnd1 (red) computed from the experimental SAXS data (Supplementary Figure S1) compared with the P(r) obtained for (MBP-Hop2)-Mnd1, Hop2-(MBP-Mnd1) and (MBP-Hop2)-(MBP-Mnd1) assemblies (light, dark gray, cyan) and Hop21–84 construct (violet). The oscillations at 21, 42, ∼70 and ∼95 Å distances are indicated (black arrow). (b) Two views of the average SAXS envelope of the Hop2–Mnd1 assembly. The average envelope of the Hop21-84 construct (violet) is shown in proximity to the predicted N-terminal region in the Hop2–Mnd1 envelope. (c) Four (MBP-Hop2)-Mnd1 and Hop2-(MBP-Mnd1) multiphase envelopes calculated by MONSA are shown as a surface indicating the calculated phase for MBP (red) and Hop2–Mnd1 (yellow). Corespondiong multiphase fits to the experimental curves are shown in Supplementary Figure S1b. (d) Four (MBP-Hop2)-(MBP-Mnd1) multiphase envelopes calculated by MONSA are shown as a surface indicating the calcualted phase for MBP (red) and Hop2–Mnd1 (yellow). Corespondiong multiphase fits to the experimental curves are shown in Supplementary Figure S1b.
Comparison of the Hop2–Mnd1 envelopes with those of Hop2–Mnd1 species in which either protein is N-terminally tagged with MBP enabled us to identify the positions of the protein N-termini (Figure 1c). Specifically, the MBP tags were located using multiphase modeling (see Methods) at the tips of the V-shaped Hop2–Mnd1 complex (Figure 1c). Examination of multiphase envelopes for the singly MBP-tagged Hop2–Mnd1 complexes to those of Hop2–Mnd1 in which both subunits were N-terminally MBP-tagged confirmed the locations of the protein complex’s N-termini (Figure 1c and d). SAXS analysis of a Hop2 fragment encompassing the N-terminal 84 residues further verified the location of the Hop2 N-terminus (Figure 1b). We wished to confirm the ab initio SAXS model of Hop2–Mnd1 by negative stain electron microscopy, which showed that the protein molecule is indeed a V-shaped molecule (Supplementary Figure S1d).
Evidence for three distinct DNA binding domains in the Hop2–Mnd1 complex
With the overall shape of Hop2–Mnd1 having been defined (Figure 1 and Supplementary Figure S1) and the knowledge that both Hop2 and Mnd1 bind DNA (28,30), we set out to define the DNA binding domains within the Hop2–Mnd1 complex and also the functional significance of DNA binding in the DMC1-mediated HR reaction.
A coiled-coil region located in the middle part of both Hop2 and Mnd1 mediates protein complex formation (28). Being guided by this information, deletions of either the N- or C-terminus of Hop2 and Mnd1 that would preserve the protein complex were constructed. These truncation mutants, namely, Hop21−143–Mnd1, Hop2126−217–Mnd1, Hop2–Mnd11−113 and Hop2–Mnd1100−205, were purified (Supplementary Figure S2a and b) and analyzed for DNA binding with 80-mer ssDNA and dsDNA, as before (30). As shown in Supplementary Figure S2c, deletion of the N-terminal 125 residues of Hop2 affects dsDNA and ssDNA binding, whereas deletion of the C-terminal 74 residues compromises ssDNA binding significantly and dsDNA binding slightly. In the case of Mnd1, deletion of the N-terminal 99 residues attenuates dsDNA binding only, whereas deletion of the C-terminal 92 residues has no measurable effect on DNA binding (Supplementary Figure S2c). Thus, these analyses provide evidence that three regions of Hop2–Mnd1 are concerned with DNA binding, with both N-terminal arms of the complex being needed for maximal dsDNA binding and the Hop2 C-terminus possessing a preference for ssDNA.
Capitalizing on the above results, we isolated and tested the properties of the two DNA binding domains in Hop2 by purifying protein fragments encompassing residues 1–125 (Hop21−125) or residues 144–217 (Hop2144−217) (Supplementary Figure S3a). Consistent with other results (Supplementary Figure S2), Hop21−125 binds dsDNA with a higher affinity, whereas Hop2144−217 slightly prefers ssDNA (Supplementary Figure S3b). Because Mnd1 alone cannot be expressed in soluble form, we instead tested the Mnd1 fragments harboring either residues 1–113 (Mnd11−113) or residues 100–205 (Mnd1100−205) in complex with Hop2 by Southwestern analysis, as we have done earlier (30). The results revealed that Mnd11−113, but not Mnd1100−205, is able to bind dsDNA (Supplementary Figure S3c, middle) and that neither fragment binds ssDNA (Supplementary Figure S3c, right). We note that, in the Southwestern analysis, Mnd1 appears to bind dsDNA with a lower affinity than Hop2 (Supplementary Figure S3c and S4f) and that the Mnd11−113 fragment is slightly attenuated for dsDNA binding compared with the full-length protein (Supplementary Figure S3c). These differences could stem from varying efficiencies of protein transfer onto the nitrocellulose membrane or of in situ renaturation of the protein species. Alternatively, or in addition, the results might reflect genuine differences in the inherent affinities of the protein species for the DNA ligand.
Point mutants of Hop2 and Mnd1 compromised for DNA binding
To further dissect the complex’s DNA binding sites, we next constructed Hop2–Mnd1 point mutants that are impaired for DNA binding. To guide our effort, we aligned Hop2 orthologs to identify conserved regions that are enriched in basic and aromatic residues, which could contribute to DNA engagement via ionic interactions with the phosphodiester backbone and stacking interactions with DNA bases, respectively. The most conserved regions in the N-terminus and C-terminus of Hop2 that fit this criterion reside within residues 59–71 and residues 171–178, respectively (Supplementary Figure S4A). Interestingly, secondary structure prediction reveals similarities of the Hop2 N-terminal segment to the winged helix family of DNA binding proteins (43,44) (Supplementary Figure S4b). We changed the highly conserved K63, Y65 and K67 residues located within the predicted β-hairpin of the putative winged helix motif to alanine (the KYK mutation) in the Hop21−125 fragment. The KYK Hop21−125 mutant is clearly impaired for DNA binding (Supplementary Figure S4c). To inactivate the Hop2 C-terminal DNA binding domain, we changed the highly conserved R176 residue to alanine in the Hop2144−217 fragment. The R176A mutant of Hop2144−217 is ∼2–3-fold less capable of ssDNA binding (Supplementary Figure S4d).
The same strategy was used to isolate a DNA binding deficient mutant of Mnd1. Interestingly, secondary structure prediction of the Mnd1 N-terminal portion also shows a potential winged helix motif between residues 10–89 (Supplementary Figure S4b). Accordingly, the highly conserved residues R64, Y70 and Y71 within the predicted β-hairpin region of the Mnd1 motif were changed to alanine (the RYY mutation), and the mutant protein was co-purified with Hop2 (Supplementary Figure S4e and f). Importantly, the Mnd1 RYY mutant is defective in dsDNA binding as shown in the Southwestern analysis (Supplementary Figure S4f). Notably, a triple point mutation harboring the change of R82, K83 and R84, residues that are located outside of the predicted winged helix motif, to alanine (the RKR mutation) has no effect on the DNA binding activity of Mnd1 (Supplementary Figure S5a and c).
Effects of point mutations on DNA binding by the Hop2–Mnd1 complex
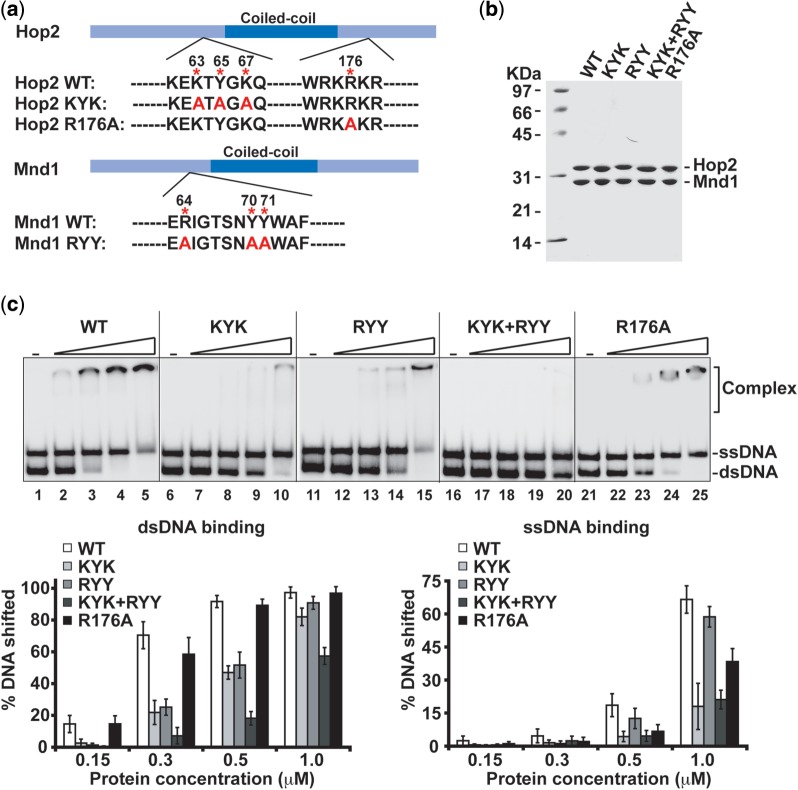
Next, we expressed and purified four mutant Hop2–Mnd1 complexes, namely, those that harbor the Hop2-KYK, Hop2-R176A or Mnd1-RYY mutation alone, or the combination of the Hop2-KYK and Mnd1-RYY mutations (Figure 2a and b). Compared with the wild-type (WT) counterpart, all four mutant complexes were expressed to the same level in E. coli, exhibited the same chromatographic behavior and could be purified with the same procedure and a similar yield (Figure 2b). Results from DNA binding experiments revealed that the Hop2-KYK mutation compromises both ssDNA and dsDNA binding by Hop2–Mnd1, whereas the Hop2-R176A mutation affects mostly its ssDNA binding activity (Figure 2c). The Mnd1-RYY mutation reduces dsDNA binding severalfold (Figure 2c). Combining the Hop2-KYK and Mnd1-RYY mutations has an additive effect on DNA binding (Figure 2c). Notably, as determined by an affinity pull-down assay, the four mutant Hop2–Mnd1 complexes have no overt deficiency in DMC1 interaction (Figure 3a). We note that the Hop2-Mnd1-RKR mutant complex harbors the WT level of DNA binding and DMC1 interaction activities (Supplementary Figure S5b).
Figure 2.
DNA binding properties of point mutants of the Hop2–Mnd1 complex. (a) Mutants of Hop2 and Mnd1 made in this study. (b) SDS-PAGE analysis of purified Hop2–Mnd1 and mutant complexes. (c) Analysis of WT and mutant variants of Hop2–Mnd1 (KYK, RYY, KYK + RYY and R176A) (0.15, 0.3, 0.5 and 1.0 µM) for DNA binding. The mean values ± SD from three independent experiments were plotted.
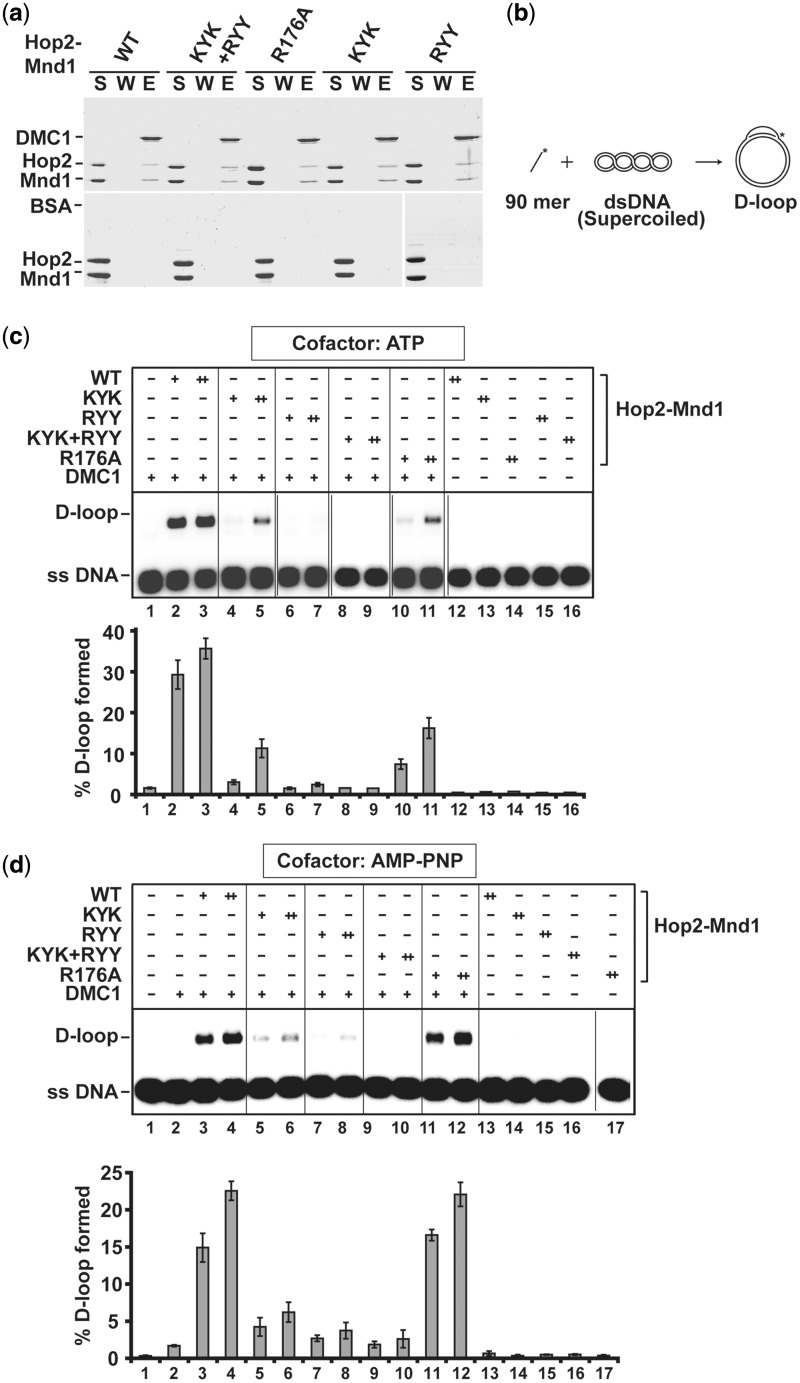
Figure 3.
Examination of DNA binding mutants of Hop2–Mnd1 in the D-loop reaction. (a) Pulldown assay making use of Affi-Gel 15 Agarose beads coupled to DMC1 or BSA was conducted to test for the interaction with WT or mutant Hop2–Mnd1 complexes. The supernatant (S) containing unbound proteins, the wash (W) and the eluate (E) fractions were analyzed by SDS-PAGE and Coomassie Blue staining. (b) Schematic representation of the D-loop reaction. (c) WT and mutant variants of the Hop2–Mnd1 complex (60 nM or 120 nM) were tested for their ability to promote D-loop formation with ATP as nucleotide cofactor. The mean values ± SD from three independent experiments were quantified and plotted. (d) WT and mutant variants of the Hop2–Mnd1 complex (60 nM or 120 nM) were tested for their ability to promote D-loop formation with AMP-PNP as nucleotide cofactor. The mean values ± SD from three independent experiments were quantified and plotted.
Examination of the Hop2–Mnd1 mutant complexes in the D-loop reaction
We tested the mutant Hop2–Mnd1 complexes for the ability to enhance the DMC1-mediated D-loop reaction (Figure 3b) with ATP as the nucleotide cofactor (40). Importantly, the Hop2-KYK and Hop2-R176A mutations each diminished the efficiency of the D-loop reaction severalfold, whereas little or no D-loop was made when the Mnd1-RYY or Hop2-KYK/Mnd1-RYY mutant was used (Figure 3c). Thus, the functional integrity of the Hop2–Mnd1 complex is reliant on its three DNA binding domains. In contrast, the Hop2-Mnd1-RKR mutant behaved like the WT counterpart in the D-loop reaction (Supplementary Figure S5d and e). As will be described later, the mutant complex harboring Hop2-R176A is proficient in the D-loop reaction when the DMC1 presynaptic filament is stabilized by the use of a non-hydrolyzable ATP analog (Figure 3d).
Dependence of DMC1 presynaptic filament stability on the Hop2 C-terminal DNA binding domain
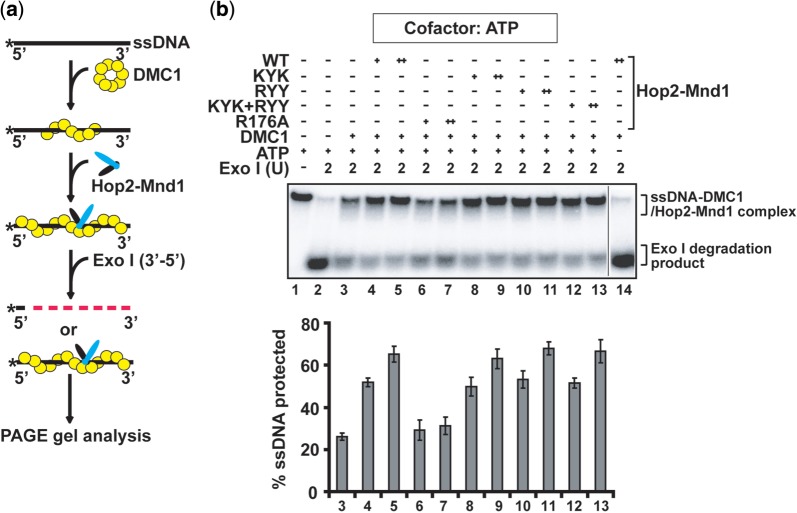
Hop2–Mnd1 stabilizes the DMC1 presynaptic filament (31). We used the previously devised assay involving protection of DNA against E. coli Exo I digestion (31) (Figure 4a) to test whether any of the Hop2–Mnd1 mutant complexes is defective in this presynaptic filament stabilization function. Neither Hop2–Mnd1 nor any of the mutant complexes could protect the DNA against digestion in the absence of DMC1 (Supplementary Figure S6a and b). As shown in Figure 4B, while the Hop2-KYK, Mnd1-RYY and the Hop2-KYK/Mnd1-RYY mutant complexes were just as effective as the WT complex in stabilizing the DMC1 presynaptic filament, the Hop2-R176A mutant complex (compromised for ssDNA binding mostly) was inactive (Figure 4b and Supplementary Figure S6b). Thus, among the mutations that affect ssDNA binding (Figure 2c), only the Hop2-R176A mutation impairs presynaptic filament stabilization (Figure 4b and Supplementary Figure S6b).
Figure 4.
Role of the Hop2 C-terminal DNA binding domain in DMC1 filament stabilization. (a) Schematic representation the exonuclease I protection assay to examine DMC1 presynaptic filament stability. (b) WT and mutant variants of the Hop2–Mnd1 complex (0.3 or 0.6 µM) were tested for their ability to protect the DMC1 presynaptic filament from Exo I digestion. The mean values ± SD from three independent experiments were plotted.
To further ascertain the role of the Hop2 C-terminal DNA binding domain in DMC1 presynaptic filament stabilization, we tested the Hop2-R176A mutant complex in the D-loop reaction using the non-hydrolyzable AMP-PNP as the nucleotide cofactor to enhance the stability of the DMC1 presynaptic filament (40). The Hop2-R176A mutant complex was just as capable as the WT counterpart in the enhancement of D-loop formation when AMP-PNP was used (Figure 3d). In contrast, the use of AMP-PNP could not overcome the deficiency of the Hop2-KYK, Mnd1-RYY or Hop2-KYK/Mnd1-RYY mutant Hop2–Mnd1 complex in the D-loop reaction (Figure 3d).
Dependence of synaptic complex assembly on the N-terminal DNA binding domains
We used two separate assays to test the Hop2–Mnd1 mutants in synaptic complex assembly. First, the duplex capture step was examined (Figure 5a) with ATP present. The results showed that all four mutant complexes are deficient in the capture of duplex DNA (Supplementary Figure S7a). When AMP-PNP replaced ATP as the nucleotide cofactor, however, the Hop2-R176A complex was adept at capturing duplex DNA, even though the functional deficiency in the other mutants remained just as severe (Figure 5b).
Figure 5.
Role of the N-terminal DNA binding domains of Hop2–Mnd1 in duplex DNA capture and synaptic complex assembly. (a) Schematic representation of the duplex capture assay. (b) WT and mutant variants of the Hop2–Mnd1 complex (0.5 µM) were tested for their ability to promote duplex capture with AMP-PNP as nucleotide cofactor. The mean values ± SD from three independent experiments were plotted. (c) Schematic representation the restriction enzyme protection assay to examine synaptic complex formation. (d) WT and mutant variants of the Hop2–Mnd1 complex (0.3 µM) were tested for their ability to promote synaptic complex formation with AMP-PNP as nucleotide cofactor. The mean values ± SD from three independent experiments were plotted.
We next determined the impact of the Hop2–Mnd1 mutations on the assembly of the synaptic complex. The assay (45) involves monitoring the protection of linear dsDNA by the DMC1 presynaptic filament against digestion by a restriction enzyme (Figure 5c). We conducted the reactions with ATP or AMP-PNP as the nucleotide cofactor. As expected (31,41), the DMC1 presynaptic filament alone did not efficiently assemble the synaptic complex, but the addition of Hop2–Mnd1 led to a marked stimulation (Figure 5d and Supplementary Figure S7b). Importantly, and consistent with results from other functional assays above, we found that the Hop2-KYK, Mnd1-RYY and Hop2-KYK/Mnd1-RYY mutants are all deficient in synaptic complex formation (Figure 5d and Supplementary Figure S7b). As we had anticipated, the Hop2-R176A mutant was just as active as the WT counterpart with AMP-PNP (Figure 5d), but much less active with ATP (Supplementary Figure S7b). Altogether, the results revealed a dependence of presynaptic filament stabilization on the Hop2 C-terminal ssDNA binding activity and that facilitation of synaptic complex assembly requires the N-terminal dsDNA-binding function of both Hop2 and Mnd1.
DISCUSSION
Previous biochemical studies have furnished evidence for a dual function of the mammalian Hop2–Mnd1 complex in the homologous DNA pairing reaction mediated by either the RAD51 or DMC1 recombinase (30,31). Here, we have examined the role of Hop2–Mnd1 as a DMC1 cofactor, and applied a combinatorial approach to help elucidate the mechanistic basis for the dual function of Hop2–Mnd1 in DMC1 presynaptic filament stabilization and synaptic complex assembly.
Published studies have suggested that both Hop2 and Mnd1 harbor a DNA binding function (28,30). By biochemical, bioinformatic and mutagenic analyses, we provide evidence for the existence of three distinct DNA binding domains in the Hop2–Mnd1 complex. Specifically, the N-terminal region of Hop2 and Mnd1 each possesses such a domain, with both having a preference for dsDNA. Interestingly, the C-terminal portion of Hop2 also harbors a DNA binding domain, but that has a preference for ssDNA. Importantly, a mutation that compromises the Hop2 ssDNA binding domain affects only the DMC1 presynaptic filament stabilization attribute of Hop2–Mnd1, whereas mutations that ablate the dsDNA binding activity of Hop2 or Mnd1 impair the complex’s ability to promote duplex DNA capture and synaptic complex assembly specifically.
Combined SAXS and electron microscopy show that Hop2–Mnd1 is a V-shaped molecule. By biochemical mapping, we have revealed that the dsDNA-specific binding domains of Hop2–Mnd1 are situated within the arms of the V-shaped complex, but we do not yet have enough information to locate the Hop2 ssDNA binding domain within the bundled, stem region of the complex. Our published work has shown that Mnd1 interacts with RAD51 (30), and we now know that the last 20 residues of Mnd1 are indispensable for DMC1 interaction (Supplementary Figure S8). Based on this information, we hypothesize that the V-shaped Hop2–Mnd1 complex associates with the DMC1 presynaptic filament through its bundled region (Figure 6). Within this structural arrangement, Hop2–Mnd1 is well positioned to capture duplex DNA to bring it into proximity of the secondary binding site of the presynaptic filament, wherein the duplex DNA is sampled for homology and DNA joint formation occurs (46). Moreover, based on the biochemical analyses of the Hop2-R176A mutant, it is tempting to suggest that the C-terminal DNA binding domain of Hop2 may access ssDNA within the DMC1 presynaptic filament, and that ssDNA engagement by this Hop2 DNA binding domain is indispensable for presynaptic filament stabilization.
Figure 6.
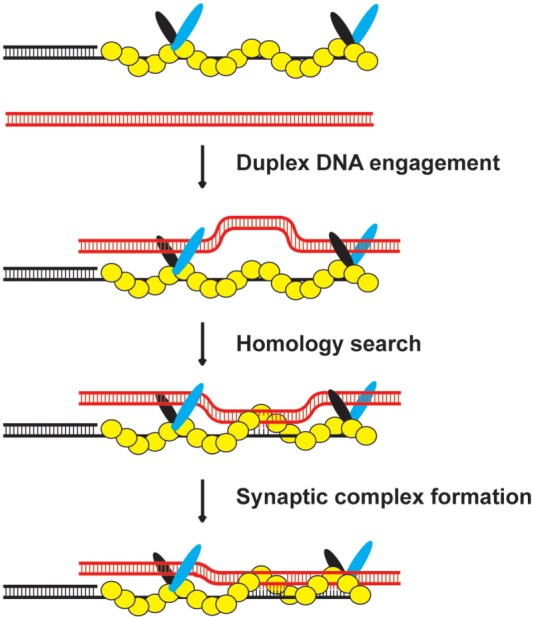
Model for the mechanism of the Hop2-Mnd1-DMC1-ssDNA ensemble. Our model posits that Hop2–Mnd1 stabilizes the DMC1 presynaptic filament by its ssDNA binding activity within the C-terminus of Hop2 and helps capture duplex DNA by its dsDNA binding function located in the N-termini of Hop2 and Mnd1, thus increasing the efficiency at which the duplex could be engaged and sampled for DNA homology by the DMC1 presynaptic filament.
Hop2–Mnd1 is most effective in synaptic complex assembly and in the D-loop reaction when present at amounts ∼1/4–1/15 the concentration of DMC1 (26,27,29,31). Cytological analysis has revealed that the majority of Hop2 and Mnd1 proteins do not co-localize with DMC1 in S. cerevisiae and A. thaliana cell nuclei during meiosis (16,17,19,47). The cytological results are consistent with the premise that only a fraction of the intracellular Hop2–Mnd1 pool is associated with the DMC1 presynaptic filament in meiotic cells. Based on the cytological observations and our biochemical analyses, we suggest that a DMC1 presynaptic filament lightly decorated with Hop2–Mnd1 is most optimally poised to conduct the homologous DNA pairing reaction, as depicted in Figure 6. According to this mechanistic framework, an excess of Hop2–Mnd1 would impede homologous DNA pairing by restricting access of the captured duplex DNA to the secondary DNA binding site of the DMC1 presynaptic filament (26,27,29).
ACCESSION CODES
The SAXS data including curves, P(r) functions and reconstructed models will be deposited in BIOISIS database during revision of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health [RO1ES015252, RO1CA168635, RO1ES007061 and PO1CA092584]; the National Science Council of Taiwan [NSC 99-2311-B-002-012 and NSC 100-2311-B-002-009]; and also by the National Basic Research Program of China [2010CB912401] and the National Center for Protein Sciences Beijing. Funding for open access charge: [RO1ES015252,RO1CA168635, RO1ES007061 and PO1CA092584].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the Yale CryoEM and High Performance Computational facilities for support.
REFERENCES
- 1.Roeder GS. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 2.McKee BD. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta. 2004;1677:165–180. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 5.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 6.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 8.Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 9.Raynard S, Niu H, Sung P. DNA double-strand break processing: the beginning of the end. Genes Dev. 2008;22:2903–2907. doi: 10.1101/gad.1742408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein KA, Rothstein R. At loose ends: resecting a double-strand break. Cell. 2009;137:807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radding CM. Homologous pairing and strand exchange in genetic recombination. Annu. Rev. Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- 14.Gerton JL, DeRisi JL. Mnd1p: an evolutionarily conserved protein required for meiotic recombination. Proc. Natl Acad. Sci. USA. 2002;99:6895–6900. doi: 10.1073/pnas.102167899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry JM, Camahort R, Rice DA, Florens L, Swanson SK, Washburn MP, Gerton JL. Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol. Cell. Biol. 2006;26:2913–2923. doi: 10.1128/MCB.26.8.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leu JY, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 17.Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 19.Zierhut C, Berlinger M, Rupp C, Shinohara A, Klein F. Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 2004;14:752–762. doi: 10.1016/j.cub.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 20.Domenichini S, Raynaud C, Ni DA, Henry Y, Bergounioux C. Atmnd1-delta1 is sensitive to gamma-irradiation and defective in meiotic DNA repair. DNA Repair (Amst.) 2006;5:455–464. doi: 10.1016/j.dnarep.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Kerzendorfer C, Vignard J, Pedrosa-Harand A, Siwiec T, Akimcheva S, Jolivet S, Sablowski R, Armstrong S, Schweizer D, Mercier R, et al. The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J. Cell. Sci. 2006;119:2486–2496. doi: 10.1242/jcs.02967. [DOI] [PubMed] [Google Scholar]
- 22.Panoli AP, Ravi M, Sebastian J, Nishal B, Reddy TV, Marimuthu MP, Subbiah V, Vijaybhaskar V, Siddiqi I. AtMND1 is required for homologous pairing during meiosis in Arabidopsis. BMC Mol. Biol. 2006;7:24. doi: 10.1186/1471-2199-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petukhova GV, Romanienko PJ, Camerini-Otero RD. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell. 2003;5:927–936. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- 24.Schommer C, Beven A, Lawrenson T, Shaw P, Sablowski R. AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J. 2003;36:1–11. doi: 10.1046/j.1365-313x.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen YK, Leng CH, Olivares H, Lee MH, Chang YC, Kung WM, Ti SC, Lo YH, Wang AH, Chang CS, et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl Acad. Sci. USA. 2004;101:10572–10577. doi: 10.1073/pnas.0404195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enomoto R, Kinebuchi T, Sato M, Yagi H, Kurumizaka H, Yokoyama S. Stimulation of DNA strand exchange by the human TBPIP/Hop2-Mnd1 complex. J. Biol. Chem. 2006;281:5575–5581. doi: 10.1074/jbc.M506506200. [DOI] [PubMed] [Google Scholar]
- 27.Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson JY, Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- 28.Pezza RJ, Petukhova GV, Ghirlando R, Camerini-Otero RD. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J. Biol. Chem. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- 29.Ploquin M, Petukhova GV, Morneau D, Dery U, Bransi A, Stasiak A, Camerini-Otero RD, Masson JY. Stimulation of fission yeast and mouse Hop2-Mnd1 of the Dmc1 and Rad51 recombinases. Nucleic Acids Res. 2007;35:2719–2733. doi: 10.1093/nar/gkm174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi P, San Filippo J, Sehorn MG, Petukhova GV, Sung P. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–1757. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pezza RJ, Voloshin ON, Vanevski F, Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–1766. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 33.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, II, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, Dyer K, Hammel M, Meigs G, Frankel KA, et al. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guinier A, Fournet F. Small angle scattering of X-rays. New York: Wiley Interscience; 1955. [Google Scholar]
- 36.Svergun DI. Determination of the regularization Parameter in Indirect-Transform Methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 37.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J.Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst.) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Dray E, Dunlop MH, Kauppi L, Filippo JS, Wiese C, Tsai MS, Begovic S, Schild D, Jasin M, Keeney S, et al. Molecular basis for enhancement of the meiotic DMC1 recombinase by RAD51 associated protein 1 (RAD51AP1) Proc. Natl Acad. Sci. USA. 2011;108:3560–3565. doi: 10.1073/pnas.1016454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 43.Gajiwala KS, Burley SK. Winged helix proteins. Curr. Opin. Struct. Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 44.Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 45.Ferrin LJ, Camerini-Otero RD. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991;254:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- 46.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 47.Vignard J, Siwiec T, Chelysheva L, Vrielynck N, Gonord F, Armstrong SJ, Schlogelhofer P, Mercier R. The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 2007;3:1894–1906. doi: 10.1371/journal.pgen.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.