Abstract
Clostridium thermocellum polynucleotide kinase (CthPnk), the 5′ end-healing module of a bacterial RNA repair system, catalyzes reversible phosphoryl transfer from an NTP donor to a 5′-OH polynucleotide acceptor. Here we report the crystal structures of CthPnk-D38N in a Michaelis complex with GTP•Mg2+ and a 5′-OH oligonucleotide and a product complex with GDP•Mg2+ and a 5′-PO4 oligonucleotide. The O5′ nucleophile is situated 3.0 Å from the GTP γ phosphorus in the Michaelis complex, where it is coordinated by Asn38 and is apical to the bridging β phosphate oxygen of the GDP leaving group. In the product complex, the transferred phosphate has undergone stereochemical inversion and Asn38 coordinates the 5′-bridging phosphate oxygen of the oligonucleotide. The D38N enzyme is poised for catalysis, but cannot execute because it lacks Asp38—hereby implicated as the essential general base catalyst that abstracts a proton from the 5′-OH during the kinase reaction. Asp38 serves as a general acid catalyst during the ‘reverse kinase’ reaction by donating a proton to the O5′ leaving group of the 5′-PO4 strand. The acceptor strand binding mode of CthPnk is distinct from that of bacteriophage T4 Pnk.
INTRODUCTION
Polynucleotide kinases (Pnk) comprise a widely distributed family of nucleic acid repair enzymes that heal broken RNA or DNA ends by converting 5′-OH termini into 5′-PO4 ends that can be sealed by RNA or DNA ligases. The founding member of this family is bacteriophage T4 polynucleotide kinase-phosphatase (T4 Pnkp), a bifunctional enzyme with 5′-OH kinase and 3′-phosphatase activities that collaborates with T4 RNA ligase 1 to repair tRNA damage inflicted during the Escherichia coli antiviral response (1). Whereas E. coli has no endogenous RNA end-healing enzymes, many other bacteria do. For example, the Pnkp•Hen1 RNA repair system is encoded in an operon present in diverse bacteria from many phyla (2–5). Bacterial Pnkp is the end-healing and end-sealing component and comprises three catalytic domains: N-terminal kinase, central phosphatase and C-terminal ligase (2,6–8). The kinase domain catalyzes phosphoryl transfer to the 5′-OH RNA end. The phosphatase domain releases Pi from 2′-PO4, 3′-PO4 or 2′,3′-cyclic-PO4 ribonucleotides (9–12). The ligase domain, in a 1:1 complex with the N-terminal half of the Hen1 protein, is an ATP-dependent RNA sealing enzyme (13). The C-terminal half of Hen1 is a Mn2+-dependent methyltransferase that installs a 2′-OCH3 mark at the RNA repair junction before ligation and thereby protects the junction from recurrent damage (3,4,14,15).
The N-terminal 170-amino acid segment of Clostridium thermocellum (Cth) Pnkp is an autonomous polynucleotide kinase (Pnk) domain. CthPnk phosphorylates 5′-OH single-stranded RNA or DNA using ATP, GTP, CTP, UTP or dATP as the phosphate donor (16). The enzyme also catalyzes the reverse reaction, in which a polynucleotide 5′-PO4 group is transferred to ADP, GDP, CDP, UDP or dADP to form the corresponding NTP (16). CthPnk requires a divalent cation cofactor; this requirement is satisfied by magnesium, manganese or cobalt (2).
Initial crystal structures of CthPnk bound to ATP•Mg2+ and ADP•Mg2+, reflective of substrate and product nucleotide complexes, respectively, showed that CthPnk is a homodimer and a member of the P-loop phosphotransferase superfamily (6). CthPnk shares with phage T4 Pnk a core phosphotransferase fold (a central 4-strand parallel β sheet flanked by α helices) (17,18), but is embellished by a distinctive homodimerization module composed of secondary structure elements that have no counterparts in T4 Pnk.
The ATP phosphate donor binds to CthPnk within a crescent-shaped groove formed by the P-loop (15GSSGSGKST23) and an overlying ‘lid’ composed of helices α6 and α7 and the connecting 120RTDRQVE126 peptide. The ATP α and β phosphates are engaged by a network of hydrogen bonds from Thr23 and the P-loop main-chain amides; the γ phosphate is anchored by the lid residues Arg120 and Arg123 and the P-loop Ser17. The P-loop lysine (Lys21) and the catalytic Mg2+ bridge the ATP β and γ phosphates. The P-loop serine (Ser22) is the sole enzymic constituent of the octahedral metal coordination complex, which also includes nonbridging β and γ phosphate oxygens of ATP and three waters. Mutational analysis by alanine scanning underscored the essentiality of P-loop residues Lys21 and Ser22 in the NTP donor site (6). The lid residues Arg120 and Arg123 provide important, but functionally redundant, contacts to the γ phosphate oxygens (16). Based on the structural and mutational data, we proposed a catalytic mechanism whereby Lys21 and Mg2+ stabilize the transition state on the ATP γ phosphorus, assisted by the lid arginines (6,16).
Subsequent crystal structures of CthPnk in complexes with GTP, CTP, UTP and dATP phosphate donors (16) revealed the principles of nucleobase nonspecific NTP substrate utilization, whereby the kinase establishes an identical network of ionic and hydrogen-bonding contacts to the α, β and γ phosphates of whichever NTP donor is bound, while the interactions of the nucleoside moiety are limited to a π-cation stack of the nucleobase on the Arg116 side chain in helix α6 of the lid.
Having explored the architecture of the NTP donor site, we aimed here to define the structural basis for engagement of the phosphoacceptor strand and the mechanism by which the polynucleotide 5′-OH nucleophile is oriented and activated for its attack on the NTP γ phosphorus. To that end, we have co-crystallized CthPnk mutant D38N with GTP•Mg2+ and a 5′-OH oligonucleotide and with GDP•Mg2+ and a 5′-PO4 oligonucleotide and determined the atomic structures of the Michaelis complex and product complex.
MATERIALS AND METHODS
CthPnk purification and mutagenesis
The pET28b-Smt3CthPnkp-(1–170) expression plasmid encoding the Pnk domain was described previously (6). Alanine mutations were introduced into the expression vector by the two-stage polymerase chain reaction (PCR)-based overlap extension method. The Pnk inserts were sequenced to confirm the presence of the desired mutation and the absence of unwanted coding changes. The plasmids were transformed into E. coli BL21(DE3). Recombinant protein production was induced with IPTG and the CthPnk proteins were purified as described (16). In brief, the His10Smt3-CthPnk proteins were recovered from soluble bacterial lysates by Ni-agarose chromatography. The tag was removed by treatment with Smt3 protease Ulp1 and the tag-free CthPnk was separated from the cleaved His10Smt3 tag by a second round of Ni-agarose chromatography. The CthPnk preparations were then adjusted to 10 mM EDTA, concentrated by centrifugal ultrafiltration, and gel-filtered through a column of Superdex-200 equilibrated in 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA. The enzyme preparations were stored at −80°C. Protein concentrations were determined by using the BioRad dye reagent with bovine serum albumin as the standard.
Kinase assay
Reaction mixtures (10 μl) containing 50 mM Tris–HCl (pH 7.0), 10 mM MgCl2, 5 mM DTT, 100 μM [γ32P]ATP, 100 pmol of a 10-mer 5′-OH DNA oligonucleotide phosphoacceptor (HOCCTGTATGAT) and wild-type or mutant kinase as specified were incubated for 30 min at 45°C. The reactions were quenched by adding an equal volume of 90% formamide, 50 mM EDTA and 0.01% bromophenol blue/xylene cyanol, and the mixtures were analyzed by electrophoresis (at 7 W constant power) through a 15-cm 20% polyacrylamide gel containing 8 M urea in 45 mM Tris borate, 1.2 mM EDTA. The 32P-labeled DNAs were visualized and quantified by scanning the gel with a Fuji Film BAS-2500 imager.
Crystallization, diffraction data collection and structure determination
A D38N mutation was introduced into the pET28b-His10Smt3-CthPnk-L137M expression vector by the two-stage PCR-based overlap extension method. The tag-free CthPnk-D38N protein was purified as described (6,16). The protein solution (10.5 mg/ml) was adjusted to either 2 mM GTP, 10 mM MgCl2 and 1.1 mM 5′-OH oligonucleotide (HOCCTGT) or 2 mM GDP, 10 mM MgCl2 and 1.1 mM 5′-PO4 oligonucleotide (pCCTGT) and incubated for 10 min before aliquots of the protein solution (2 µl) were mixed on a coverslip with an equal volume of precipitant solution containing 100 mM sodium citrate, pH 5.0, 100 mM MgCl2, 18–24% (v/v) PEG-6000. Crystals were grown at 22°C by hanging drop vapor diffusion against a reservoir of the same precipitant solution. Single crystals were transferred to a solution containing 100 mM sodium citrate, pH 5.0, 2 mM GTP/GDP, 100 mM MgCl2, 18–24% PEG-6000, 1.1 mM 5′-OH/5′-PO4 oligonucleotide and 15% glycerol before freezing the crystals in liquid nitrogen. Diffraction data were collected at NSLS beamline X25 equipped with a Pilatus 6M detector in 370 continuous increments of 0.5° each for the Michaelis complex and in 440 continuous increments of 0.2° each for the product complex. The data were integrated and reduced with MOSFLM and SCALA. Diffraction statistics are compiled in Table 1. The crystals were in space group P212121 and had two CthPnk protomers per asymmetric unit. To solve the structure of the Michaelis complex with 5′-OH oligonucleotide, phases were obtained via molecular replacement in PHASER using a search model constructed by removing waters from the CthPnk•GTP•Mg2+ dimer structure reported previously (16). Following the placement of the dimer by PHASER and rigid body refinement, the active sites of both protomers revealed clear and continuous Fo–Fc density for the 5′-OH oligonucleotide. To determine the structure of the product complex with GDP, Mg2+ and 5′-PO4 oligonucleotide, the diffraction data were refined against the model of the Michaelis complex, from which the GTP γ-phosphates and the 5′-OH oligonucleotides had been removed. The 5′-PO4 oligonucleotides were then modeled into the Fo–Fc maps of the active site. The models were iteratively rebuilt by hand in COOT (25) and refined in PHENIX (26). The final models had excellent geometry, no Ramachandran outliers, and no large Fo–Fc difference Fourier peaks (Table 1).
Table 1.
Crystallographic data and refinement statistics
| CthPnk•GTP•Mg2+•HODNA | CthPnk•GDP•Mg2+•pDNA | |
|---|---|---|
| Space group | P212121 | P212121 |
| Unit cell dimensions (Å) @ 130 K | a = 53.0 | a = 54.4 |
| b = 72.8 | b = 73.1 | |
| c = 119.5 | c = 119.0 | |
| Diffraction data quality | ||
| Resolution (Å) | 53.0–1.73 (1.76–1.73) | 46.2–1.8 (1.9–1.8) |
| Radiation source | NSLS X25 | NSLS X25 |
| Wavelength | 1.10 Å | 1.10 Å |
| Processing software | Mosflm/Scala | Mosflm/Scala |
| Rsyma, % | 8.4 (38.7) | 4.5 (19.8) |
| Unique reflections | 49 311 (2645) | 43 723 (6304) |
| Mean redundancy | 11.9 (7.4) | 3.0 (3.0) |
| Completeness, % | 100.0 (99.2) | 97.8 (97.6) |
| Mean I/σI | 20.1 (4.7) | 13.6 (4.7) |
| Phasing statistics | ||
| Phasing method | Phenix AutoMR | |
| Resolution (Å) | 53.0–2.0 | |
| Signal to noiseb | RFZ = 27.6, TFZ = 38.2 | |
| Refinement and model statistics (F > 0) | ||
| Resolution (Å) | 48.5–1.73 (1.75–1.73) | 41.0–1.8 (1.82–1.80) |
| Completeness, % | 99.5 (97.2) | 97.3 (98.0) |
| Rfreec/Rwork, % | 19.6/16.3 (27.7/22.3) | 20.6/16.4 (27.1/22.3) |
| RMSD bonds/angles | 0.006 Å/1.13° | 0.006 Å/1.09° |
| Protomers/ASU | 2 | 2 |
| Ramachandran plot | 99.4% favored, no outliers | 99.1% favored, no outliers |
| B-factors, Å2 | ||
| Overall/Wilson | 21.3/17.8 | 21.8/20.0 |
| PDB ID | 4MDF | 4MDE |
| Model contents | ||
| Protein residues | 342 | 342 |
| Heteroatoms | 2 GTP, 2 Mg2+, 1 citrate, 2 oligonucleotides | 2 GDP, 2 Mg2+, 2 oligonucleotides |
| Water | 395 | 440 |
Standard definitions are used for all parameters. Figures in parentheses refer to data in the highest resolution bin. The refinement and geometric statistics come from PHENIX.
aRsym output as as Rmerge by SCALA.
bSignal to noise ratios for final translation function Z-score (TFZ) and rotation function Z-score (RFZ) as output by PHASER for CthPnk•GTP•Mg2+•HODNA data set.
cRfree sets consisted of 8% of data chosen at random against which structures were not refined.
RESULTS AND DISCUSSION
Strategy for crystallizing a Michaelis complex of CthPnk
Bacterial, bacteriophage and baculovirus Pnk enzymes implicated in RNA repair all share a conserved motif Asp-X-X-Arg (38DFCR41 in CthPnk; 35DDYR38 in T4 Pnk) located downstream of the P-loop in the primary structure (6,19,20). Mutational analysis of T4 Pnkp had identified the Asp35 and Arg38 side chains as essential for catalysis (21,22). The initial crystal structure of the T4 Pnkp kinase domain homodimer implicated Arg38 in binding the terminal phosphodiester of the HONpN– acceptor strand, as surmised from its bidentate coordination of a sulfate anion in the acceptor site (17). The essential Asp35 side chain was proposed to coordinate the 5′-OH and activate its attack on the NTP γ phosphorus (17). Consistent with the proposed general base mechanism, replacing Asp35 with Asn abolished T4 Pnkp kinase activity (21). However, other investigators invoked a role for Asp35 in binding a magnesium ion (18). Subsequent crystal structures obtained by soaking short oligonucleotides into preformed T4 Pnkp crystals revealed that Asp35 does indeed coordinate the 5′-OH and that Arg38 contacts the first phosphodiester (23). However, none of the available T4 Pnk structures have a metal cofactor in the kinase active site and none have an NTP substrate in the phosphate donor site.
The missing link in the RNA-repairing Pnk saga is a structure of the Michaelis complex containing NTP donor, metal cofactor and 5′-OH polynucleotide acceptor. Capturing this state along the reaction pathway requires that phosphoryl transfer reaction chemistry be precluded, e.g. by a minimally perturbing modification of the enzyme or the substrates. Our mutational analysis of CthPnk had shown that the Asp38 and Arg41 residues in the putative acceptor site are essential for kinase activity (6). We reasoned that replacing Asp38 with asparagine might arrest the kinase reaction as a stable Michaelis complex without compromising substrate binding. Hence, we grew crystals of CthPnk-D38N in the presence of GTP, Mg2+ and oligonucleotide HOCpCpTpGpT. The crystals diffracted X-rays to 1.73 Å, were in space group P212121 and had two protomers per asymmetric unit organized as a kinase homodimer. The electron density maps revealed occupancy of the acceptor sites of both protomers by oligonucleotide. The refinement statistics and model contents of the CthPnk•GTP•Mg2+•HODNA structure are provided in Table 1. Unless specified otherwise, the images and descriptions of the Michaelis complex refer to the A protomer.
Overview of the structure
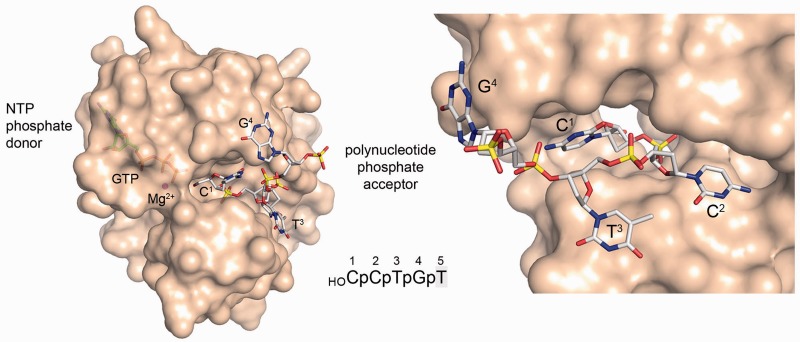
A semitransparent surface view of the CthPnk protomer is shown in Figure 1 (left panel) with stick models of GTP in the phosphate donor site and of the 5′-OH oligonucleotide in the phosphoacceptor site. A single Mg2+ ion (magenta sphere) is present in the donor site, where it bridges the GTP β and γ phosphates. The Fo–Fc electron density in the acceptor site (shown in stereo in Figure 2A) permitted modeling of the first four nucleotides HOC1pC2pT3pG4p of the pentanucleotide ligand used for co-crystallization. All four nucleobases adopt an anti nucleoside conformation. The first two nucleotides of the 5′-OH acceptor strand bind in a deep groove on the enzyme surface so as to place the terminal 5′-OH group next to the GTP γ phosphate (Figure 1, left panel). The sequential nucleobases are splayed out in different directions from the phosphodiester backbone, i.e. there is no base stacking. The C1 and C2 nucleobases are sandwiched between protein surfaces that comprise the deep groove. The T3 and G4 nucleobases each pack on one face against an exposed surface of the kinase (Figure 1, right panel). The opposite faces of T3 and G4 are solvent exposed. The structure indicates that the 5′-OH end of the oligonucleotide phosphoacceptor must be single-stranded to access the kinase active site.
Figure 1.
Surface views of the Michaelis complex. The left panel shows a semitransparent surface view of CthPnk with stick models of the GTP phosphate donor (at left) and the oligonucleotide phosphoacceptor strand (at right). A Mg2+ ion (magenta sphere) bridges the GTP β and γ phosphates. The 5′-OH of the acceptor strand is adjacent to the GTP γ phosphate. The right panel shows a close-up view of the first four nucleotides of the phosphoacceptor strand, highlighting the splayed out trajectories of the consecutive nucleobases and their packing against the enzyme surface.
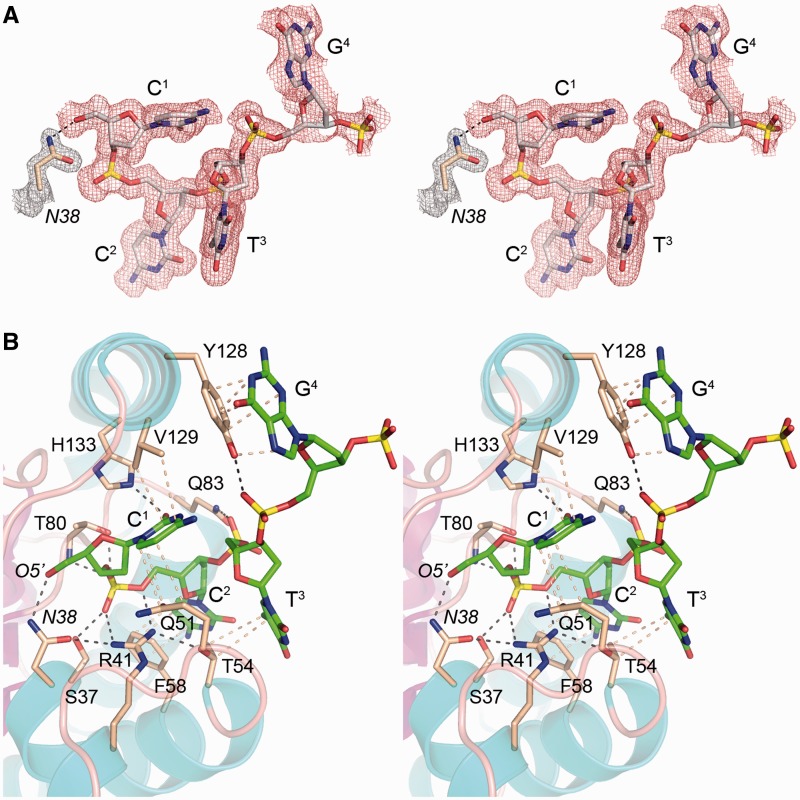
Figure 2.
The phosphoacceptor binding site. (A) Stereo view of the Fo–Fc electron density map (colored red) of the 5′-OH oligonucleotide contoured at 3.0 σ. The component nucleotides are shown as stick models with gray carbons. The 2Fo–Fc density for the Asn38 side chain is colored gray and contoured at 1.5 σ. (B) Stereo view of the architecture of the phosphoacceptor site and pertinent atomic contacts. The kinase fold is depicted with cyan helices and magenta strands. Selected amino acids are shown as stick models with beige carbons. The oligonucleotide is rendered as a stick model with green carbons. Electrostatic and hydrogen bonding interactions are denoted by black dashed lines. Van der Waals contacts and π stacking interactions are denoted by beige dashed lines.
The phosphoacceptor binding site
A stereo view of the atomic contacts of the HOC1pC2pT3pG4p acceptor strand is shown in Figure 2B. Electrostatic and hydrogen-bonding interactions are denoted by black dashed lines; van der Waals contacts are depicted as beige dashed lines. The first phosphodiester HOC1pC2 of the acceptor strand is fixed in place by a network of contacts with three of the phosphodiester oxygens. The terminal guanidinium nitrogens of the essential Arg41 residue make bidentate contacts with the nonbridging OP2 and bridging O5′ atoms of the first phosphodiester. The OP2 atom receives an additional hydrogen bond from Ser37-Oγ. The nonbridging OP1 oxygen is coordinated by hydrogen bonds from Thr80-Oγ and the Thr80 main-chain amide nitrogen. The second phosphodiester C2pT3 receives a hydrogen bond to a nonbridging oxygen from Gln83-Nε. The third phosphodiester T3pG4 accepts a hydrogen bond to a nonbridging oxygen from the Tyr128 hydroxyl group.
The kinase engages each of the four nucleobases via interactions with aliphatic and/or aromatic side chains. Val129 and Gln51 sandwich the first pyrimidine base (C1). Val129 sits above the C1 base in the view shown in Figure 2B and makes van der Waals contacts from Cγ1 (3.5 Å) and Cγ2 (3.9 Å) to the cytosine C2 and C4 atoms, respectively. Gln51 makes van der Waals contacts to the underside of the C1 base, from Cγ (3.8 Å), Cδ (3.6 Å) and Oε (3.9 Å) to the cytosine C5, C6 and N1 atoms, respectively. Phe58 makes a π stack on the second pyrimidine base (C2). Thr54 makes van der Waals contacts from Cγ (3.7 Å) and Cβ (3.9 Å) to the C7 and C5 atoms, respectively, of the third pyrimidine base (T3). Tyr128 makes a π stack on the fourth purine base (G4) and a van der Waals contact of the phenolic oxygen (3.6 Å) to the guanosine sugar O4′ atom. These interactions are nonspecific with respect to the identity of the nucleobase. The C1 base does make two hydrophilic interactions via the base edge: (i) an intramolecular hydrogen bond from N4 to a nonbridging oxygen of the third phosphodiester, and (ii) accepting a hydrogen bond to O2 from His133-Nε.
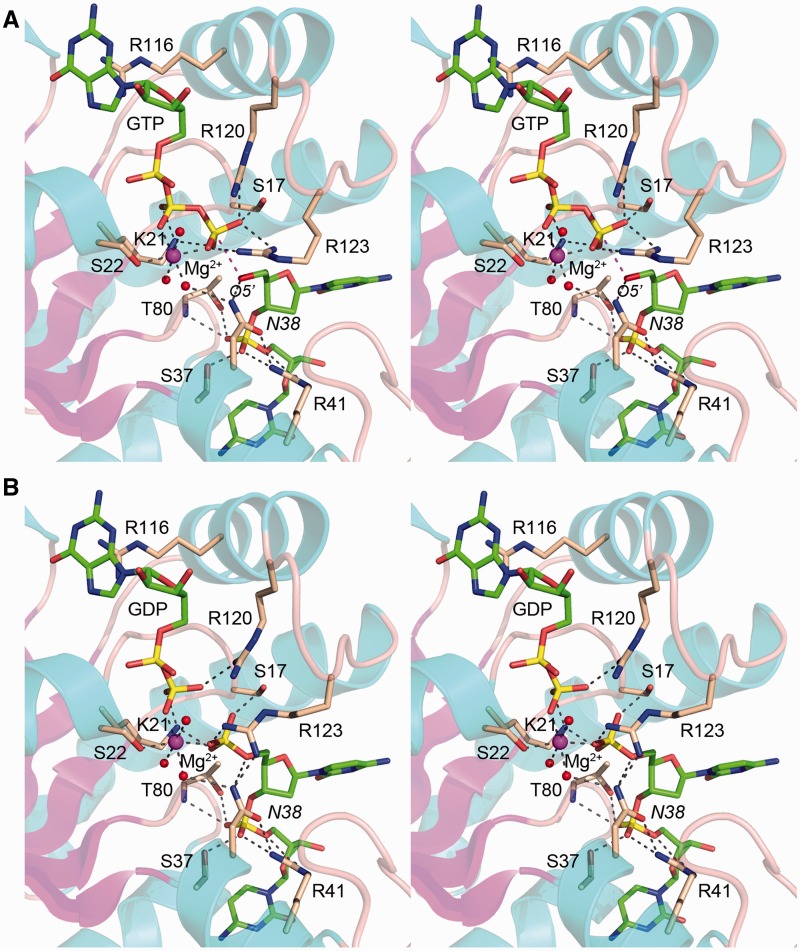
Active site of the Michaelis complex: geometry and general base catalysis
A detailed stereo view of the active site is shown in Figure 3A, which highlights the orientation and pertinent atomic contacts of GTP and the first two nucleotides HOC1pC2 of the acceptor strand. GTP and Mg2+ occupy the same positions in the Michaelis complex as in the kinase•GTP•Mg2+ donor complex reported previously (16). The guanine nucleobase makes a π-cation stack over Arg116. Lys21 makes a bifurcated ionic interaction with the GTP β and γ phosphates. The GTP γ phosphate is engaged by P-loop residue Ser17 and by lid residues Arg120 and Arg123. The octahedral coordination complex of the Mg2+ ion includes Ser22-Oγ, three waters and two nonbridging β and γ phosphate oxygen atoms (those not engaged by Lys21).
Figure 3.
Active sites of the Michaelis complex and product complex. Stereo views of the Michaelis complex (A) and product complex (B). The kinase fold is depicted with cyan helices and magenta strands. Selected amino acids are shown as stick models with beige carbons. The guanine nucleotides and oligonucleotides are rendered as stick models with green carbons. Mg2+ is a magenta sphere; waters in the metal coordination complex are red spheres. Electrostatic and hydrogen bonding interactions are denoted by black dashed lines. The magenta dashed line indicates the path of the O5′ nucleophile of the acceptor strand toward the GTP γ phosphorus.
The O5′ nucleophile of the acceptor strand is situated 3.0 Å from the GTP γ phosphorus atom (see the magenta dashed line in Figures 3A and 4A) and is almost perfectly apical to the bridging β phosphate oxygen of the GDP leaving group (O5′–P–O angle = 168°). Asn38-Nδ coordinates the 5′-OH (2.8 Å). We surmise that the kinase is ‘all revved up with no place to go’ because Asp38 is the essential general base catalyst that abstracts a proton from the 5′-OH and activates the nucleophile for attack on the NTP donor. Orientation of the aspartate general base is aided by atomic contacts of Oδ1 to Arg41 and of Oδ2 (Nδ in the structure) with a water in the metal coordination complex (Figure 3A). The position and conformation of the Asn38 side chain in the Michaelis complex mimics that of the native Asp38 side chain seen in the structure of the wild-type CthPnk•GTP•Mg2+ donor complex (16). Although the electron density does not discriminate Asn38 Nδ from Oδ, our assignment of the orientation of the amide group is consistent with the 2.8 Å distance from Arg41-NH1 to a terminal atom of the Asn38 amide, whereby Arg41-NH1 donates a hydrogen bond to Asn38-Oδ. The reverse orientation of Asn38 would not favor this close contact, insofar as the Nδ atom is not the preferred hydrogen bond acceptor.
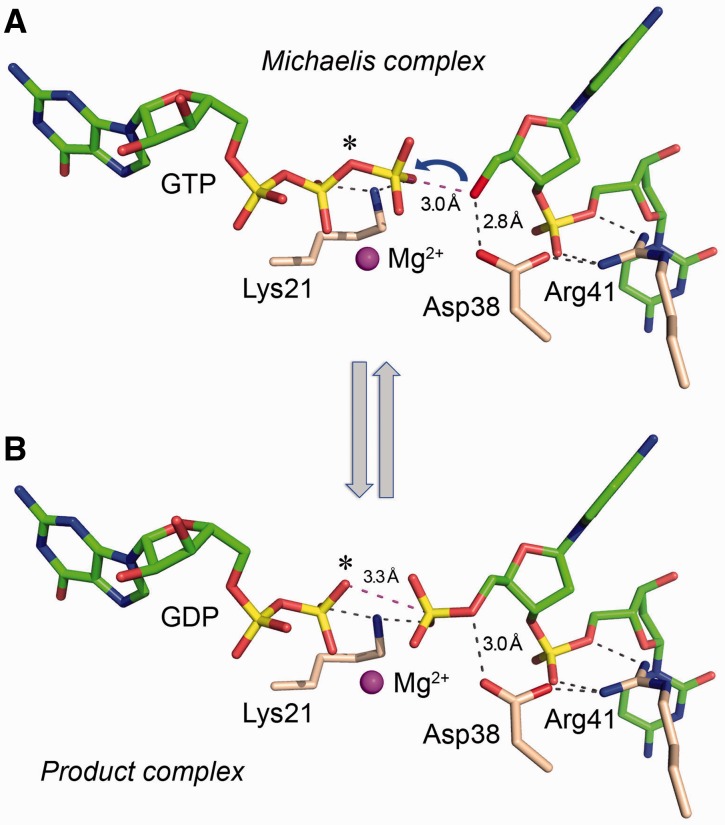
Figure 4.
Michaelis complex and product complex structures illuminate the mechanism and stereochemistry of the Pnk reaction. The guanine nucleotide, Mg2+ and oligonucleotide moieties of the superimposed Michaelis complex (A) and product complex (B) are shown, indicative of the states immediately before and following the phosphoryl transfer reaction. For illustrative purposes, Asn38 was changed to the native Asp38 by substituting a red oxygen atom for Nδ. Atomic contacts of the Lys21, Arg41 and Asp38 residues are indicated by dashed lines. (A) Attack of the oligonucleotide O5′ nucleophile on the NTP γ phosphorus is depicted by the blue arrow. The bridging β-γ oxygen of the GTP substrate (denote by the asterisk) is the leaving group and is situated apical to the O5′ nucleophile. (B) The transferred phosphate has undergone stereochemical inversion. The GDP β phosphate OP3 atom (denoted by asterisk), which was the leaving atom in the forward kinase reaction, is poised to act as the attacking nucleophile in the ‘reverse kinase’ reaction. Lys21 and Mg2+ stabilize the putative phosphorane transition state of the γ phosphate during the forward kinase reaction and of the 5′-PO4 during the reverse reaction. Asp38 is a proposed general base catalyst of the forward kinase reaction and a general acid catalyst of the reverse reaction.
Figure 4A shows a stripped-down model of the Michaelis complex in which the Asn38 side chain is changed back to the native aspartate. This image shows only the enzymic functional groups Lys21, Asp38 and Arg41 that are essential per se for catalysis, as judged by the ablation of activity when they are replaced by alanine. The model summarizes the key features of the proposed chemical mechanism of the kinase reaction: (i) Lys21 and Mg2+ stabilize an associative phosphorane transition state on the NTP γ phosphate; (ii) Arg41 positions the 5′-OH acceptor strand proximal to the NTP γ phosphate; and (iii) Asp38 orients and activates the O5′ nucleophile by general base catalysis.
Capturing a product complex of CthPnk with GDP•Mg2+ and 5′-PO4 strand
We grew co-crystals of CthPnk-D38N with GDP•Mg2+ and a 5′-PO4-terminated oligonucleotide, pCpCpTpGpT and solved the structure at 1.8 Å resolution (Table 1). All five nucleotides of the DNA strand were modeled into Fo–Fc electron density (Supplementary Figure S1). A detailed stereo view of the product complex is shown in Figure 3B, while Figure 4B shows a stripped-down model of the product complex in which the Asn38 side chain is changed back to the native aspartate. In the product complex, the GDP β phosphate and oligonucleotide 5′-PO4 are 2.5 Å apart at their closest point and are bridged by Lys21 (2.7 Å) and the Mg2+ ion (Figure 4B), which ameliorate the electrostatic repulsion of the adjacent β phosphate and 5′ phosphate groups. Asn38-Nδ (i.e. Asp38 in the native kinase) coordinates the 5′-bridging oxygen of the terminal phosphate (3.0 Å) (Figure 4B). We surmise that this structure captures the forward kinase reaction in the state immediately following phosphoryl transfer. Note that the 5′-PO4 has undergone stereochemical inversion in the product complex compared with the GTP γ phosphate in the substrate complex, consistent with an in-line mechanism (24). Moreover, it is clear that the GDP β phosphate O3 atom of the product complex (denoted by the asterisk in Figure 4B) corresponds to the bridging oxygen-leaving group of the Michaelis complex (asterisk in Figure 4A). The product complex can be viewed as synonymous with the Michaelis complex of the ‘reverse kinase’ reaction. This inference is supported by the proximity of the GDP β phosphate O3 atom (the putative nucleophile in the reverse kinase reaction; denoted by the asterisk in Figure 4B) to the 5′ phosphorus of the oligonucleotide (3.3 Å) and its near apical orientation with respect to the 5′-bridging phosphate oxygen (O–P–O5′ angle = 146°). In this state, the D38N mutant kinase is arrested in place because it lacks the Asp38 general acid that donates a proton to the O5′ leaving group.
In the product complex, Arg120 coordinates the GDP β phosphate. Ser17 and Arg123 engage the oligonucleotide 5′-PO4 (Figure 3B). Otherwise, the trajectory and enzymic contacts of the first four nucleotides of the 5′-PO4 strand are virtually identical to those observed for the 5′-OH strand in the forward Michaelis complex. The fifth thymidine nucleotide in the product complex makes no atomic contacts with the kinase protomer to which it is bound (though the T5 nucleosides do make lattice contacts with kinase protomers in neighboring asymmetric units).
Mutational analysis of the phosphoacceptor site
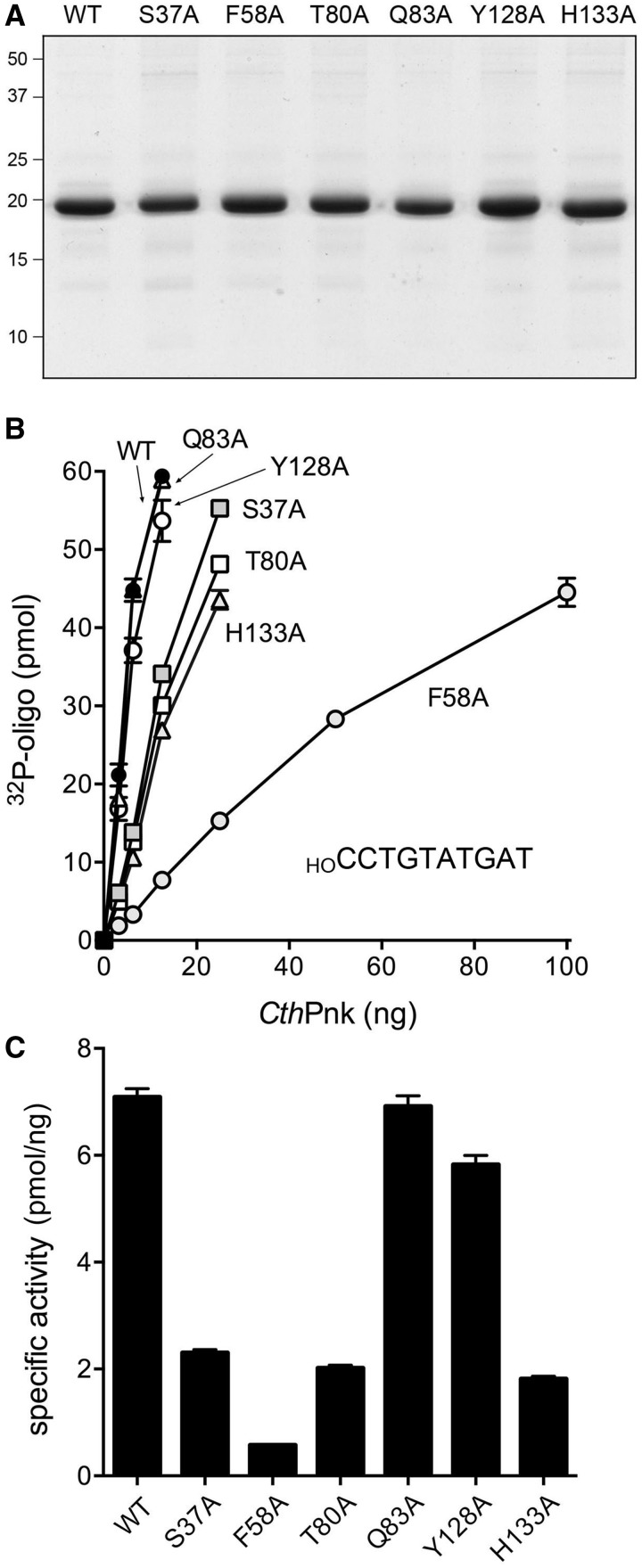
To gauge the contributions of selected amino acids identified by the structure as constituents of the phosphoacceptor site, we mutated Ser37, Phe58, Thr80, Gln83, Tyr128 and His133 to alanine. The wild-type and mutant CthPnk proteins were produced in E. coli and purified as described previously (16). Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the preparations verified that they were similarly pure with respect to the 19 kDa CthPnk polypeptide (Figure 5A). We assayed the extent of label transfer from 100 µM [γ32P]ATP to 10 µM 10-mer HODNA acceptor as a function of input protein (Figure 5B). (The nucleobase sequence of the 5′ half of the acceptor substrate was identical to that of the ligand used for co-crystallization of the Michaelis complex.) Specific activities of the wild-type and mutant kinases were derived by linear regression curve fitting in Prism and are plotted in bar graph format in Figure 5C. Mutational effects fell into three categories. Alanine substitutions for Gln83 and Tyr128 were benign, i.e. the specific activities of the Q83A and Y128A kinases were 98 and 82% of the wild-type value. Alanines in lieu of Ser37, Thr80 or His133 elicited moderate decrements in specific activity: to 33, 28 and 26% of wild-type, respectively. Replacing Phe58 with alanine was singularly deleterious, the specific activity of the F58A mutant being only 8% of the wild-type value (Figure 5C). Thus, of the individual phosphoacceptor contacts probed here by alanine scanning, the π-π interaction of Phe58 with the penultimate nucleobase appears to be the most important for kinase function.
Figure 5.
Structure-guided mutagenesis of the phosphoacceptor site. (A) Aliquots (3 µg) of the CthPnk-(1–170) proteins were analyzed by SDS-PAGE. The polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (kDa) of marker polypeptides are indicated. (B) Kinase reactions were performed as described in ‘Materials and Methods’ section. The reaction mixtures included 100 μM [γ32P]ATP, 100 pmol of a 10-mer 5′-OH DNA oligonucleotide phosphoacceptor (HOCCTGTATGAT) and wild-type or mutant kinase as indicated. The extents of DNA phosphorylation are plotted as a function of input protein. Each datum is the average of three titration experiments ± SEM. (C) Specific activities of the wild-type and mutant kinases derived by linear regression curve fitting in Prism. The error bars indicate the standard deviation of the curve fit.
Comparison of phosphoacceptor binding modes in CthPnk versus T4 Pnk
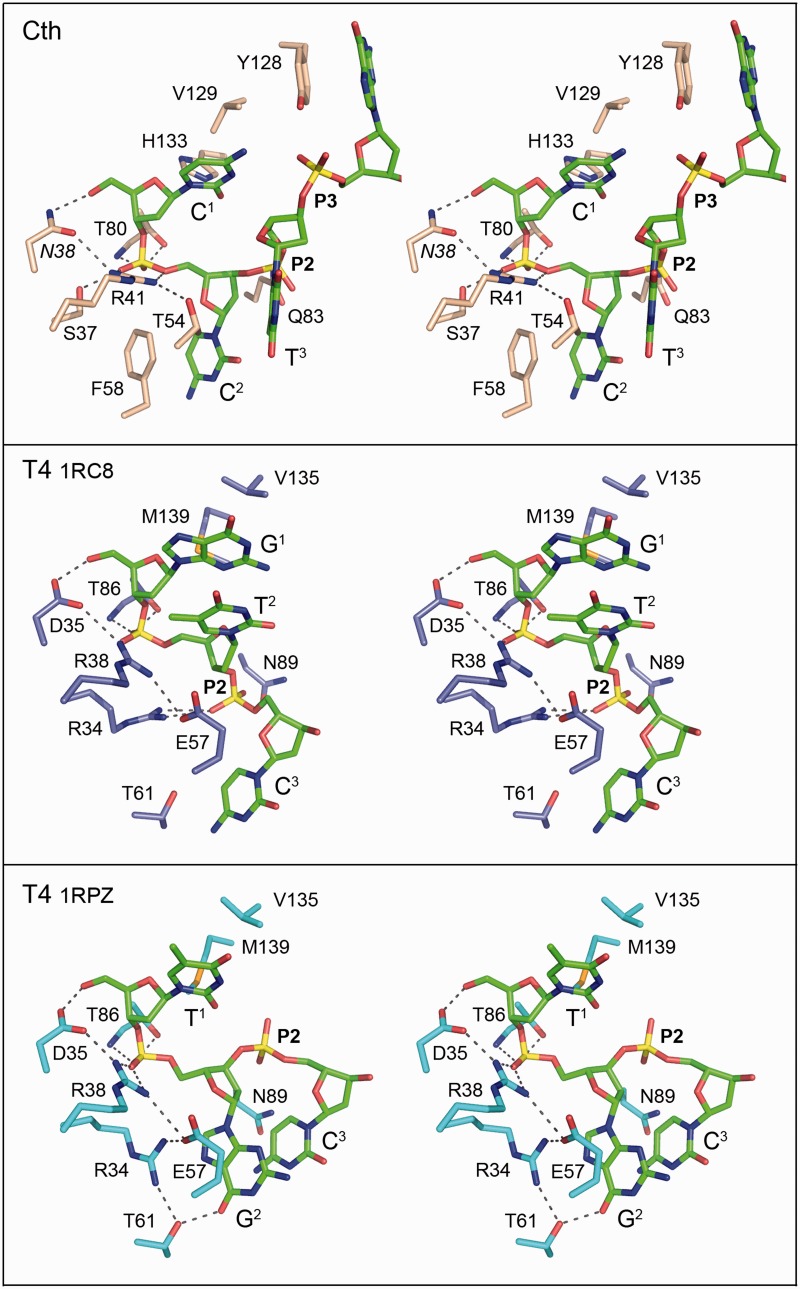
Eastberg et al. (23) obtained structures of T4 Pnk with short 5′-OH DNAs in the kinase acceptor site by soaking two different 5-mer oligonucleotides (HOGTCAC and HOTGCAC) into preformed kinase crystals. In both cases (pdb 1RC8 and 1RPZ), only the first three nucleotides of the acceptor strand could be modeled into electron density. The positions and contacts of the terminal HONp nucleotide are conserved in the two T4 Pnk structures, but the path and contacts of the second and third nucleotides are different (Figure 6, compare middle and lower panels) (23). To query shared and distinctive aspects of acceptor recognition by the bacterial and phage kinases, we aligned the acceptor sites of the two T4 Pnk•DNA structures to that of the CthPnk Michaelis complex (Figure 6). That the variant Asn38 side chain in the CthPnk Michaelis complex coordinates the oligonucleotide O5′ in the same manner as does the Asp35 side chain in the T4 Pnk•oligo complexes supports our contention that Asn38 faithfully mimics the atomic interactions of the native Asp38 residue, with Asp38 and Asp35 acting as general base catalysts for CthPnk and T4 Pnk, respectively. Other conserved features of acceptor recognition include (i) anchoring of the first phosphodiester by arginine and threonine side chains (Arg41/Thr80 in CthPnk; Arg38/Thr86 in T4 Pnk); (ii) a salt bridge between Asp38/Asp35 and the essential Arg41/Arg38 side chain that orients the general base; and (iii) packing of the first nucleobase against aliphatic side chains that line the acceptor pocket (Val129/His133 in CthPnk; Val135/Met139 in T4 Pnk). The position of the terminal HONp nucleotide is almost identical in the bacterial and phage Pnk acceptor sites, notwithstanding that the nucleobases are different (cytosine in CthPnk versus guanine or thymine in the T4 Pnk structures). A notable distinction between the interactions of the two kinases with the terminal HONp nucleotide is that the contact of CthPnkp Ser37 with the first phosphodiester (loss of which impacts kinase activity) is not conserved in T4 Pnk. Rather, the corresponding Arg34 side chain in T4 Pnk either (i) coordinates the second phosphodiester of the HOGpTpC ligand (P2 in Figure 6, middle panel) or (ii) makes a cation-π stack over the C8-N7 edge of the penultimate guanine nucleobase of the HOTpGpC ligand (G2 in Figure 6, bottom panel) while also engaging in a bonding network with Glu57 and Thr61.
Figure 6.
Comparison of acceptor binding modes in CthPnk versus T4 Pnk. Stereo views of the aligned and vertically offset phosphoacceptor binding sites of CthPnk bound to oligonucleotide HOCCTG (from the B protomer; top panel), T4 Pnk bound to oligonucleotide HOGTC (from pdb 1RC8; middle panel) and T4 Pnk bound to oligonucleotide HOTCG (from pdb 1RPZ; bottom panel). The oligonucleotides are depicted as stick models with green carbons. Amino acid carbons are colored beige (top panel), blue (middle panel) or cyan (bottom panel). Atomic contacts are indicated by black dashed lines.
The structural alignment highlights extreme divergence of the CthPnk and T4 Pnk acceptor sites starting from the second nucleotide of the acceptor strand. From the perspective of the nucleic acid substrate, the salient difference is that there is no stacking of consecutive nucleobases in the CthPnk structure. By contrast, both T4 Pnkp•DNA structures reveal acceptor base stacking, albeit different patterns of stacking according to the sequence of the acceptor strand. In the HOGpTpC complex, the G1 and T2 bases stack on one another while the C3 base is splayed out almost orthogonal to the first two bases (Figure 6, middle panel). In the HOTpGpC complex, the G2 base projects in the opposite direction and stacks over the C3 base (Figure 6, bottom panel). The G2 base in T4 Pnkp•HOTpGpC adopts a position analogous to the C2 base of the CthPnk Michaelis complex, but the enzymic interactions of the base differ: to wit, the G2 base in T4 Pnk stacks on the Arg34-Glu57 ion pair (Figure 6, bottom panel), while the C2 base in CthPnk makes a π stack on Phe58 (Figure 6, top panel). CthPnkp Phe58 occupies the same space as the Arg34 guanidinium moiety in T4 Pnkp•HOTpGpC. The T4 Pnk residue corresponding to CthPnk Phe58 is Thr61, which either (i) makes van der Waals contacts with the C3 base in the HOGpTpC complex (Figure 6, middle panel), or (ii) donates a hydrogen bond to the G2 base in the HOTpGpC complex (Figure 6, bottom panel).
The Gln83 position in CthPnkp that donates a hydrogen bond to the second phosphodiester of the acceptor strand is occupied by a shorter Asn89 side chain in T4 Pnkp. Asn89 makes van der Waals contacts to the second phosphodiester in the HOGpTpC complex and a van der Waals contact to the C3 base in the HOTpGpC complex. Although neither of the T4 Pnkp•DNA structures has a modeled fourth nucleobase akin to G4 in CthPnk, the positions of the third ribose are such that the trajectory of the fourth nucleoside will be different than what we see in CthPnk, where G4 stacks on Tyr128. (The T4 Pnkp equivalent of Tyr128 is Asp134.)
In summary, the conservation of the terminal HONp site in the T4 and Cth kinases attests to the shared catalytic mechanism via fixation of the first phosphodiester and deployment of an aspartate general base. The acceptor sites of these two enzymes are otherwise distinct, with respect to the path of the acceptor strand and the enzymic functional groups that interact with the single strand nucleic acid.
ACCESSION NUMBERS
The coordinates for the Michaelis complex and product complex have been deposited in the RCSB protein structure database (pdb ID codes 4MDF and 4MDE).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM42498]. Funding for open access charge: NIH [GM42498].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Amitsur M, Levitz R, Kaufman G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martins A, Shuman S. An end-healing enzyme from Clostridium thermocellum with 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. RNA. 2005;11:1271–1280. doi: 10.1261/rna.2690505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan CM, Zhou C, Huang RH. Reconstituting bacterial RNA repair and modification in vitro. Science. 2009;326:247. doi: 10.1126/science.1179480. [DOI] [PubMed] [Google Scholar]
- 4.Jain R, Shuman S. Bacterial Hen1 is a 3′ terminal RNA ribose 2′O-methyltransferase component of a bacterial RNA repair cassette. RNA. 2010;16:316–323. doi: 10.1261/rna.1926510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C, Chan CM, Wang P, Huang RH. Probing the substrate specificity of the bacterial Pnkp/Hen1 RNA repair system using synthetic RNAs. RNA. 2012;18:335–344. doi: 10.1261/rna.030502.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang LK, Das U, Smith P, Shuman S. Structure and mechanism of the polynucleotide kinase component of the bacterial Pnkp-Hen1 RNA repair system. RNA. 2012;18:2277–2286. doi: 10.1261/rna.036061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang LK, Smith P, Shuman S. Structure and mechanism of the 2′,3′ phosphatase component of the bacterial Pnkp-Hen1 RNA repair system. Nucleic Acids Res. 2013;41:5864–5873. doi: 10.1093/nar/gkt221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith P, Wang LK, Nair PA, Shuman S. The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family. Proc. Natl Acad. Sci. USA. 2012;109:2296–2301. doi: 10.1073/pnas.1116827109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keppetipola N, Shuman S. Mechanism of the phosphatase component of Clostridium thermocellum polynucleotide kinase-phosphatase. RNA. 2006;12:73–82. doi: 10.1261/rna.2196406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keppetipola N, Shuman S. Distinct enzymic functional groups are required for the phosphomonoesterase and phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase/phosphatase. J. Biol. Chem. 2006;281:19251–19259. doi: 10.1074/jbc.M602549200. [DOI] [PubMed] [Google Scholar]
- 11.Keppetipola N, Shuman S. Characterization of the 2′,3′ cyclic phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase and bacteriophage λ phosphatase. Nucleic Acids Res. 2007;35:7721–7732. doi: 10.1093/nar/gkm868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keppetipola N, Nandakumar J, Shuman S. Reprogramming the tRNA splicing activity of a bacterial RNA repair enzyme. Nucleic Acids Res. 2007;35:3624–3630. doi: 10.1093/nar/gkm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P, Chan CM, Christensen D, Zhang C, Selvadurai K, Huang RH. Molecular basis of bacterial protein Hen1 activating the ligase activity of bacterial protein Pnkp for RNA repair. Proc Natl Acad. Sci. USA. 2012;109:13248–13253. doi: 10.1073/pnas.1209805109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan CM, Zhou C, Brunzelle JS, Huang RH. Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc. Natl Acad. Sci. USA. 2009;106:17699–17704. doi: 10.1073/pnas.0907540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain R, Shuman S. Active site mapping and substrate specificity of bacterial Hen1, a manganese-dependent 3′ terminal RNA ribose 2′O-methyltransferase. RNA. 2011;17:429–438. doi: 10.1261/rna.2500711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das U, Wang LK, Smith P, Shuman S. Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial Pnkp•Hen1 RNA repair system. Biochemistry. 2013;52:4734–4743. doi: 10.1021/bi400412x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang LK, Lima CD, Shuman S. Structure and mechanism of T4 polynucleotide kinase—an RNA repair enzyme. EMBO J. 2002;21:3873–3880. doi: 10.1093/emboj/cdf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galburt EA, Pelletier J, Wilson G, Stoddard BL. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure. 2002;10:1249–1260. doi: 10.1016/s0969-2126(02)00835-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, Yin S, Shuman S. Characterization of polynucleotide kinase/phosphatase enzymes from mycobacteriophages Omega and Cjw1 and vibriophage KVP40. J. Biol. Chem. 2004;279:26358–26369. doi: 10.1074/jbc.M403200200. [DOI] [PubMed] [Google Scholar]
- 20.Martins A, Shuman S. Characterization of a baculovirus enzyme with RNA ligase, polynucleotide 5′ kinase and polynucleotide 3′ phosphatase activities. J. Biol. Chem. 2004;279:18220–18231. doi: 10.1074/jbc.M313386200. [DOI] [PubMed] [Google Scholar]
- 21.Wang LK, Shuman S. Domain structure and mutational analysis of T4 polynucleotide kinase. J. Biol. Chem. 2001;276:26868–26874. doi: 10.1074/jbc.M103663200. [DOI] [PubMed] [Google Scholar]
- 22.Wang LK, Shuman S. Mutational analysis defines the 5′ kinase and 3′ phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res. 2002;30:1073–1080. doi: 10.1093/nar/30.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eastberg JH, Pelletier J, Stoddard BL. Recognition of DNA substrates by bacteriophage T4 polynucleotide kinase. Nucleic Acids Res. 2004;32:653–660. doi: 10.1093/nar/gkh212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvest RL, Lowe G. The stereochemical course of the phosphoryl transfer reaction catalyzed by polynucleotide kinase (bacteriophage-T4-infected Escherichia coli B) Biochem. J. 1981;199:273–276. doi: 10.1042/bj1990273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.