Abstract
The HIV-1 transactivator of transcription (Tat) protein is thought to stimulate reverse transcription (RTion). The Tat protein and, more specifically, its (44–61) domain were recently shown to promote the annealing of complementary DNA sequences representing the HIV-1 transactivation response element TAR, named dTAR and cTAR, that plays a key role in RTion. Moreover, the kinetic mechanism of the basic Tat(44–61) peptide in this annealing further revealed that this peptide constitutes a representative nucleic acid annealer. To further understand the structure–activity relationships of this highly conserved domain, we investigated by electrophoresis and fluorescence approaches the binding and annealing properties of various Tat(44–61) mutants. Our data showed that the Tyr47 and basic residues of the Tat(44–61) domain were instrumental for binding to cTAR through stacking and electrostatic interactions, respectively, and promoting its annealing with dTAR. Furthermore, the annealing efficiency of the mutants clearly correlates with their ability to rapidly associate and dissociate the complementary oligonucleotides and to promote RTion. Thus, transient and dynamic nucleic acid interactions likely constitute a key mechanistic component of annealers and the role of Tat in the late steps of RTion. Finally, our data suggest that Lys50 and Lys51 acetylation regulates Tat activity in RTion.
INTRODUCTION
The transactivator of transcription (Tat) is crucially required for efficient transcription of the integrated HIV viral genome (1,2). Tat increases the efficiency of the transcription complex via the recruitment of various cellular factors on binding to the transactivating response (TAR) element (3–5), leading to the active production of full-length viral RNA (6–10). The sequence of Tat that mediates specific TAR binding has been mapped to the basic (49–57) domain composed mostly of Arg and Lys residues (11,12), in line with the ability of arginine-rich peptides to bind specifically to RNA (13–15). Moreover, Tat is thought to regulate Rev-dependent mRNA transport (16), mRNA capping (17), splicing (18) and translation (19), and to interfere with the cellular RNA interference machinery through interactions with DICER and RNA (20,21).
In addition, several lines of evidence also suggest a role of Tat in the stimulation of reverse transcription (RTion). HIV-1 mutants deleted of the Tat gene display a 3- to 5-fold defect in RTion compared with wild-type HIV-1 in infected T cells and in endogeneous RTion reactions (22,23), whereas full restoration of RTion can be achieved by transfecting the producer cells by a Tat expression plasmid. Moreover, recombinant Tat also stimulates 2- to 3-fold in vitro DNA synthesis directed by reverse transcriptase (24). Ex vivo studies on Tat mutants further reveal that Tat activities in RTion and transactivation are not correlated, suggesting that Tat directly stimulates RTion (23). The stimulatory activity of Tat in RTion is mainly supported by its 60 first amino acids (22), with a critical role for the basic (49–57) domain and the Tyr47 residue of the core domain (24).
The possible stimulatory effect of Tat in RTion within HIV-1 virions was substantiated by the detection of Tat in HIV-1 virions produced by macrophages (25). Alternatively, by permeating the plasma membrane, extracellular Tat could enter in newly infected cells to stimulate RTion after disassembly of the viral core or the natural endogeneous RTion that occurs during assembly (26–28).
Part of the stimulatory effect of Tat in RTion likely results from its nucleic acid annealing activity. Tat can promote the annealing of (i) the primer tRNA onto the viral HIV-1 RNA (29), (ii) complementary viral DNA sequences representing the HIV-1 TAR element, named dTAR and cTAR (30,31), (iii) the HIV-1 (±) primer binding sequences (32) and (iv) complementary RNA model sequences of 21 nt (33). Moreover, based on mechanistic studies with the three last systems, it appears that Tat can efficiently promote annealing of complementary sequences, without showing any nucleic acid destabilizing property. Therefore, Tat does not constitute a canonical chaperone protein, but rather belongs to the family of nucleic acid annealers. The annealer activity of Tat is mainly mediated through its 44–61 domain, as demonstrated by the potent DNA and RNA annealing activities of the Tat(44–61) peptide, which corresponds to the smallest known peptide endowed with these activities (30).
The eight basic residues of the Tat basic domain are critical for both binding to TAR (12) and accelerating the annealing of model complementary RNA sequences (33). Moreover, the distribution of the basic amino acids appears more important than their nature for the nucleic acid annealing (33) and transactivation activities (12). A further illustration of the importance of these basic amino acids is that specific acetylation of Lys50 and Lys51 residues, altering the positive charge distribution, allows a fine regulation of Tat activity (34–39). Interestingly, mutation of the aromatic Tyr47 residue induces only limited defects in transactivation, but strongly delays replication (40,41), probably as a result of a RTion defect (24,42). A further difference in the structure–activity relationship of the roles of Tat in transactivation and RTion is that only the former correlates with the nucleic acid binding affinity of Tat mutants (12,24,33).
In this context, to further delineate the relationship between the amino acid sequence of the Tat(44–61) domain and its nucleic acid annealing activity in RTion, we investigated by fluorescence techniques and gel electrophoresis the binding and annealing activities of various mutants of the Tat(44–61) peptide on the complementary cTAR and dTAR DNA sequences involved in RTion (Figure 1). The binding of Tat(44–61) to cTAR DNA appears largely governed by electrostatic interactions with the basic residues, and further stabilized by the stacking of the Tyr47 residue with the cTAR bases. Though all the tested peptides promoted cTAR/dTAR annealing, our data clearly evidenced that their efficiency is associated with their ability to rapidly associate and dissociate the intermediate cTAR/dTAR complex that nucleates the final cTAR/dTAR duplex. Moreover, comparison of our data with the literature further highlighted a good correlation between the ability of the Tat-mutated peptides to induce transient and dynamic interactions between the peptide-coated nucleic acids and the efficiency in RTion of the corresponding mutations in Tat. Thus, promotion of transient and dynamic nucleic acid interactions likely constitutes a key mechanistic component of the role of Tat in RTion and for nucleic acid annealers, in general. Finally, our data further suggest that acetylation of Lys50 and Lys51 residues may regulate Tat activity in RTion.
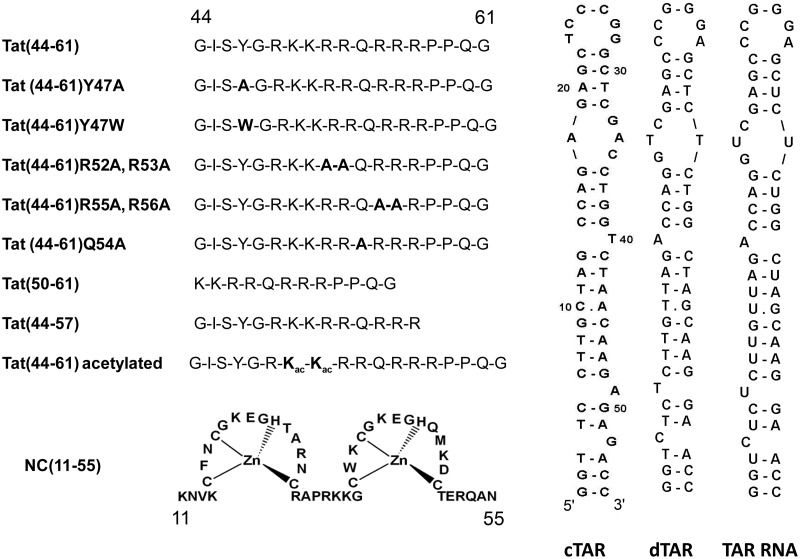
Figure 1.
Sequences of the peptides and oligonucleotides used.
MATERIALS AND METHODS
Tat(44–61), its truncated forms Tat(50–61) and Tat(44–57), its mutants Tat(44–61)R52A R53A, Tat(44–61)R55A R56A, Tat(44–61)Y47A, Tat(44–61)Q54A and the 50, 51ω-acyl-Lys form of Tat(44–61) [Tat(44–61)ac] were synthesized by solid-phase peptide synthesis on a 433 A synthesizer (ABI, Foster City, CA, USA) and purified by HPLC (43). The peptides were stored lyophilized. Their purity was >98% as judged from the HPLC elution profiles. The concentration of Tat(44–61), Tat(44–57), Tat(44–61)R52A R53A, Tat(44–61)R55A R56A, Tat(44–61)ac and Tat(44–61)Q54A was measured using an extinction coefficient at 280 nm of 1300 M−1cm−1. The concentration of Tat(44–61)Y47W was measured using an extinction coefficient at 280 nm of 5400 M−1 cm−1. The concentration of Tat(50–61) and Tat(44–61)Y47A was calculated from the absorbance of the peptide solutions at 214 nm as described in reference (44). Unless otherwise mentioned, experiments were done in freshly prepared and degassed 25 mM Tris buffer (pH 7.5) containing 30 mM NaCl and 0.2 mM MgCl2.
The oligodeoxynucleotides corresponding to sense and anti-sense HIV-1 TAR and oligodeoxynucleotides selectively labeled at different positions with 2′-deoxyribosyl-2-aminopurine (2-Ap) were purchased from IBA GmbH Nucleic Acids Product Supply (Göttingen, Germany). Doubly labeled cTAR was modified at its 5′ terminus with 5 (and-6) carboxytetramethylrhodamine (TMR) and at its 3′ terminus with 5 (and 6)-carboxyfluorescein (Fl), via an amino-linker with a six carbon spacer arm. The oligonucleotides were purified by the manufacturer by reverse-phase HPLC and polyacrylamide gel electrophoresis. An extinction coefficient at 260 nm of 515 070 M−1 cm−1 was used to calculate the concentrations of cTAR and dTAR.
For gel electrophoresis experiments, cTAR and dTAR DNAs were 5′-end labeled using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and [γ-32P] ATP (Perkin Elmer, Waltham, MA). For the footprinting experiments, the cTAR DNA was labeled at the 3′-end with [α-32P] dATP (Perkin Elmer, Waltham, MA) and Taq DNA polymerase (New England Biolabs, Ipswich, MA), as described previously (45). The 32 P-labeled nucleic acids were purified by electrophoresis on a 12% denaturing polyacrylamide gel and isolated by elution followed by ethanol precipitation.
Investigating the TAR/peptide interactions
Fluorescence monitoring of the peptide binding to cTAR
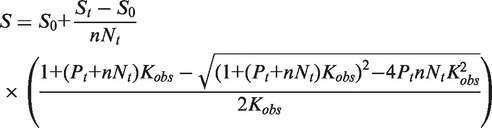
Equilibrium binding experiments aimed at determining the apparent association constant, Kobs, of the complexes between cTAR and Tat peptides were performed by adding small aliquots (2.5–5 µl) of the peptide stock solution to 10 nM cTAR-3′-Fl. Steady-state fluorescence anisotropy (with excitation at 480 nm) of each data point was monitored at 20°C using a T-format SLM 8000 spectrofluorometer equipped with a thermostated cell holder. A home-made device ensured the automatic rotation of the excitation polarizer. The light emitted by the reaction mixture was monitored through high-pass filters (Kodak, 520 nm). The best estimate of Kobs was obtained by fitting the fluorescence data to Equation (1) (30), which assumes that one molecule of cTAR can bind n molecules of peptide on independent and equivalent binding sites:
 |
(1) |
where So and St are the initial and final anisotropy, respectively, Nt the oligonucleotide concentration and Pt the protein concentration. The value of n was determined to be 10 for the cTAR/Tat(44–61) interaction (30), which corresponds to an occluded binding site of 5.5 nt per peptide. This stoichiometry was used to calculate the Kobs values for all the complexes studied here. The salt dependence of the binding constants was determined from titrations performed at salt concentrations, varying from 30 to 150 mM NaCl.
Gel retardation assays
Assays were carried out in a final volume of 10 µl. The 5′-end labeled cTAR DNA (2.5 pmol) at 8 × 103 cpm/pmol was dissolved in 6 µl of water, heated at 90°C for 2 min and chilled for 2 min on ice. Then, 2 µl of renaturation buffer was added [final concentrations: 30 mM NaCl, 0.2 mM MgCl2 and 25 mM Tris–HCl (pH 7.5)] and the sample was incubated for 15 min at 20°C in the absence or presence of peptide at various concentrations. Gel loading buffer (final concentrations: 10% w/v glycerol, 0.01% w/v bromophenol blue, 0.01% w/v xylene cyanol) was added and the samples were analyzed by electrophoresis on a 10% polyacrylamide gel (Acrylamide:Bis-acrylamide = 29:1) at 4°C in 0.5 × TBE buffer [45 mM Tris-borate (pH 8.3), 1 mM EDTA]. After electrophoresis, the gel was fixed, dried and autoradiographed.
Footprinting assays
Mung bean (MB) nuclease and DNase I were purchased from New England Biolabs (Ipswich, MA) and Promega (Madison, WI), respectively. Structural probing of cTAR DNA was carried out in a final volume of 10 µl. The 3′-end labeled cTAR DNA (2.5 pmol at 2×104 cpm/pmol) in 5.5 µl of water was heated at 90°C for 2 min and chilled for 2 min on ice. Then, 2.5 µl of renaturation buffer (final concentrations: 30 mM NaCl, 0.2 mM MgCl2 and 25 mM Tris-HCl pH 7.5 for probing with DNase I; 30 mM NaCl, 0.2 mM MgCl2 and 50 mM sodium cacodylate pH 6.5 for probing with MB nuclease) were added and the sample was incubated for 15 min at 20°C in the absence or presence of peptide at various concentrations. The samples were then incubated with either 2 units of MB nuclease for 15 min at 20°C or 0.1 unit of DNase I for 7 min at 20°C. The cleavage reactions were stopped by phenol-chloroform extraction followed by ethanol precipitation. The dried pellets were resuspended in 10 µl of loading buffer (7 M urea, 0.03 w/v% bromophenol blue and 0.03 w/v% xylene cyanol). The G, G+A and T+C sequence markers of the labeled cTAR were produced by the Maxam–Gilbert method (46). To identify all cleavage sites at the nucleotide level, the samples were analyzed by electrophoresis on denaturing 14% polyacrylamide gels using short and long migration times.
Investigating the kinetics of the cTAR/dTAR annealing reaction
The kinetics of cTAR/dTAR annealing was investigated at 20° C under pseudo first-order conditions by reacting 10 nM doubly labeled cTAR with, at least, a 10-fold higher concentration of unlabeled dTAR (30,31,47) in the presence of Tat(44–61) or one of its derivative added to a molar ratio of three peptides per oligonucleotide. The reaction was triggered by mixing 400 µl of a solution of dTAR and peptide with the same volume of a solution of TMR-5'-cTAR-3′-Fl and peptide, a process that avoids aggregation resulting from local high concentration of reagents. The annealing process was followed by continuously monitoring the Fl fluorescence intensity at 520 nm (with excitation at 480 nm). All reported concentrations of the reagents correspond to those after mixing. Emission spectra and kinetic traces were recorded with a FluoroMax 3 spectrofluorometer (Jobin Yvon Instruments) equipped with a thermostated cell holder. All fluorescence intensities were corrected for buffer emission and lamp fluctuations.
Non-linear least square fits were performed with the Levenberg–Marquardt algorithm using the Origin software (Microcal). Numerical resolution of the kinetic parameters was performed by fitting simultaneously the set of progress curves with the Dynafit software (BioKin Ltd.) (48). Numerical tests (Kolmogorov, Durbin–Wattson, Tukey statistics) and the reduced χ2 values were used to evaluate goodness of fit.
RESULTS AND DISCUSSION
Identification of the determinants for Tat(44–61) binding to cTAR
To map the critical amino acids of Tat(44–61) for binding to cTAR, comparative quantitative affinity measurements of Tat(44–61) mutants (Figure 1) were assessed using fluorescence anisotropy by titrating a fixed amount of cTAR-Fl with increasing peptide concentrations (Supplementary Figure S1, Supplementary Data). The values of the observed equilibrium association constants Kobs for the various peptides were obtained by fitting the data with Equation (1), assuming that the n = 10 binding sites on TAR DNA were identical and independent. This approach was shown to be reasonable to comparatively assess the affinities of other viral proteins and their mutants with cTAR DNA (30,49,50). The Kobs value of 9(±2) × 107 M−1 obtained for the native Tat(44–61) peptide was close to the 1.7 × 108 M−1 and 5.1 × 107 M−1 values (Table 1) reported for slightly shorter peptides, namely, Tat(47–58) and Tat(46–60), respectively, to their specific binding site on the upper part of TAR RNA (12,51). This similarity in the binding constants clearly indicates that the Tat(44–61) peptide binds to cTAR DNA with high affinity, giving further credit to the relevance of cTAR as a target of the Tat protein.
Table 1.
Equilibrium association constant for the peptide: cTAR complexes
| Peptide | Kobs (M-1) |
|---|---|
| Tat(44–61) | 9 ± 2 × 107a |
| Tat(44–61)Y47A | 3.5 ± 1 × 107 |
| Tat(44–61)Y47W | 9 ± 2 × 107 |
| Tat(44–57) | 1.2 ± 0.3 × 108 |
| Tat(50–61) | 2.4 ± 0.4 × 107 |
| Tat(44–61)Q54A | 5.7 ± 1 × 107 |
| Tat(44–61)R52A, R53A | 8 ± 2 × 106 |
| Tat(44–61)R55A, R56A | 1.0 ± 0.1 × 107 |
| Tat(44–61)ac | 3.3 ± 1 × 106 |
aFrom ref. (30).
Titrations were monitored by fluorescence anisotropy at 20°C in 25 mM Tris (pH 7.5), 30 mM NaCl and 0.2 mM MgCl2, as described in Supplementary Figure S1, Supplementary Data. The values of the observed binding constants, Kobs, were calculated by fitting the titrations curves with Equation (6), assuming ten independent and equivalent binding sites per cTAR molecule.
Although replacement of the Tyr47 residue by an Ala residue in the Tat(44–61)Y47A mutant resulted in a decrease of the Kobs value by a factor of about three, nearly no change in affinity accompanied its replacement with Trp in the Tat(44–61)Y47W mutant, suggesting that an aromatic residue at position 47 is required to stabilize the complex, likely through stacking with the bases of cTAR. Interestingly, removal of the six N-terminal amino acids [Tat(50–61)] only resulted in a marginal additional decrease of the Kobs value as compared with the Tat(44–61)Y47A mutant, indicating that the Tyr47 residue plays the most important role in the six N-terminal residues for stabilizing the complex of Tat(44–61) with cTAR. As Trp conservatively replaces Tyr in the Tat(44–61)Y47W mutant, and as Trp fluorescence properties are highly sensitive to stacking interactions with bases (52–54), we investigated the steady-state and time-resolved fluorescence properties of the conservative Tat(44–61)Y47W mutant to further characterize the binding mode of the Tyr47 residue with cTAR.
In the absence of cTAR, the wavelength of the emission maximum (351 nm) and the quantum yield (0.105) of Trp47 in Tat(44–61)Y47W were typical of those of fully solvent-exposed Trp residues in unfolded peptides (55). This conclusion was further substantiated by the similarity of the time-resolved fluorescence parameters of Tat(44–61)Y47W (Supplementary Table S1 in Supplementary Materials) with those of N-acetyl tryptophanamide in buffer (55). Addition of cTAR induced a dramatic 3.2-fold decrease in the quantum yield, associated with an only 1.37-fold decrease in the mean lifetime. The differential behavior of these two parameters clearly suggests the presence of a population of ‘dark’ species, with a lifetime shorter than the detection limit of our equipment (≈ 30 ps) and an α0 amplitude that could be calculated by:
| (2) |
where <τ>T and <τ>T-cT are the mean lifetime of Tat(44–61)Y47W in the absence and presence of cTAR, respectively, and Rm is the ratio of their quantum yields. An α0 value of 0.46 was obtained. In addition, a short-lived lifetime of 100 ps associated with an amplitude of 0.14 was also observed. Both the short-lived component and the dark species could be attributed to a stacking interaction of Trp47 with the cTAR bases (52–54). According to the amplitudes of these components, the Trp47 residue appears to be stacked in 60% of the Tat(44–61)Y47W/cTAR complexes, so that stacking is the dominant interaction mode for this residue. In the remaining 40% of the complexes, the lifetimes of Trp47 were only slightly smaller than those of the free peptides, suggesting that in these populations, the Trp47 residue exhibits only limited contacts with the cTAR bases.
In contrast to the N-terminal residues, deletion of the four C-terminal amino acids [Tat(44–57)] induced a modest increase of the Kobs value, suggesting a marginal contribution of the 58PPQG61 residues in the binding of the peptide to cTAR. Similarly, mutation of the Gln54 residue located within the cluster of basic residues to an Ala was only accompanied by a moderate decrease of Kobs (< 2-fold), indicating a limited role of this residue in the binding process. The contribution of the positively charged and highly conserved basic residues of Tat was also investigated. Double mutation of pairs of basic residues (Arg52 and Arg53, Arg55 and Arg56) to Ala residues or acetylation of Lys50 and Lys51 residues was accompanied by a 9- to 30-fold decrease of the Kobs value, indicating a major role of these residues in the stability of the cTAR/Tat(44–61) complexes. Interestingly, acetylation of Lys50 and Lys51 resulted in an even stronger decrease (∼30-fold) of the binding affinity than substitution of the (Arg52, Arg53) or (Arg55, Arg56) pairs by Ala residues, suggesting that in addition to the neutralization of charges, acetylation may further hinder the binding through a steric effect of the acetyl groups. Irrespective of its exact mechanism, acetylation appears efficient to reduce the affinity of Tat for its nucleic acid target. As previously described for the interaction with RNAs (12,33), ionic interactions of the basic residues with the negatively charged phosphate groups of cTAR may well explain their role in the stability of the cTAR/Tat(44–61) complex.
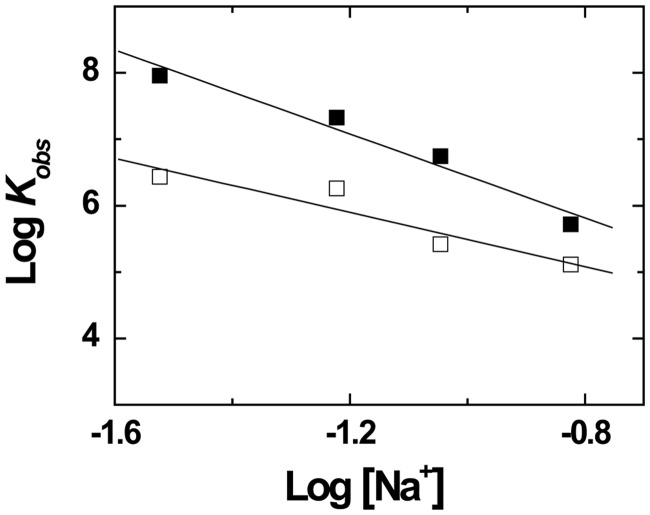
To further evaluate the contribution of ionic interactions to the binding of Tat (44–61) to cTAR, we analyzed the salt dependence of the Kobs values according to (56):
| (3) |
where K(1M) is the equilibrium association constant at 1 M Na+ (which represents the non-ionic contribution to the interaction), ΨNa+ is the fraction of cation thermodynamically bound per phosphate group and m′ is the number of ion pairs between the peptide and the oligonucleotide. As cTAR contains both single-stranded and double-stranded regions, we took for ψNa+, a value of 0.8, intermediate to the values reported for single-strand DNA (0.71) and double-strand DNA (0.88) (56). From the fit with Equation (3) of the linear dependence of logKobs on log[Na+], we obtained m′ = 4.0 and K(1M) = 1900 M−1 (Figure 2). The m’ value indicated that half of the basic residues of the Tat(44–61) peptide were involved in the binding to cTAR, whereas only one basic residue was involved in the binding of the Tat(46–60) peptide to its high-affinity binding site on TAR RNA (51). In contrast, application of Equation (3) to the binding data of Tat(46–60) to TAR RNA (51) revealed that its K(1M) value was about two orders of magnitude higher than the K(1M) value of Tat(44–61) for cTAR. As non-electrostatic interactions are a hallmark for specific binding (56), it appears that despite their comparable binding constants at 30 mM NaCl, the multiple binding sites of cTAR for the Tat peptides are of less specific nature than the high-affinity binding site of TAR.
Figure 2.
Salt dependence of the binding of Tat(44–61) and Tat(44–61)ac to cTAR. The binding experiments were performed by adding increasing concentrations of Tat(44–61) (closed squares) or Tat(44–61)ac (open squares) to 10 nM cTAR-3'-Fl in 25 mM Tris buffer (pH 7.5), 0.2 mM MgCl2 and the NaCl concentrations indicated on the X-axis. The binding process was monitored by steady-state fluorescence anisotropy, as described in ‘Materials and Methods’ and Supplementary Figure S1 in the Supplementary Materials. The apparent association constant, Kobs, for each titration was obtained using Equation (1). The solid lines correspond to the fits of the data points to Equation (3), using the parameters given in the text.
To get further insight in the involvement of Tat(44–61) basic residues in cTAR binding, we characterized the salt dependence of Kobs for the Tat(44–61)ac peptide. The obtained m′ value (2.6) was significantly lower than the value (4.0) for the native peptide, indicating that Lys50 and Lys51 residues contribute to the electrostatic interactions between the native peptide and cTAR. In contrast, the K(1M) value (2400 M−1) being close to that for the native peptide indicates that Lys50 and Lys51 residues do not significantly contribute to non-ionic interactions. Interestingly, the decrease of the m’ value by a value of <2 for the acetylated derivative, as well as the stabilization of the Tat(44–61)/cTAR complexes by only half of the basic residues of the peptide were somewhat in variance with the fact that replacement of any combination of two basic residues of Tat(44–61) by Ala residues strongly decreased the binding constant. This apparent contradiction suggests that due to the high flexibility of the peptide (57) and the highly heterogeneous structure of cTAR (that contains double-stranded regions, a central and an internal loop as well as bulges and mismatches), different combinations of four basic residues likely bind to the various cTAR binding sites.
Taken together, our data show that the Tat(44–61) domain binds to cTAR DNA with an affinity comparable with that for TAR RNA, suggesting that cTAR DNA is a likely target for Tat in the viral life cycle. Moreover, the clear relationship between the positively charged amino acids of the Tat(44–61) domain and the affinity of the peptide for cTAR evidenced a major role of electrostatic interactions in the binding process. A further stabilization can be achieved by the Tyr47 residue, mainly through stacking with the DNA bases. Finally, our data suggest that both the flexible structure of Tat and the redundancy of basic residues in its (44–61) domain allow Tat to adapt to a large number of DNA and RNA sequences, and thus to coat them efficiently.
Gel retardation analysis of the binding of Tat(44–61) derivatives to cTAR
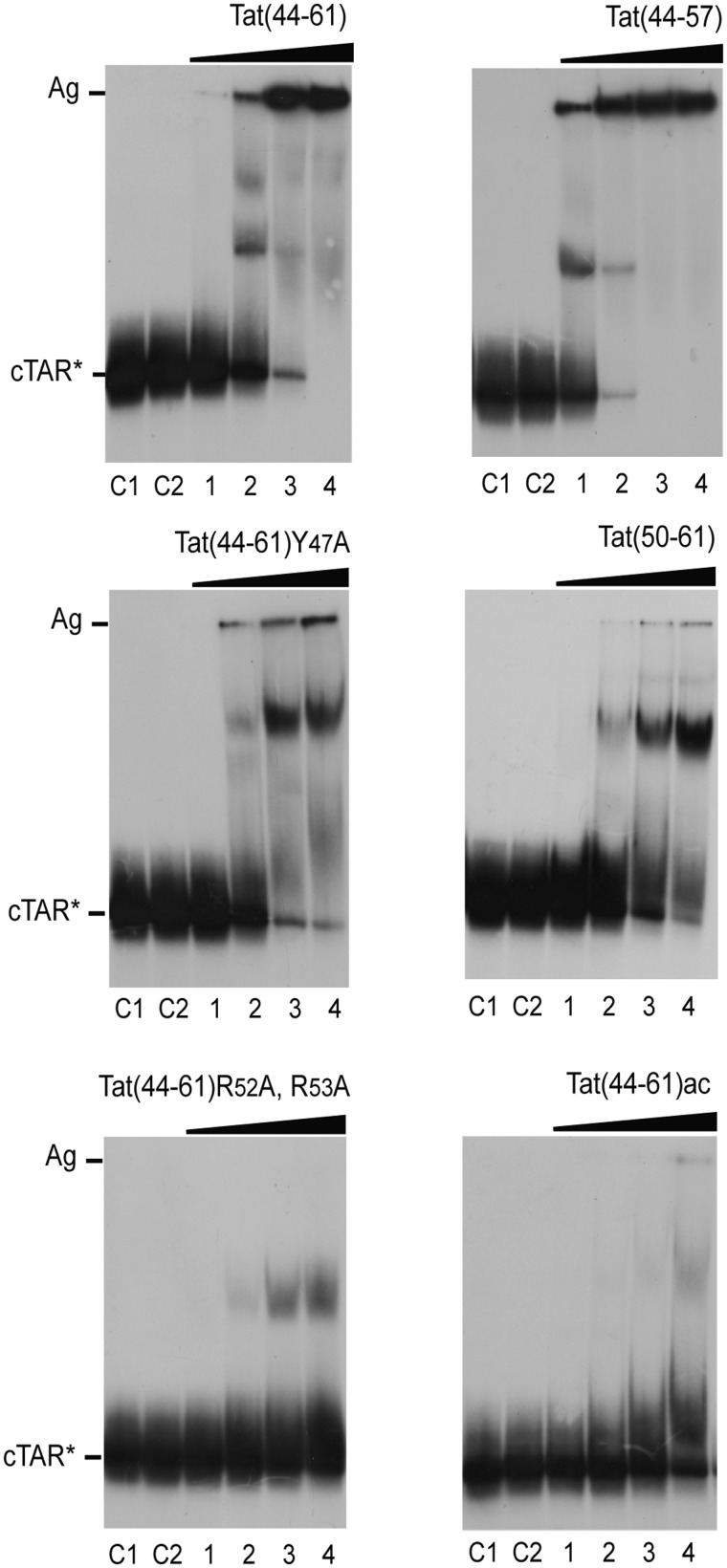
As the anisotropy technique does not allow discriminating the different species in solution, the binding properties of Tat(44–61), Tat(44–61)Y47A, Tat(44–57), Tat(50–61), Tat(44–61)ac and Tat(44–61)R52, R53A to cTAR were further investigated by gel retardation experiments (Figure 3). Controls C1 and C2 were used to locate the position of free cTAR DNA that has been heat-denatured in water and incubated in the binding buffer, respectively. The binding of the native Tat(44–61) peptide at a 1:2 ratio resulted in a decrease of the band of the free peptide to the benefit of a discrete band and smears with lower mobility as well as a band (Ag) in the wells that corresponds to high molecular weight peptide:cTAR complexes (aggregates) [Figure 3, Tat(44–61), lane 2]. In the latter, the cTAR molecules were likely largely coated by Tat(44–61) molecules, giving neutral complexes prone to aggregate. The discrete band corresponds probably to complexes where a limited number of Tat peptides are bound to cTAR. These data confirmed that cTAR contains several binding sites for Tat(44–61) and that complexes with various levels of coating by Tat(44–61) coexisted. Finally, at the highest peptide to nucleotide ratio (2:1), the free cTAR and the discrete band disappeared completely, so that most complexes were aggregates [Figure 3, Tat(44–61), lane 4]. Interestingly, the distributions of complexes with the Tat(44–57) derivative at the various peptide to nucleotide ratios were close to those of the wild-type peptide. Nevertheless, at the two lowest peptide to nucleotide ratios (compare in Figure 3, lanes 1 and 2 of the two peptides), a higher concentration of complexes as well as a lower concentration of free cTAR were observed with the Tat(44–57) derivative, indicating that this peptide shows a higher affinity to cTAR, as compared with the wild-type peptide. This higher affinity was also perceived in the binding parameters (Table 1) from the anisotropy titrations, but did not appear clearly, due to the rather large error bars.
Figure 3.
Gel retardation assays of the binding of the Tat peptides to cTAR DNA. The cTAR 32P-DNA was incubated in the absence or the presence of peptide and analyzed by electrophoresis on a 10% polyacrylamide gel as described in ‘Materials and Methods’. Lanes C1, heat-denatured cTAR DNA. Lanes C2, controls without peptide; lanes 1–4, peptide to nucleotide molar ratios were 1:4, 1:2, 1:1 and 2:1. The high molecular mass peptide:cTAR complexes (aggregates) are indicated by Ag.
In line with the anisotropy data, the binding patterns obtained with Tat(44–61)Y47A and Tat(50–61) were similar. In both cases, we observed a progressive accumulation of complexes with intermediate mobility and high molecular weight complexes with increasing peptide concentrations. The affinities of these two peptides for cTAR were clearly lower than those of the Tat(44–61) and Tat(44–57) peptides, as the concentrations of the high molecular weight complexes were much higher for the latter at the various peptide concentrations. Noticeably, the discrete band of intermediate mobility suggests an accumulation of a cTAR complex with a given number of Tat peptides. This observation is in variance with our assumption of identical and independent binding sites in anisotropy data, as this should have led to a distribution of complexes of various stoichiometries at sub-saturating concentrations of Tat peptides. Whether this discrete band corresponds to a cooperative binding of a few Tat peptides remains an open question.
In full line with their reduced affinities, Tat(44–61)R52A,R53A and Tat(44–61)ac did only produce limited amounts of complexes with intermediate mobility and nearly, no high molecular weight complexes, confirming the key roles of the basic residues of Tat in its affinity for cTAR. The absence of high molecular weight complexes could be due in part to the lower affinity of these peptides for cTAR, but it is also likely that all positive charges of the Tat peptide are needed to ensure the appropriate neutralization of the complexes with cTAR, and thus allow their aggregation. Moreover, comparison of the gels with the two peptides confirmed that Tat(44–61)ac is of lower affinity than Tat(44–61)R52A,R53A, as the former only induced a faint complex of intermediate mobility at the highest peptide to nucleotide ratio.
Taken together, these observations showed that the electrophoresis patterns of the various Tat derivatives are different and correlate well with their binding affinities determined by fluorescence anisotropy. The Tat derivatives associated with the highest binding constants form efficiently high molecular weight complexes, in which cTAR molecules are thought to be largely coated by Tat molecules. Though smears are observed in nearly all lanes, the discrete bands seen with all peptides suggest that cTAR molecules coated by a defined number of molecules can accumulate. The equilibrium between these complexes and the high molecular mass complexes that cannot enter the gel appeared finely tuned by the basic and Tyr residues of the Tat(44–61) sequence, as enlightened by the significant changes in the distribution of the complexes in the mutational binding pattern analysis.
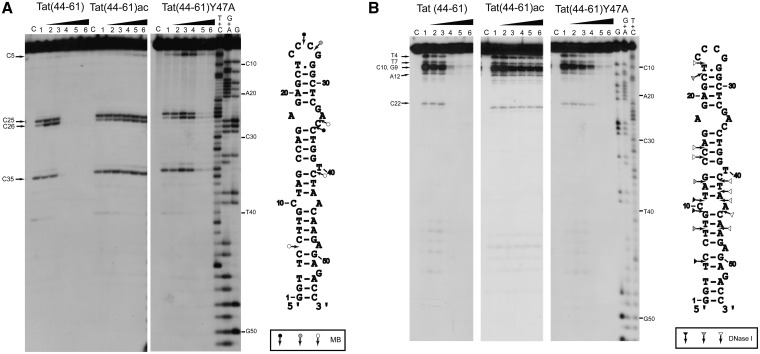
Analysis of cTAR protection and conformational changes on interaction with Tat peptides
Next, to further characterize the interaction of the Tat peptides with cTAR, we investigated the peptide-induced protection of cTAR to enzymatic digestion to identify the cTAR nucleotides coated by the peptides. To this end, we performed a footprinting analysis, using MB nuclease and DNase I. MB is highly selective for single-stranded sequences and domains (58), whereas DNase I is a double-strand-specific endonuclease that produces single-strand nicks (59). The protections induced by Tat(44–61) in the cTAR hairpin were identified by comparing the cleavage patterns of cTAR in the absence and in the presence of increasing concentrations of Tat(44–61) (Figure 4). Consistent with the ability of Tat(44–61) to efficiently coat and aggregate the cTAR molecules at high peptide to nucleotide molar ratios, we observed a complete abolishment of cTAR cleavage by MB (Figure 4A, lanes 4–6) and DNase I (Figure 4B, lanes 4–6) at peptide to nucleotide ratios ≥1:2. In contrast, the Tat(44–61)Y47A and Tat(44–61)ac peptides induced limited and nearly no protection, respectively, to the enzymes (Figure 4). Thus, the protection induced by the various peptides correlates well with their ability to induce high molecular weight complexes (Figure 3). At the lowest peptide to nucleotide molar ratios [≤1:4 for Tat(44–61) and ≤1:2 for Tat(44–61)Y47A], only modest changes in the accessibility to MB were observed at the level of C26 of the apical loop, and C5 of the lower part of the stem (Figure 4A). For DNase I, moderate but significant decreases in the accessibility to this enzyme were observed at the same peptide to nucleotide molar ratios for positions T4, T7, G9, C10 and A12, in the double-stranded regions of the lower part of cTAR (Figure 4B). The moderate protection observed at the lowest peptide to nucleotide ratios suggests that either the nucleotides targeted by the two enzymes may not be included in preferential binding sites for the Tat peptides or that Tat peptides can be rapidly out-competed by the enzymes from their binding sites. Moreover, the absence of additional bands in the presence of the various Tat peptides likely excludes that the Tat peptides induce major conformational changes in the cTAR molecules. Perhaps, the only exception is the significant increase in MB accessibility at the level of the C5 nucleotide, observed only for the Tat(44–61)Y47A peptide. Together with the decrease of DNaseI accessibility at the neighbor T4 nucleotide, the change of accessibility of C5 to MB suggests that Tat(44–61)Y47A may destabilize the penultimate double-strand segment of cTAR. However, this destabilization may not extend up to the cTAR termini, as no destabilization was observed by fluorescence techniques, using the doubly labeled TMR-5′-cTAR-3′-Fl (data not shown).
Figure 4.
Footprinting analysis of the complexes of cTAR with Tat peptides using MB nuclease (A) and DNase I (B). Footprinting experiments were performed as described in ‘Materials and Methods’. The 3′-end-labeled cTAR DNA was incubated with MB (2 units) or DNase I (0.1 unit) in the absence (lanes 1) or in the presence of the peptides (lanes 2–6). The peptide to nucleotide molar ratios were 1:8 (lanes 2), 1:4 (lanes 3), 1:2 (lanes 4) 1:1 (lanes 5) and 2:1 (lanes 6). Lanes C are controls without peptide and endonuclease. The G, G+A and T+C refer to Maxam–Gilbert sequence markers of cTAR run in parallel to identify the cleavage sites. Arrows indicate the cleavage sites. Closed, gray and open symbols indicate strong, medium and weak cleavage sites in the absence of peptide, respectively.
In a next step, we monitored the peptide-induced conformational changes of cTAR, using the cTAR Lai sequence where A residues were substituted by 2-aminopurine (2-Ap), a fluorescent analog of Adenine (60), at positions 9, 17, 21, 28, 35, 45, 49, 53 and 55 (61). Due to the exquisite sensitivity of 2-Ap fluorescence to its local environment (62,63), site-specific characterization of the structural modifications within the cTAR hairpin sequence can be determined. The Tat(44–61) peptide induced significant changes in the 2-Ap quantum yield, only at position 9 and 49, where increases of ∼ 100 and 200%, respectively, were observed (Supplementary Figure S2, Supplementary Materials). Nevertheless, these changes were significantly less that those observed with NC(11–55) (61), a peptide that corresponds to the zinc-bound finger domain of NCp7 and that shows nucleic acid destabilizing and annealing activities (47,64). In contrast, the Tat(44–61)-induced changes in 2-Ap quantum yield were similar to those induced by the unfolded (SSHS)2NC(11–55) peptide, that acts as a nucleic acid annealer, being defective in cTAR destabilization (49) but efficient in promoting cTAR/dTAR annealing (61). Therefore, by analogy to the (SSHS)2NC(11–55) peptide, the basic amino acids of Tat(44–61) peptide are thought to interact mainly through electrostatic interactions with the DNA phosphate groups, so that the structure of the major part of cTAR is only marginally affected by Tat(44–61), in line with the footprinting data. Moreover, the changes in 2-Ap quantum yield at positions 9 and 49 are consistent with a peptide-induced base tilting, as was reported for arginine/lysine oligopeptides (65) and Tat(44–61) (33,66)
with other DNA or RNA sequences. As both residues are in the lower part of the cTAR stem that plays a key role in cTAR/dTAR annealing (31), the tilting of these two residues is thought to favor the intermolecular base pairings required for nucleating the cTAR/dTAR duplex.
Identification of the determinants of the nucleic acid annealing activity of Tat(44–61)
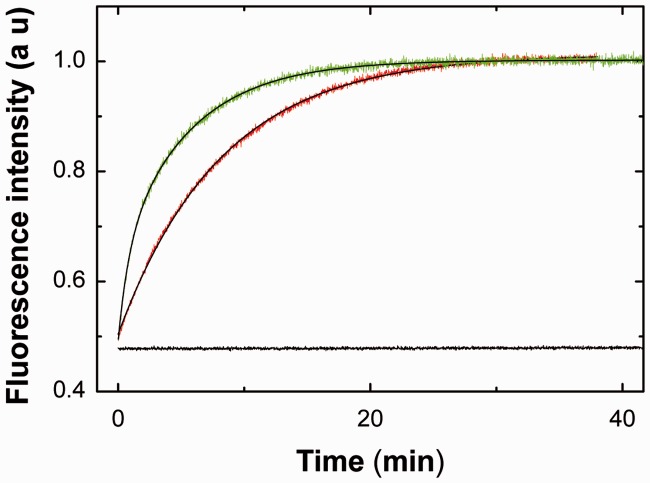
As a preliminary step in the identification of the determinants of the Tat(44–61) domain in its nucleic acid annealing activity during RTion, we investigated whether the Tat(44–61) peptide is able to direct the annealing of the TAR RNA and cTAR DNA sequences involved in the first strand transfer. To this end, we used a previously reported annealing assay based on the fluorescence restoration of a doubly labeled TMR-5′-cTAR-3′-Fl sequence (47,67). In contrast to NC(11–55), used as a positive control (Figure 5, red trace), no significant fluorescence enhancement could be detected during the time course of the experiment in the presence of 3 molecules of Tat(44–61) per oligonucleotide, showing that Tat(44–61) was unable to promote the annealing of TAR RNA to the doubly labeled cTAR (Figure 5, black trace). The inability of Tat(44–61) to promote cTAR/TAR annealing was confirmed by gel electrophoresis, using radiolabeled RNA (data not shown). In contrast, when TAR RNA was replaced by dTAR, the DNA equivalent of TAR RNA, the annealing reaction was fully achieved in ∼1500 s (Figure 5, green curve). These data can be explained by the fact that the promotion of TAR/cTAR annealing involves a critical destabilization step (61) that can be induced by NC(11–55) (68,69) but not by Tat(44–61) (31). Moreover, in contrast to TAR, the existence of a small amount of thermally melted and reactive dTAR at 20°C (64,68,70,71) can facilitate its annealing with cTAR, already in the absence of peptide.(47) As a consequence, the strong acceleration of the annealing reaction between cTAR and dTAR but not TAR by Tat(44–61) confirms that this peptide promotes only annealing reactions that can already occur in the absence of peptide. Therefore, Tat(44–61) is confirmed as a nucleic acid annealer rather than a true chaperone (33).
Figure 5.
Evidence that Tat(44–61) does not promote cTAR/TAR RNA annealing in vitro. The 10 nM TMR-5′-cTAR-3′-Fl was reacted with 100 nM TAR RNA (black trace) or with 100 nM dTAR (green trace) using a peptide to oligonucleotide molar ratio of 3. For comparison, cTAR/TAR RNA annealing was also monitored in the presence of NC(11–55) at a molar ratio of 11 peptides per oligonucleotide (red trace). The solid lines correspond to the best fit of the kinetic curves to Equation (4).
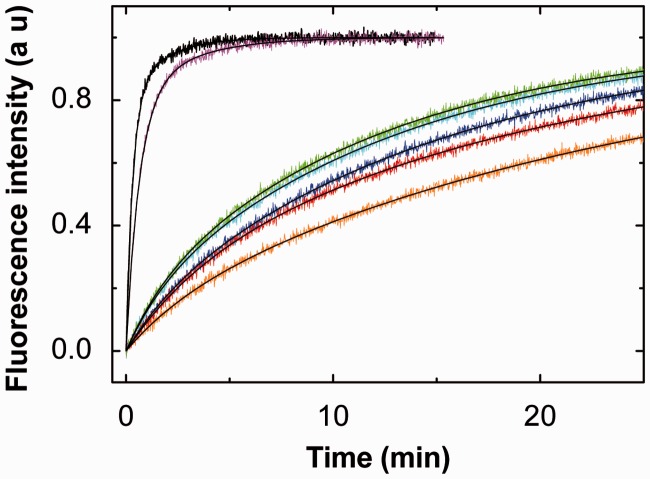
To investigate the determinants of the nucleic acid annealing properties of Tat(44–61), we characterized the cTAR/dTAR annealing reaction in the presence of the various Tat(44–61) mutants. As Tat is probably present at low concentrations during RTion both in the virus and the cytosol of infected cells, the annealing reactions were examined at the low molar ratio value of three peptides per oligonucleotide. Moreover, this low ratio corresponds also to aggregation-free conditions (31), which are critical to avoid artifacts in the fluorescence techniques. For all the tested mutants, we observed a large fluorescence increase resulting from the conversion of the doubly labeled cTAR stem-loop to the final cTAR/dTAR duplex (Figure 6). The fluorescence intensity reached a final plateau value in 400–8000 s, depending on the peptide. The plateau value by itself was not influenced by the dTAR concentration or by the peptide nature, indicating that the doubly labeled cTAR was always fully converted to an extended duplex (ED) that cannot be dissociated by the peptides. As cTAR/dTAR annealing is a long lasting process in the absence of peptide, taking more than one day at room temperature (47), the kinetic curves (Figure 6) clearly indicated that all tested peptides promoted annealing, although with different efficiency. As reported for the Tat(44–61)-directed annealing (30,31), all traces were best described by a double exponential function:
| (4) |
where I(t) is the fluorescence intensity at time t, kobs1 and kobs2 are the apparent pseudo first-order rate constants governing the two kinetic components, a is the relative amplitude of the fast component, and t0 is a dead time. I0 and If stand for the fluorescence intensities of TMR-5′-cTAR-3′-Fl in its free form and in the final ED, respectively. The If, I0 and t0 were determined independently.
Figure 6.
Kinetics of cTAR/dTAR annealing in the presence of Tat(44–61) derivatives. The annealing of TMR-5′-cTAR-3′-FL (10 nM) with dTAR (500 nM) was monitored in 25 mM Tris buffer (pH 7.5), 0.2 mM MgCl2, 30 mM NaCl, in the presence of Tat(44–61) and its various derivatives used at a molar ratio of three peptides per oligonucleotide. Black trace: Tat(44–61), violet: Tat(44–57), green: Tat(44–61)Y47A, cyan: Tat(44–61)R52A,R53A, blue: Tat(44–61)R55A,R56A, red: Tat(44–61)ac and orange: Tat(50–61). The kinetic traces of Tat(44–61)Q54A and Tat (44–61)Y47W were nearly superimposable with the Tat(44–61) one, and thus were not represented. The solid lines correspond to the best fit of the kinetic curves to Equation (4).
For most of the Tat(44–61) peptides, the fast component kobs1 linearly varies with [dTAR], whereas kobs2 shows a hyperbolic dependence on [dTAR]. This suggests that the various peptides likely promote the formation of an intermediate complex (IC) that further converts into the final ED, in a two-step reaction scheme (Scheme 1) similar to that of the native peptide (31):
Scheme 1.
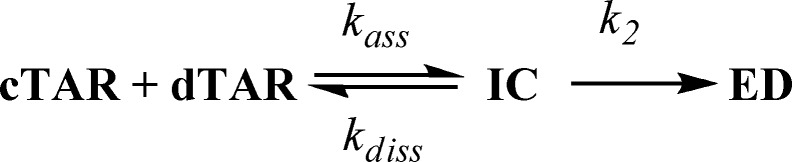
Mechanism for the Tat(44–61)-promoted cTAR/dTAR annealing.
where kass is the second-order association rate constant for the formation of IC, and kdiss and k2 are the first-order rate constants for the dissociation of IC and for its conversion into ED, respectively. The values of the different parameters (Table 2) were determined using the Dynafit numerical resolution software (48), which allows to simultaneously fit the experimental progress curves obtained at different dTAR concentrations. These values were in excellent agreement with those obtained by fitting the kobs1 and kobs2 dependence on the dTAR concentration (data not shown), giving good confidence in the retrieved values and the proposed reaction scheme. Comparison of (i) the kinetic curves (Figure 6), (ii) the kass, kdiss and k2 values (Table 2) and iii) the gel electrophoresis data of the various peptides in cTAR/dTAR annealing (Supplementary Figure S3, Supplementary Data) clearly revealed the existence of two classes of peptides. These two classes could not be related to differences in the number of bound peptides per oligonucleotide, because based on the binding data of Table 1 and the experimental conditions used for the annealing experiments, we calculated that at least 80% of the added peptides were bound in these experiments. The first class, composed of Tat(44–61), Tat(44–57), Tat(44–61)Y47W and Tat(44–61)Q54A was characterized by high values for both the kinetic association (kass = 0.41 − 1.2 × 105 M−1s−1) and dissociation rate constants (kdiss = 1.5 – 2.0 × 10−2 s−1) that were respectively up to 48- and 9-fold higher than the corresponding values of the peptides of the second class [Tat(50–61), Tat(44–61) R52A,R53A, Tat(44–61) R55A,R56A, Tat(44–61)ac and Tat(44–61)Y47A]. Thus, efficient cTAR/dTAR annealing requires the interactions between the peptide-coated reactants to be highly dynamic. Interestingly, the kass values of the first class of peptides were comparable with those observed for the NCp7-promoted cTAR/TAR annealing at nearly saturating concentrations of NCp7 (72), which likely allow the oligonucleotides to rapidly explore various interaction modes, and thus find the appropriate one that will nucleate the ED. Moreover, the high kdiss values likely avoid cTAR and dTAR to be kinetically trapped in misfolded ICs that would be unable to lead to the ED. As the full length Tat was reported to exhibit kinetic rate constants close to those of Tat(44–61) in promoting the cTAR/dTAR annealing (31), these highly dynamic interactions constitute likely also a major component of the annealer properties of Tat. Noticeably, as the decrease in the kass values for the second class of peptides as compared with the first class was partly compensated by their decreased kdiss values, the values of the IC equilibrium constant, Ka, were comparable for both classes, suggesting that the IC stability per se does not constitute a key factor in the different annealing properties of the two classes.
Table 2.
Parameters for the cTAR fret/dTAR annealing in the presence of Tat(44–61) and its variants at 20°Ca
| Parameter | Tat(44–61) | Tat(44–57) | Tat(44–61) Y47W | Tat(44–61) Q54A | Tat(50–61) | Tat(44–61) R52A, R53A | Tat(44–61) R55A, R56A | Tat(44–61)ac | Tat(44–61) Y47A |
|---|---|---|---|---|---|---|---|---|---|
| kass (M−1.s−1 × 10−5) | 1.2b | 0.8 | 1.1 | 0.41 | 0.025 | 0.036 | 0.031 | 0.025 | 0.039 |
| kdiss (s−1 × 100) | 1.6b | 2 | 1.5 | 1.9 | 0.38 | 0.29 | 0.22 | 0. 40 | 0.24 |
| k2 (s−1 × 100) | 1.3b | 2.2 | 1 | 1.3 | 0.59 | 0.13 | 0.17 | 0.46 | 0.56 |
| Ka (M−1 × 10−6)c | 7.5 | 4 | 7.3 | 2.2 | 0.66 | 1.2 | 1.4 | 0.65 | 1.6 |
| ΔG‡ (kcal.mol−1)d | 6.3 | 6.6 | 6.4 | 7 | 8.6 | 8.4 | 8.5 | 8.6 | 8.3 |
| ΔG nuc (kcal.mol−1)e | 5.2 | 5.5 | 5.3 | 5.9 | 7.5 | 7.3 | 7.4 | 7.5 | 7.2 |
| Activity in RTionf | 100 | 100 | 100 | ∼30 | ∼30 | 35 | 20 |
aThe annealing reactions were performed as described in Figure 6. The values of the kinetic parameters, kass, kdiss and k2 were calculated by Dynafit from the experimental progress curves on the basis of a two-step mechanism.
bValues from (31).
ckass/kdiss ratio.
dcalculated using ΔG‡ = −RTln(kass/kdiff) with kdiff = 6.5 × 109 M−1s−1.70.
eΔGnuc values were calculated from Equation (5).
Peptides of the first class were also characterized by k2 values (1.3–2.2 × 10−2 s−1) that are ∼2- to 16-fold higher than the k2 values of the second class, indicating that they more efficiently convert the IC into the final ED. Moreover, comparison of the kinetic parameters of the two classes further revealed that the Gln54 residue and the 58PPQG61 sequence of Tat(44–61) marginally contribute to the annealing activity of the peptide. In sharp contrast, both the basic residues and the Tyr47 residue were critical for efficient cTAR/dTAR annealing by contributing both to the highly dynamic interaction between the oligonucleotides and the efficient interconversion of the IC into the ED. Moreover, although replacement of Tyr47 with Ala led to strong changes of the kinetic parameters, its replacement with Trp preserved the full activity, indicating that the aromatic nature of the residue at position 47 is crucial for nucleic acid annealing.
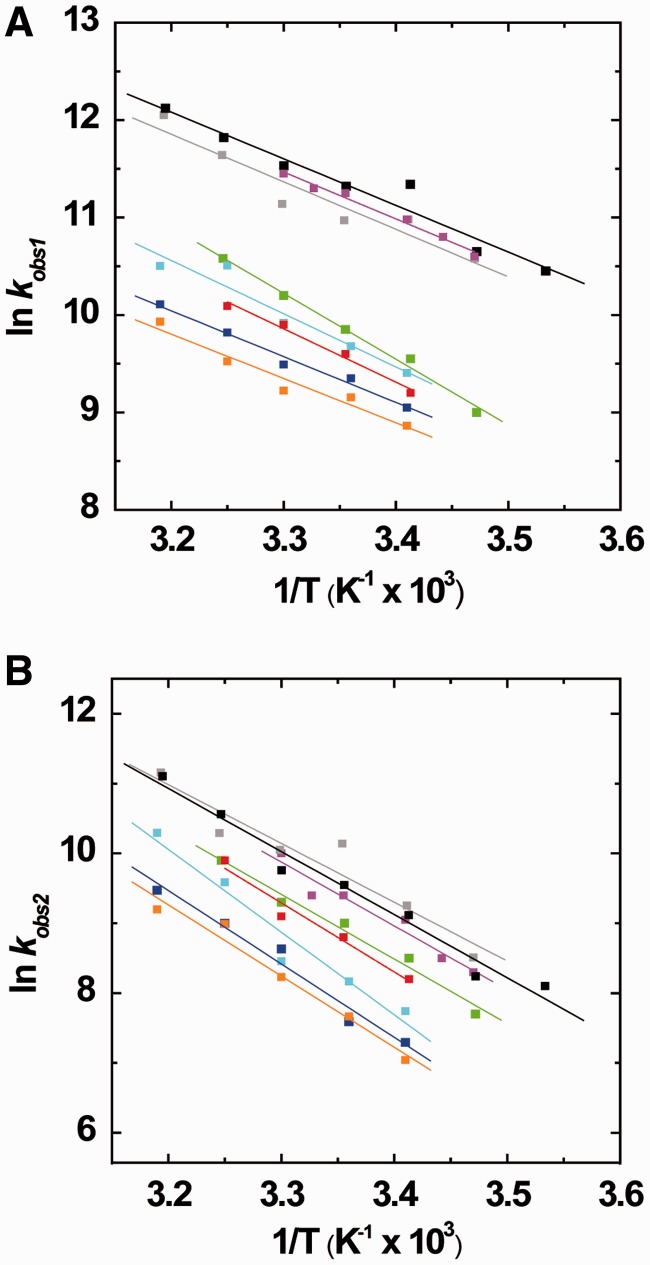
To further dissect the effect of the Tat mutants on the cTAR/dTAR annealing reaction, we plotted the dependence of the kobs1/2 values on the temperature in the range 5–50°C through an Arrhenius plot (Figure 7). According to the Arrhenius model, the transition state thermodynamic parameters can be derived from the reaction rates using:
| (5) |
where the rate constant k1/2 is given by kobs1/2/[cTAR], A is a pre-exponential factor, Ea is the activation energy, R the universal gas constant and T is the absolute temperature in Kelvin. Mutations and truncations of the Tat(44–61) peptide marginally influenced the slopes of the straight lines fitted to the data in Figure 7A (fast component) and 7B (slow component). The values of Ea and the corresponding transition state enthalpy ΔH‡ (Supplementary Table S2, Supplementary Materials) showed that the different peptides comparatively influenced the energy barriers for the transition states of the cTAR/dTAR annealing reaction. As previously reported (31), the ΔH‡ values of ∼10 and 18 kcal.mol−1 for the fast and slow components, respectively, indicate that the cTAR/dTAR annealing promoted by the Tat peptides involves premelting of 2–3 base pairs for the fast component and 4–5 base pairs for the slow component.
Figure 7.
Arrhenius analysis of the annealing of cTAR to dTAR in the presence of Tat (44–61) derivatives. Temperature dependence of kobs1 (A) and kobs2 (B) for the reaction of 10 nM TMR-5′-cTAR-3′-FL with 100 nM dTAR in the presence of Tat(44–61) (black), Tat(44–61)Q54A (gray), Tat(44–57) (magenta), Tat(44–61)Y47A (green), Tat(44–61)R52A,R53A (cyan), Tat R55A,R56A (blue), Tat(50–61) (orange) or Tat(44–61) acetylated (red). The straight lines represent the best fits of the data to Equation (5).
To further delineate the differences between the two classes of Tat peptides in their ability to promote cTAR/dTAR annealing, the kass parameter, which is largely responsible for the different behaviors of the two classes, was expressed as (73):
| (6) |
where kdiff (∼6.5 × 109 M−1s−1) is the theoretical diffusional collision rate of annealing molecules (73,74), whereas exp(−ΔGm/RT) and exp(−ΔGnuc/RT) reflects the probability of the colliding molecules to be in the melted reactive species and the probability of the collision to be productive, and thus nucleate the IC, respectively. As none of the Tat peptides could destabilize the cTAR termini (data not shown), the probability of the cTAR termini to be in a melted reactive form was set to the 0.15 value of the equilibrium dissociation constant that results from thermal fraying (68). From this value and Equation (6), we deduced for Tat(44–61) and the peptides of the first class, a ΔGnuc value of ∼5 kcal.mol−1. This value for the transition free energy for nucleation is significantly smaller than the >7 kcal.mol−1 values for the peptides of the second class, indicating that the peptides from the first class substantially increase the probability of productive collisions for nucleating the IC. These efficient productive collisions likely result from the ability of the basic residues of Tat(44–61) to efficiently screen the negatively charged cTAR and dTAR sequences and create short range interactions between them, as well as from the base tilting promoted by Tyr47 stacking that probably favors intermolecular base pairing between the complementary sequences.
Finally, our objective was to correlate our kinetic and thermodynamic data with previously reported RTion data obtained using HIV-1 viruses prepared from cell lines expressing HIV-1 Δtat viruses (in which the tat gene has been functionally deleted) and stably transcomplemented with wild-type or mutant Tat proteins (42). The ability of these Tat proteins to restore RTion in HIV-1 Δtat viruses was evaluated in natural endogenous RTion assays, comparing HIV-1 viruses transcomplemented with Tat proteins mutated in the (44–57) domain relatively to viruses transcomplemented with the wild-type Tat protein. Interestingly, a perfect correlation was observed between the efficiency of Tat mutants to promote RTion and their ability to promote fast association and dissociation of the IC (Table 2). A dramatic decrease in RTion efficiency was seen with mutation of Tyr47 in Ala and replacement of pairs of basic residues by Ala, whereas in contrast, replacement of Gln54 with Ala or replacement of Tyr47 with Trp had no effect on Tat-promoted RTion.
Taken together, our data show that the efficiency of the Tat peptides to promote cTAR/dTAR annealing is associated with their ability to favor highly dynamic interactions between the complementary oligonucleotides to nucleate the IC. Moreover, the ability of a given Tat mutant to promote these dynamic interactions closely correlates with its ability to promote RTion, suggesting that it constitutes a key feature of the annealer properties of Tat.
CONCLUSION
In this work, we showed that the basic (44–61) domain of Tat binds to cTAR DNA with an affinity comparable with that for TAR RNA, suggesting that cTAR DNA is a likely target for Tat in the viral life cycle. This strong binding affinity was found to be mediated through electrostatic interactions between on the average 4 of the 8 basic residues of the Tat(44–61) domain and the DNA phosphate groups as well as through a stacking of the Tyr47 residue with the DNA bases. Interestingly, our data further suggest that both the high flexibility of Tat and the presence of more basic residues than needed for tight binding enable Tat to bind to nearly any nucleic acid sequence, by adjusting its structure and selecting the most appropriate combination of basic residues. Moreover, the Tat basic domain was found to induce only limited conformational changes in cTAR, with the exception of nucleotides in the lower part of the cTAR stem that are thought to be tilted by the Tat(44–61) domain, and thus become annealing competent. As similar observations were reported for the binding of the fingerless and unfolded (SSHS)2NC(11–55) derivative to cTAR (61) and for the binding of the basic domains of the core protein of the Hepatitis C virus to its nucleic acid targets (75), it is likely that these basic nucleic acid annealers share common binding modes and mechanisms.
One major aspect of this work is the demonstration that efficient cTAR/dTAR annealing requires highly dynamic interactions between the peptide-coated oligonucleotides, probably to efficiently nucleate the IC and prevent trapping of misfolded intermediates. As the Tat(44–61) peptide and the full-length Tat protein induce similar cTAT/dTAR kinetics, these highly dynamic interactions are believed to be also promoted by the native Tat protein and constitute a general property of basic nucleic acid annealers. Structure–activity relationship studies further indicated that these dynamic interactions are supported by the basic residues and the Tyr47 residue of the (44–61) domain, and that they correlate well with the ability of Tat to promote RTion (42). As annealing between complementary DNA sequences occurs during the late steps of RTion, our data suggest that Tat may intervene as a nucleic acid annealer role in these late steps. This conclusion is in line with the much higher (30-fold) defect in the synthesis of full-length DNA as compared with the first strand (10-fold) observed with Δtat HIV-1 virions deleted of the tat gene (22), indicating a probable role of Tat during the late steps of RTion. Moreover, due to its significantly lower affinity for double-stranded DNA as compared with single-stranded DNA and RNA (76), NCp7, the major nucleic acid chaperone in RTion is thought to partially dissociate from the nucleic acids during the late steps of RTion (77–79), so that Tat may partly replace NC during these late steps. This hypothesis is further reinforced by the fact that the two proteins were shown to not only exhibit comparable annealing activities but also to act synergistically (31). Thus, together with the likely dissociation of NC from double-stranded DNA due to its low affinity for this substrate, the synergistic annealing activity of the two proteins reinforces the hypothesis that Tat may play a major role in the presence of residual NC molecules during the late steps of RTion. Furthermore, our data indicate that the Tat(44–61) peptide is unable to promote the annealing of cTAR with TAR RNA that occurs during the first obligatory strand transfer of RTion. Again, these data are consistent with the fact that similar defects were observed for the synthesis of the first strand and the (−) strong stop DNA with Δtat HIV-1 virions, indicating that in contrast to NCp7 [for review, see refs (69,80,81)], Tat probably plays a marginal role in the first strand transfer. The inability of the Tat(44–61) peptide to promote cTAR/TAR annealing is reminiscent of the poor efficiency of the (SSHS)2NC(11–55) peptide to promote the same reaction (61), suggesting that the absence of nucleic acid destabilizing component in annealers likely prevents them to promote annealing reactions involving stable sequences, such as TAR RNA. Moreover, as single-point mutations of the Tyr47 residue were shown to be unable to affect the activity of Tat in transcription of the integrated HIV viral genome (42) as well as the ability of the Tat basic domain to promote the annealing of two short single-stranded RNA sequences (33), it is believed that Tat exhibits different interaction modes and mechanisms on RNA and DNA sequences. Finally, the efficient decrease in the binding affinity to cTAR and the strong decrease in the Tat-promoted cTAR/dTAR kinetics induced by acetylation of Lys50 and Lys51 residues indicate that this in vivo post-translational modification of Tat (34,82) may efficiently down-regulate the role of Tat in RTion.
As active Tat peptides are responsible for highly dynamic interactions between oligonucleotides, it is likely that these peptides have to be in fast and transient equilibrium with their target nucleic acids as well. Such a fast nucleic acid binding and dissociation was described for NCp7, and shown to be a key component of the nucleic acid chaperone properties of this protein (83). Moreover, by analogy to the Tat(44–61) peptide, the aromatic residues of NCp7 were found essential in these fast kinetics (84). Thus, it may be speculated that this rapid binding and dissociation may also be a hallmark of Tat(44–61), in particular, and nucleic acid annealers, in general. This point is under current investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [85–87].
FUNDING
Agence Nationale de la Recherche [ANR-10-BLAN-1529 to Y. M.] and Agence Nationale de Recherches sur le Sida et les hépatites virales (to O. M., J. G. and Y. M.). Funding for open access charges: Centre National de la Recherche Scientifique (CNRS) and University of Strasbourg.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Anny Wund, Fanny Courte, Roman Storchak, Pauline Rémy and Florian Moser for their technical help.
REFERENCES
- 1.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 3.Feng S, Holland EC. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 4.Weeks KM, Ampe C, Schultz SC, Steitz TA, Crothers DM. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990;249:1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- 5.Churcher MJ, Lamont C, Hamy F, Dingwall C, Green SM, Lowe AD, Butler JG, Gait MJ, Karn J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J. Mol. Biol. 1993;230:90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- 6.Jones KA. HIV trans-activation and transcription control mechanisms. New Biol. 1989;1:127–135. [PubMed] [Google Scholar]
- 7.Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 8.Jeang KT. Tat, Tat-associated kinase, and transcription. J. Biomed. Sci. 1998;5:24–27. doi: 10.1007/BF02253352. [DOI] [PubMed] [Google Scholar]
- 9.Jeang KT, Xiao H, Rich EA. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 10.Karn J. Tackling tat. J. Mol. Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- 11.Selby MJ, Peterlin BM. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990;62:769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- 12.Calnan BJ, Biancalana S, Hudson D, Frankel AD. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 13.Weiss MA, Narayana N. RNA recognition by arginine-rich peptide motifs. Biopolymers. 1998;48:167–180. doi: 10.1002/(SICI)1097-0282(1998)48:2<167::AID-BIP6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Puglisi JD, Chen L, Frankel AD, Williamson JR. Role of RNA structure in arginine recognition of TAR RNA. Proc. Natl Acad. Sci. USA. 1993;90:3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Arginine-mediated RNA recognition: the arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 16.Meredith LW, Sivakumaran H, Major L, Suhrbier A, Harrich D. Potent inhibition of HIV-1 replication by a Tat mutant. PLoS One. 2009;4:e7769. doi: 10.1371/journal.pone.0007769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell. 2002;10:585–597. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 18.Berro R, Kehn K, de la Fuente C, Pumfery A, Adair R, Wade J, Colberg-Poley AM, Hiscott J, Kashanchi F. Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32. J. Virol. 2006;80:3189–3204. doi: 10.1128/JVI.80.7.3189-3204.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charnay N, Ivanyi-Nagy R, Soto-Rifo R, Ohlmann T, López-Lastra M, Darlix J-L. Mechanism of HIV-1 Tat RNA translation and its activation by the Tat protein. Retrovirology. 2009;6:74. doi: 10.1186/1742-4690-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennasser Y, Jeang K-T. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology. 2006;3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennasser Y, Le S-Y, Benkirane M, Jeang K-T. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Harrich D, Ulich C, García-Martínez LF, Gaynor RB. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulich C, Dunne A, Parry E, Hooker CW, Gaynor RB, Harrich D. Functional domains of Tat required for efficient human immunodeficiency virus type 1 reverse transcription. J. Virol. 1999;73:2499–2508. doi: 10.1128/jvi.73.3.2499-2508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apolloni A, Meredith LW, Suhrbier A, Kiernan R, Harrich D. The HIV-1 Tat protein stimulates reverse transcription in vitro. Curr. HIV Res. 2007;5:473–483. doi: 10.2174/157016207781662443. [DOI] [PubMed] [Google Scholar]
- 25.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lori F, di Marzo Veronese F, de Vico AL, Lusso P, Reitz MS, Jr, Gallo RC. Viral DNA carried by human immunodeficiency virus type 1 virions. J. Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Dornadula G, Pomerantz RJ. Natural endogenous reverse transcription of HIV-1. J. Reprod. Immunol. 1998;41:255–260. doi: 10.1016/s0165-0378(98)00062-x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Dornadula G, Pomerantz RJ. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J. Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kameoka M, Morgan M, Binette M, Russell RS, Rong L, Guo X, Mouland A, Kleiman L, Liang C, Wainberg MA. The Tat protein of human immunodeficiency virus type 1 (HIV-1) can promote placement of tRNA primer onto viral RNA and suppress later DNA polymerization in HIV-1 reverse transcription. J. Virol. 2002;76:3637–3645. doi: 10.1128/JVI.76.8.3637-3645.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuciak M, Gabus C, Ivanyi-Nagy R, Semrad K, Storchak R, Chaloin O, Muller S, Mély Y, Darlix JL. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boudier C, Storchak R, Sharma KK, Didier P, Follenius-Wund A, Muller S, Darlix JL, Mély Y. The mechanism of HIV-1 Tat-directed nucleic acid annealing supports its role in reverse transcription. J. Mol. Biol. 2010;400:487–501. doi: 10.1016/j.jmb.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 32.Godet J, Boudier C, Humbert N, Ivanyi-Nagy R, Darlix J-L, Mély Y. Comparative nucleic acid chaperone properties of the nucleocapsid protein NCp7 and Tat protein of HIV-1. Virus Res. 2012;169:349–360. doi: 10.1016/j.virusres.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doetsch M, Fürtig B, Gstrein T, Stampfl S, Schroeder R. The RNA annealing mechanism of the HIV-1 Tat peptide: conversion of the RNA into an annealing-competent conformation. Nucleic Acids Res. 2011;39:4405–4418. doi: 10.1093/nar/gkq1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, et al. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaehlcke K, Dorr A, Hetzer-Egger C, Kiermer V, Henklein P, Schnoelzer M, Loret E, Cole PA, Verdin E, Ott M. Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol. Cell. 2003;12:167–176. doi: 10.1016/s1097-2765(03)00245-4. [DOI] [PubMed] [Google Scholar]
- 36.Col E, Caron C, Seigneurin-Berny D, Gracia J, Favier A, Khochbin S. The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator. Tat. J. Biol. Chem. 2001;276:28179–28184. doi: 10.1074/jbc.M101385200. [DOI] [PubMed] [Google Scholar]
- 37.Ott M, Schnölzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol. 1999;9:1489–1492. doi: 10.1016/s0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- 38.Deng L, de la Fuente C, Fu P, Wang L, Donnelly R, Wade JD, Lambert P, Li H, Lee CG, Kashanchi F. Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology. 2000;277:278–295. doi: 10.1006/viro.2000.0593. [DOI] [PubMed] [Google Scholar]
- 39.Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 1998;273:24898–24905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- 40.Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- 41.Hooker CW, Scott J, Apolloni A, Parry E, Harrich D. Human immunodeficiency virus type 1 reverse transcription is stimulated by tat from other lentiviruses. Virology. 2002;300:226–235. doi: 10.1006/viro.2002.1554. [DOI] [PubMed] [Google Scholar]
- 42.Apolloni A, Hooker CW, Mak J, Harrich D. Human immunodeficiency virus type 1 protease regulation of tat activity is essential for efficient reverse transcription and replication. J. Virol. 2003;77:9912–9921. doi: 10.1128/JVI.77.18.9912-9921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Rocquigny H, Ficheux D, Gabus C, Fournié-Zaluski MC, Darlix JL, Roques BP. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 1991;180:1010–1018. doi: 10.1016/s0006-291x(05)81166-0. [DOI] [PubMed] [Google Scholar]
- 44.Kuipers BJH, Gruppen H. Prediction of molar extinction coefficients of proteins and peptides using UV absorption of the constituent amino acids at 214 nm to enable quantitative reverse phase high-performance liquid chromatography-mass spectrometry analysis. J. Agric. Food Chem. 2007;55:5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- 45.Kanevsky I, Chaminade F, Chen Y, Godet J, René B, Darlix JL, Mély Y, Mauffret O, Fossé P. Structural determinants of TAR RNA-DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2011;39:8148–8162. doi: 10.1093/nar/gkr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Meth. Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 47.Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix JL, Mély Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 48.Kuzmic P. DynaFit—a software package for enzymology. Meth. Enzymol. 2009;467:247–280. doi: 10.1016/S0076-6879(09)67010-5. [DOI] [PubMed] [Google Scholar]
- 49.Beltz H, Clauss C, Piémont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix JL, de Rocquigny H, Mély Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 50.Egelé C, Piémont E, Didier P, Ficheux D, Roques B, Darlix JL, de Rocquigny H, Mély Y. The single-finger nucleocapsid protein of moloney murine leukemia virus binds and destabilizes the TAR sequences of HIV-1 but does not promote efficiently their annealing. Biochemistry. 2007;46:14650–14662. doi: 10.1021/bi7012239. [DOI] [PubMed] [Google Scholar]
- 51.Suryawanshi H, Sabharwal H, Maiti S. Thermodynamics of peptide-RNA recognition: the binding of a Tat peptide to TAR RNA. J. Phys. Chem. B. 2010;114:11155–11163. doi: 10.1021/jp1000545. [DOI] [PubMed] [Google Scholar]
- 52.Montenay-Garestier T, Hélène C. Molecular interactions between tryptophan and nucleic acid components in frozen aqueous solutions. Nature. 1968;217:844–845. doi: 10.1038/217844a0. [DOI] [PubMed] [Google Scholar]
- 53.Mély Y, Piémont E, Sorinas-Jimeno M, de Rocquigny H, Jullian N, Morellet N, Roques BP, Gérard D. Structural and dynamic characterization of the aromatic amino acids of the human immunodeficiency virus type I nucleocapsid protein zinc fingers and their involvement in heterologous tRNA(Phe) binding: a steady-state and time-resolved fluorescence study. Biophys. J. 1993;65:1513–1522. doi: 10.1016/S0006-3495(93)81222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bombarda E, Ababou A, Vuilleumier C, Gérard D, Roques BP, Piémont E, Mély Y. Time-resolved fluorescence investigation of the human immunodeficiency virus type 1 nucleocapsid protein: influence of the binding of nucleic acids. Biophys. J. 1999;76:1561–1570. doi: 10.1016/S0006-3495(99)77315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lakowicz JR. Principles of Fluorescence Spectroscopy. Science + Business Media LLC, New York: Springer; 2006. [Google Scholar]
- 56.Record MT, Jr, Lohman ML, De Haseth P. Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 57.Shojania S, O’Neil JD. HIV-1 Tat is a natively unfolded protein: the solution conformation and dynamics of reduced HIV-1 Tat-(1-72) by NMR spectroscopy. J. Biol. Chem. 2006;281:8347–8356. doi: 10.1074/jbc.M510748200. [DOI] [PubMed] [Google Scholar]
- 58.Desai NA, Shankar V. Single-strand-specific nucleases. FEMS Microbiol. Rev. 2003;26:457–491. doi: 10.1111/j.1574-6976.2003.tb00626.x. [DOI] [PubMed] [Google Scholar]
- 59.Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. Footprinting: a method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods. 2007;42:128–140. doi: 10.1016/j.ymeth.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 60.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 61.Godet J, Kenfack C, Przybilla F, Richert L, Duportail G, Mély Y. Site-selective probing of cTAR destabilization highlights the necessary plasticity of the HIV-1 nucleocapsid protein to chaperone the first strand transfer. Nucleic Acids Res. 2013;41:5036–5048. doi: 10.1093/nar/gkt164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jean JM, Hall KB. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc. Natl Acad. Sci. USA. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guest CR, Hochstrasser RA, Sowers LC, Millar DP. Dynamics of mismatched base pairs in DNA. Biochemistry. 1991;30:3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]
- 64.Bernacchi S, Stoylov S, Piémont E, Ficheux D, Roques BP, Darlix JL, Mély Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 65.Helene C, Maurizot JC. Interactions of oligopeptides with nucleic acids. CRC Crit. Rev. Biochem. 1981;10:213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- 66.Doetsch M, Schroeder R, Fürtig B. Transient RNA-protein interactions in RNA folding. FEBS J. 2011;278:1634–1642. doi: 10.1111/j.1742-4658.2011.08094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernacchi S, Piémont E, Potier N, van Dorsselaer A, Mély Y. Excitonic heterodimer formation in an HIV-1 oligonucleotide labeled with a donor-acceptor pair used for fluorescence resonance energy transfer. Biophys. J. 2003;84:643–654. doi: 10.1016/S0006-3495(03)74884-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Azoulay J, Clamme JP, Darlix JL, Roques BP, Mély Y. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J. Mol. Biol. 2003;326:691–700. doi: 10.1016/s0022-2836(02)01430-4. [DOI] [PubMed] [Google Scholar]
- 69.Godet J, Mély Y. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biol. 2010;7:687–699. doi: 10.4161/rna.7.6.13616. [DOI] [PubMed] [Google Scholar]
- 70.Beltz H, Azoulay J, Bernacchi S, Clamme J-P, Ficheux D, Roques B, Darlix JL, Mély Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- 71.Cosa G, Harbron EJ, Zeng Y, Liu HW, O’Connor DB, Eta-Hosokawa C, Musier-Forsyth K, Barbara PF. Secondary structure and secondary structure dynamics of DNA hairpins complexed with HIV-1 NC protein. Biophys. J. 2004;87:2759–2767. doi: 10.1529/biophysj.104.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vo M-N, Barany G, Rouzina I, Musier-Forsyth K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop ‘kissing’ to ‘zipper’. J. Mol. Biol. 2009;386:789–801. doi: 10.1016/j.jmb.2008.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hargittai MRS, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 74.Bloomfield VA, Crothers DM, Tinoco I. Nucleic Acids: Structures, Properties, and Functions. Sausalito, CA: University Science Books; 2000. [Google Scholar]
- 75.Sharma K kant, Didier P, Darlix JL, de Rocquigny H, Bensikaddour H, Lavergne JP, Pénin F, Lessinger JM, Mély Y. Kinetic analysis of the nucleic acid chaperone activity of the hepatitis C virus core protein. Nucleic Acids Res. 2010;38:3632–3642. doi: 10.1093/nar/gkq094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Darlix JL, Godet J, Ivanyi-Nagy R, Fossé P, Mauffret O, Mély Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 77.Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Borde I, Reboud-Ravaux M, Restle T, Gorelick RJ, et al. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS One. 2007;2:e669. doi: 10.1371/journal.pone.0000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mirambeau G, Lyonnais S, Gorelick RJ. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010;7:85–95. doi: 10.4161/rna.7.6.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lyonnais S, Gorelick RJ, Heniche-Boukhalfa F, Bouaziz S, Parissi V, Mouscadet JF, Restle T, Gatell JM, Le Cam E, Mirambeau G. A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA. Virus Res. 2013;171:287–303. doi: 10.1016/j.virusres.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brès V, Tagami H, Péloponèse JM, Loret E, Jeang KT, Nakatani Y, Emiliani S, Benkirane M, Kiernan RE. Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF. EMBO J. 2002;21:6811–6819. doi: 10.1093/emboj/cdf669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein’s nucleic acid chaperone function. J. Mol. Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 84.Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein’s nucleic acid chaperone activity and retroviral replication. Virus Res. 2013;171:263–277. doi: 10.1016/j.virusres.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Godet J, Ramalanjaona N, Sharma KK, Richert L, de Rocquigny H, Darlix JL, Duportail G, Mély Y. Specific implications of the HIV-1 nucleocapsid zinc fingers in the annealing of the primer binding site complementary sequences during the obligatory plus strand transfer. Nucleic Acids Res. 2011;39:6633–6645. doi: 10.1093/nar/gkr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Livesey AK, Brochon JC. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys. J. 1987;52:693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brochon JC. Maximum entropy method of data analysis in time-resolved spectroscopy. Meth. Enzymol. 1994;240:262–311. doi: 10.1016/s0076-6879(94)40052-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.