Abstract
Upstream AUGs (uAUGs) and upstream open reading frames (uORFs) are common features of mRNAs that encode regulatory proteins and have been shown to profoundly influence translation of the main ORF. In this study, we employed a series of artificial 5′-untranslated regions (5′-UTRs) containing one or more uAUGs/uORFs to systematically assess translation initiation at the main AUG by leaky scanning and reinitiation mechanisms. Constructs containing either one or two uAUGs in varying contexts but without an in-frame stop codon upstream of the main AUG were used to analyse the leaky scanning mechanism. This analysis largely confirmed the ranking of different AUG contextual sequences that was determined previously by Kozak. In addition, this ranking was the same for both the first and second uAUGs, although the magnitude of initiation efficiency differed. Moreover, ∼10% of ribosomes exhibited leaky scanning at uAUGs in the most favourable context and initiated at a downstream AUG. A second group of constructs containing different numbers of uORFs, each with optimal uAUGs, were used to measure the capacity for reinitiation. We found significant levels of initiation at the main ORF even in constructs containing four uORFs, with nearly 10% of ribosomes capable of reinitiating five times. This study shows that for mRNAs containing multiple uORFs/uAUGs, ribosome reinitiation and leaky scanning are efficient mechanisms for initiation at their main AUGs.
INTRODUCTION
The regulation of mRNA translation is an important component of gene expression control. The most critical and rate limiting step in translation is initiation, which involves the formation of an elongation-competent 80S ribosome at a start codon either by the ribosome scanning mechanism or by a cap-independent mechanism (1–3). For most eukaryotic mRNAs translation occurs by the scanning mechanism, which requires the assembly of a preinitiation complex, consisting of the ribosomal 40S subunit and several initiation factors, at the 5′-cap structure of the mRNA. The 40S complex then scans linearly along the 5′-untranslated region (5′-UTR) until an AUG start codon is encountered. Recognition of an AUG leads to recruitment of the 60S subunit to complete formation of the 80S ribosome and initiation of protein synthesis. A few eukaryotic mRNAs have been shown to initiate translation in a cap-independent manner via specialized regulatory elements known as internal ribosome entry sites (IRESs) that directly recruit and bind ribosomes to an initiation codon without requiring the assembly of factors at the 5′-end of the transcript (reviewed in 1).
The regulation of initiation can be mediated by 5′-UTR elements such as stem–loop structures and upstream AUGs (uAUGs)/upstream ORFs (uORFs). The influence of stem–loops on initiation is determined by the stability of the structure, its location within the 5′-UTR and the ability of the stem–loop to bind specific regulatory proteins (4). When stable stem–loop structures are located near the 5′-cap, translation efficiency is dramatically reduced due to interference with assembly of the preinitiation complex. In mRNAs with stable stem–loops located further downstream, these structures reduce translation efficiency by opposing the unwinding activity of ribosome-associated helicase and thereby impede conventional scanning of 40S subunits. On the other hand, there are examples of 5′-UTRs containing stem–loops located near a start codon that can enhance recognition of the preceding AUG codon.
The influence of uAUGs/uORFs on initiation through modulation of the cap-dependent ribosome scanning mechanism has been recognized for some time, although their wider relevance to gene regulation has been questioned. However, the prevalence of mRNAs with complex 5′-UTRs containing one or more uAUGs and uORFs, now estimated to be ∼50% of all human mRNAs, has renewed interest in the role of these features in post-transcriptional gene regulation (5–9). The empirical evidence shows that uAUGs/uORFs usually diminish translation of the main ORF by reducing the number of ribosomes reaching and initiating at the authentic or main AUG start codons (4,10–14). Ribosomes reaching the main AUG of these mRNAs do so mainly via context-dependent leaky scanning and/or reinitiation mechanisms, although it is widely believed that these are inefficient mechanisms (12,13).
The context-dependent leaky scanning mechanism accounts for the observation that some 40S subunits will fail to initiate at AUG codons with a less than optimal context and continue scanning along the 5′-UTR. The most efficient context for initiation of protein translation is known as the Kozak sequence (GCCA/GCCAUGG), which was initially identified as a consensus sequence delineating the AUG start codon of vertebrate mRNAs (12). Two positions within this sequence, –3 and +4 (the A of the AUG codon is designated +1) are the most critical for determining the strength of the initiator and hence translation efficiency. The context of an AUG codon is regarded as strong or optimal when either one (A/GNNAUGN) or both (A/GNNAUGG) of these positions match the Kozak sequence (12). It is thought that most if not all ribosomes will initiate at these optimal AUGs. Recent bioinformatic analyses have shown that 95% of main AUGs have an optimal context, which compares to 63% of uAUGs with optimal contexts (9).
The reinitiation mechanism describes the ability of 40S subunits to continue to scan and initiate at a downstream AUG codon after translating a small independent uORF. The efficiency of this process in higher eukaryotes is governed, at least in part, by the size of uORFs and the distance between the uORF and the downstream AUG (11–14). Ribosome reinitiation is most efficient when the uORF is relatively small and the intercistronic spacer is >50 nt in length (15–18). Reinitiation is considered to be a rare event, although the incidence of this mechanism may be much greater, since only a few mRNAs with uORFs have been adequately examined (13).
Although there are many examples of mRNAs where initiation at the main AUG is influenced by uAUGs/uORFs through the mechanisms described above, there are some mRNAs where regulation of initiation by uAUGs/uORFs has been attributed to other mechanisms, including uORF peptide-dependent regulation, spacer-sequence dependent regulation, uORF-directed initiation, ribosome shunting and uORF-dependent IRES-mediated initiation. Peptide-dependent regulation has been observed for a number of mRNAs, including gpUL4 (19), CPA1 (20,21), AMD1 (22), Cx41 (23) and mdm2 (24). In these examples, specific uORF-encoded peptide products inhibit initiation of downstream start codons by stalling ribosomes at or near the termination codon of the uORF. In contrast, regulation of yeast GCN4 translation by nutrient levels is independent of the peptide sequences encoded by its uORFs. Instead it was found that translation of the main GCN4 ORF is governed by sequences adjacent to the first and fourth uORFs, with the AU-rich sequences surrounding the stop codon of uORF1 promoting high levels of reinitiation, and the GC-rich sequences flanking the stop codon of uORF4 potentiating ribosome release (11). An unknown mechanism appears to mediate glucose-regulated expression of the CD36 gene, where inhibition by a uORF is concomitantly reduced by increasing glucose levels (25). There are also several examples, including Fli1, C/EBPα, C/EBPβ and SCL, where uORFs determine the start codon that is used by directing ribosomes to an internal AUG (iAUG), thereby influencing the ratio of full-length to truncated isoforms that are produced (26–29). A ribosome shunt mechanism has been postulated to account for the observations made in studies using synthetic mRNA leaders and cauliflower mosaic virus, where translation of a uORF was required for a cap-dependent ribosome shunt across a stem–loop structure in order to initiate translation downstream (30,31). Finally, translation of the uORF in the cat1 gene was found to enhance translation of the main ORF via a cap-independent mechanism by unfolding an inhibitory structure in the mRNA leader and eliciting a conformational change that yields an active IRES (32).
While some general principles of how uAUGs/uORFs modulate initiation have been determined, our understanding of these processes is still far from complete. This is due in part to the small number of well-characterized 5′-UTRs with these features. In addition, the discovery of large numbers of mRNAs containing uAUGs/uORFs has further highlighted the need to understand how these features regulate translation so that predictive rules can be established. In order to systematically and independently assess the capability of the leaky scanning and reinitiation mechanisms and their relative contributions to the expression of the main ORF, we used two types of artificial 5′-UTR constructs, containing either one to two uAUGs with different initiator strengths or one to four small uORFs with uAUGs in either ‘weak’ or ‘strong’ contexts. The data reveals that ribosome leaky scanning and reinitiation mechanisms can produce biologically significant protein levels from mRNAs containing multiple uAUGs/uORFs.
MATERIALS AND METHODS
Construction of green fluorescent protein (GFP) vectors
Twelve pairs of complementary oligonucleotides with NheI and AgeI restriction sites were used to generate the two sets of 5′-UTR constructs containing only uAUGs (sequences detailed in Fig. 1A and B). Complementary oligonucleotide pairs were annealed and directly cloned into the corresponding sites of the GFP expression vector pEGFP-N1 (Clontech). The constructs containing one or more uORFs were generated as described previously (33). Briefly, two pairs of complementary oligonucleotides, GDq1/2 and GDq3/4 (Table 1), containing a BglII site at the 5′-end and a BamHI site at the 3′-end were used to generate the first (1–5uORFs) and second (1–4uORFsK) sets of constructs. Complementary oligonucleotides were annealed and ligated (T4 DNA ligase) in the presence of both restriction enzymes to form unidirectional concatemers. The concatemers were then used as templates for PCR amplification with primer pairs GDq5/6 and GDq7/8 (Table 1). The resultant PCR products were cloned into the pGEM-T Easy vector (Promega). All inserts were subsequently subcloned into NheI and AgeI sites of vector pEGFP-N1. The third set of constructs were similarly produced, except that the initial insert containing a single uORF with a 52 nt spacer was obtained by PCR using primers GDq7a (Table 1) and mGliR2Bgl (see table II in 33) and a GLI1 α-UTR mutant (construct αd; X.-Q. Wang and J. A. Rothnagel, in preparation) as the template. To generate construct uATGbL containing a uAUG in the most optimal context but with a longer leader sequence of 94 nt, a pair of complementary oligonucleotides (with the same sequence as uATGb but with AgeI restriction site at both ends) was annealed and directly cloned into the AgeI site of the GFP expression vector pEGFP-N1. The stem–loop sequence (ctagcggggcgcgtggtggcgggttaacccgccaccacgcgccccg) with flanking NheI sites (in bold) was cloned into the corresponding sites of uATGbL (uATGbL-SL) and pEGFP-N1 (pEGFP-N1-SL). All inserts were verified by direct sequencing.
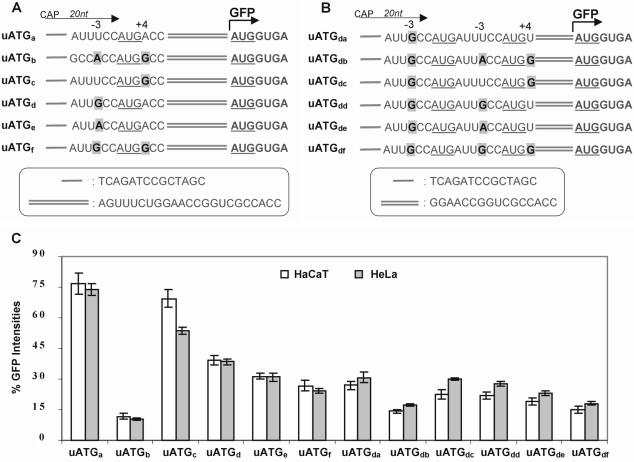
Figure 1.
Flow cytometry analysis of constructs with one or two uAUGs of various strengths and no in-frame stop codon upstream of the main AUG in transiently transfected mammalian cells. (A) Schematic of constructs containing only one uAUG with various strengths. The uORF specified by these uAUGs terminates 1 nt downstream of the GFP start codon. The 20 nt leader sequence is indicated by an arrow and includes 14 nt provided by the pEGFP-N1 vector as shown in the insert box. The uAUG is underlined and the beginning of the GFP ORF is indicated by a bent arrow. The uAUG contexts in each construct differ from each other in positions –3 and +4 as indicated in bold and highlighted. The sequence between the uAUG and main AUG is shown in the insert box. (B) Schematic of constructs containing two uAUGs within the 5′-UTR. The two uAUGs are underlined. The first uAUG has a fixed strength (G–3) and the second uAUG has differing strengths as indicated. The sequence between the second uAUG and main AUG is shown in the insert box. (C) Graphical representation of relative GFP intensities determined by flow cytometry of the constructs shown in (A) and (B). The values were normalized using a control construct containing the same sequence as uATGe but with the uAUG mutated to UUG. ANOVA statistical analysis confirmed significant differences (P < 0.001) between all constructs within each grouping.
Table 1. Primer sequences.
| Name | Nucleotide sequencesa |
|---|---|
| GDq1 | gatctatttccATGaccagtttctgag |
| GDq2 | gatcctcagaaactggtcATGgaaata |
| GDq3 | gatctgccaccATGgccagtttctgag |
| GDq4 | gatcctcagaaactggccATGgtggca |
| GDq5 | gctagcatttccATGacc |
| GDq6 | accggttcagaaactggt |
| GDq7 | gctagcgccaccATGgcc |
| GDq8 | accggttcagaaactggc |
| GDq7a | ggatccgccaccATGgccagtttctga |
aThe restriction endonuclease recognition sites for BglII, BamHI, NheI and AgeI are shown in bold.
Transfection in mammalian cells and data analysis.
Two cell lines, HaCaT (34) and HeLa (35), were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, ampicillin and streptomycin (Life Technologies). Transient transfection of all constructs was performed using LipofectAMINE 2000 reagent (Life Technologies) according to the manufacturer’s instructions. Cells were plated into 24-well or 6-well plates and the DNA–LipofectAMINE 2000 complexes added to the cultures on the second day. After incubation at 37°C for a further 20 h, the cells were harvested for flow cytometry and analysed as described previously (33). The transfections were repeated at least four times for each set of constructs. The values and error bars shown represent the mean and standard variation of four or more independent experiments.
RNA extraction and northern hybridization
Total RNA was isolated from cell cultures using TRI Reagent (Molecular Research Center) and electrophoresed (20 µg/well) on a denaturing agarose gel. The size-separated transcripts were transferred to a Hybond N nylon membrane (Amersham) by capillary diffusion and fixed by UV irradiation. The GFP probe was generated by BamHI and NotI digestion of the pEGFP-N1 vector and labeled with [α-32P]dCTP by random primer labelling using the RTS Radprime kit (Life Technologies). The membranes were hybridized in Rapid-hyb buffer (Amersham) at 65°C overnight or for 2 h, then washed and autoradiographed.
RESULTS AND DISCUSSION
Initiation at the main AUG in constructs containing uAUGs
To systematically assess the extent of leaky scanning in relation to the strength of AUG start codons, we designed two sets of 5′-UTR constructs using GFP as the reporter (Fig. 1A and B). These constructs contained a leader of 20 nt and one or two uAUGs but no in-frame terminator codon upstream of the main AUG, ensuring that initiation at the main AUG is only by ribosome leaky scanning and not by the reinitiation mechanism. The same 5′-UTRs were present in the control constructs except that the uAUGs were changed to UUG. The controls were used to set the 100% level of recognition at the main AUG. The difference in the level of GFP expression between the control and test constructs is a measure of the degree of leaky scanning past uAUGs, from which the percentage of ribosomes initiating at uAUGs can be calculated. The first set of constructs contained only one uAUG and differed from each another by the strength of their uAUG initiator codon, where either one or both nucleotide residues at positions –3 and/or +4 flanking the uAUG were the same as the Kozak consensus sequence (Fig. 1A). The second set of constructs contained two in-frame uAUGs, where the context of the first uAUG was kept invariant (suboptimal, G–3+A+4) while the strength of the second uAUG, located 6 nt downstream, was varied as shown (Fig. 1B). The influence of these 5′-UTRs on expression of the GFP reporter gene was then analysed in transiently transfected cells by flow cytometry as described previously (33). In our initial experiments, we used several different cell lines, including COS-1, CHO, BHK, HaCaT and HeLa. Although there were some differences in the levels of GFP produced in the various cell lines by individual constructs, the same trends were always observed. The results presented here are from transfected HaCaT and HeLa cells. The former is a non-tumorigenic human keratinocyte cell line (34) and the latter is a human epithelial cancer cell line (35) that has been widely used for translation studies (17,24,26).
Our results confirmed that the canonical Kozak sequence was the most efficient for translation initiation (Fig. 1C) (36,37). The strength of the AUG context sequences ranged from ‘strong’ to ‘weak’ and in descending order were A–3+G+4 > G–3+G+4 > A–3+A+4 > G–3+A+4 > U–3+G+4 > U–3+A+4 (Fig. 1C). Although this order is in general agreement with the earlier reports, the relative extent of ribosome leaky scanning of some AUG contexts observed in the present study differed from those found previously (36,37). In addition, the same relative order of initiation efficiency was also observed for the second uAUGs (Fig. 1C), thus confirming the ranking of AUG contexts in this assay. The efficiency of initiation at the second uAUG was calculated by normalizing for the total number of ribosomes available to initiate at the second uAUG by subtracting those initiating at the first uAUG. This analysis showed that for initiator codons with the same context, ribosomes were generally more likely to bypass the second uAUG than the first uAUG. It is not clear why the second uAUG was less efficient in these constructs but possible explanations include the influence of local secondary structure and/or flanking sequences. We also noticed greater initiation at the main AUG in a construct containing two uAUGs (construct uATGdb, first uAUG with G–3+A+4 and the second uAUG with A–3+G+4) than in a construct with only one uAUG (uATGb, A–3+G+4) although both constructs have one uAUG in the most optimal context (Fig. 1C). This suggests that a subset of ribosomes bypassed the second uAUG, resulting in inefficient recognition of the second uAUG in construct uATGdb. These observations suggest that the impact on a downstream AUG is not only determined by the number of uAUGs and their contextual strength but also by their relative position and the neighbouring sequence environment.
Our data show the influence of uAUGs of various strengths on initiation at a main AUG. The differences in GFP levels detected between constructs with differing contexts indicates that ribosomes initiating at the main AUG must have scanned past the uAUGs, and this could only occur by context-dependent leaky scanning. Importantly, significant leaky scanning occurred at a uAUG set in the most optimal context (uATGb) (Fig. 1A and C). However, this may be due to the relatively short leader sequence of 20 nt between the 5′-cap and the first uAUG. It has been shown previously that ribosomes will bypass an AUG codon in a good context if it is <12 nt from the 5′-cap and that this type of leaky scanning can be reduced by lengthening the leader sequence to ≥20 nt (38).
To confirm if this type of leaky scanning was occurring in the aforementioned constructs, the leader sequence was extended from 20 to 94 nt (construct: uATGbL) (Fig. 2A). Extrapolating from current paradigms, it would be predicted that translation of the main ORF of uATGbL would be almost completely inhibited by the presence of a uAUG set in an optimal context. Instead, we observed no reduction in GFP levels for the uATGbL construct, which were equivalent to those of the sibling construct containing the 20 base leader (uATGb) (Fig. 2B and C), indicating that the degree of leaky scanning was not affected by the short leader sequence. This result was unexpected and shows that leaky scanning can occur even where the leader length is optimal and the first uAUG is in the most favourable context for initiation. It is not known how the relatively high levels of leaky scanning in constructs with long leaders and strong uAUGs could occur but it has been suggested that it may be due to the absence of downstream secondary structure or perhaps by as yet unidentified mechanisms (12).
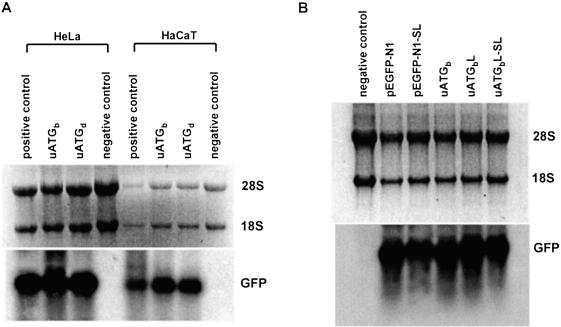
Figure 2.
Flow cytometry analysis of constructs with one uAUG in the most optimal context and a longer leader of 94 nt and constructs containing a stem–loop near the 5′-cap site in transiently transfected mammalian cells. (A) Schematic showing constructs containing one uAUG in the most optimal context with a short leader of 20 nt (uATGb), a leader of 94 nt (uATGbL) or containing a stem–loop 14 nt downstream of the 5′-cap site (uAUGbL-SL). The control constructs lacking uAUGs, with (pEGFP-N1-SL) and without (pEGFP-N1) stem–loops, are shown. The various leader lengths between the 5′-cap and first AUG in these constructs are indicated by arrows. The uAUG is underlined and the beginning of the GFP ORF is indicated by a bent arrow. The sequence between the uAUG and main AUG is shown in the insert box. (B) GFP fluorescence intensity histograms compiled from the analysis of 20 000 cells per sample for each construct in transfected HeLa cells. It shows that construct uATGbL expresses GFP fluorescence levels that are similar to construct uATGb. Both stem–loop constructs (uAUGbL-SL and pEGFP-N1-SL) display GFP fluorescence levels that are similar to the untransfected control. (C) Graphical representation of GFP intensities equating to translation efficiency for each construct in transfected HeLa cells.
Another possibility is that the observed GFP levels were not due to the translation of transcripts with full-length 5′-UTRs but instead were produced from truncated mRNAs that lack the regulatory uAUGs (39). To rule this out, we introduced a highly stable stem–loop structure 14 nt from the 5′-cap in construct uATGbL and in the control vector pEGFP-N1, creating two new constructs, uATGbL-SL and pEGFP-N1-SL (Fig. 2A). This structure consists of an 18 bp stem and a 4 nt loop (30,40) and, because of its proximity to the 5′-cap, was expected to interfere with assembly of the preinitiation complex, blocking access to the 5′-cap by 40S subunits and thus preventing downstream initiation. As can be seen in Figure 2B, the positive peaks representing GFP-expressing cells transfected with constructs uATGbL and pEGFP-N1 are completely absent in cells transfected with the corresponding stem–loop-containing constructs; uATGbL-SL and pEGFP-N1-SL. Quantitative analysis revealed that almost 99% of ribosomes were indeed blocked by the stem–loop-containing constructs compared with their corresponding controls (Fig. 2C), implying that the low level of translation observed for construct uATGbL resulted from ribosome leaky scanning rather than by initiation of anomalous truncated transcripts or by a cap-independent mechanism. In addition, northern hybridization was performed to exclude the possibility that the marked differences in GFP expression observed for different constructs were due to differences in mRNA levels. This showed that mRNA levels were similar for all constructs in both HeLa and HaCaT cell lines, indicating that the differences in GFP protein production can be attributed to differences in translational efficiency (Fig. 3). Finally, we determined that GFP expression in these constructs was initiated at the main AUG start codon since a construct corresponding to uAUGbL, but with the start codon of the GFP reporter mutated to UUG, produced a signal just above background (data not shown). Taken together our data show a degree of leaky scanning at an AUG set in the most favourable context that was hitherto not thought to occur. However, it does imply that the ‘first AUG rule’ requires modifying with the qualification that a subset of 40S subunits can scan past optimal uAUGs to allow low level translation of the main AUG. This suggests that leaky scanning is probably a very common mechanism for restricting the levels of certain regulatory proteins such as oncogenes, growth factors and signalling proteins.
Figure 3.
Northern analysis of mRNA levels in transfected cells. (A) Total RNA was prepared from cell cultures transfected with constructs uATGb, uATGd, a positive control (a construct with the same leader length but lacking a uAUG) and a negative control (untransfected control) as indicated on top of each lane. Similar intensities were detected for the positive control and uATGb and uATGd constructs when adjusted for RNA loading levels (28S and 18S RNA). (B) Northern hybridization of transcripts produced by constructs pEGFP-N1, pEGFP-N1-SL, uATGb, uATGbL and uATGbL-SL transfected into HeLa cells. Similar GFP mRNA levels were seen for all transfected constructs.
Initiation at the main AUG in constructs containing uORFs
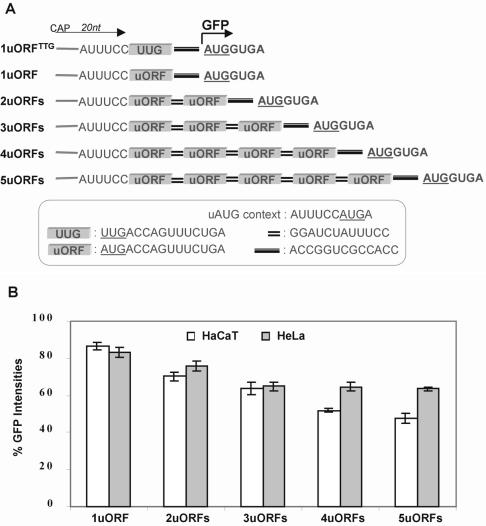
To determine the level of inhibition at a main AUG imposed by increasing numbers of uORFs, we initially used a set of constructs containing up to five copies of a uORF. We used a four amino acid uORF that is present in one of the alternative GLI1 5′-UTRs (33). These constructs consisted of a 20 nt leader sequence, one or more uORFs each with a ‘weak’ uAUG (same context as in construct uATGa, Fig. 1A) and a 12 nt spacer between uORFs (Fig. 4A). It was predicted that initiation at the main AUG in these constructs would occur not only by ribosome reinitiation but also by context-dependent leaky scanning. The small size of the uORF was predicted to give maximum capacity for reinitiation and a 12 nt spacer was included to provide a reasonable level of reinitiation recovery. The inhibitory effect of individual uORFs was determined indirectly by measuring initiation at the main AUG in comparison with a corresponding 5′-leader containing no uAUGs and uORFs. As shown in Figure 4B, initiation at the main AUG gradually declined as the number of uORFs increased. It was also observed that the inhibitory effect of an individual uORF on GFP levels reduced as its relative position from the 5′-cap site increased. Nevertheless, initiation at the main AUG remained high even in a construct containing five uORFs (Fig. 4B). From Figure 4 we calculate that 50–65% of 40S subunits entering from the 5′-cap are able to reach and initiate at a sixth AUG in a mRNA with five ‘weak’ uAUGs, by both ribosome leaky scanning and reinitiation mechanisms.
Figure 4.
Flow cytometry analysis of constructs containing multiple uORFs with weak uAUGs and a 12 nt intercistronic spacer between ORFs in transiently transfected mammalian cells. (A) Schematic showing the 5′-UTR structure of constructs containing differing numbers of uORFs with weak uAUGs. The 20 nt leader sequence is indicated by an arrow and includes 14 nt provided by the pEGFP-N1 vector. The insert box shows the context of uAUGs, the sequences of the uORFs, the intercistronic spacer between uORFs and the spacer between the ultimate uORF and the main ORF. The beginning of the GFP ORF is indicated by a bent arrow. The control construct 1uORFTTG has the same sequence and length as 1uORF but with the uAUG mutated to uUUG. (B) Graphical representation of GFP intensities determined by flow cytometry of constructs shown in (A). All values are relative to construct 1uORFTTG.
While the context of uAUGs determines the rate of leaky scanning and the length of both uORFs and intercistronic spacers affects the efficiency of reinitiation, the peptides encoded by some uORFs have been shown to have regulatory properties. In these cases the peptides block ribosome movement by targeting the components of the translation machinery, thereby preventing leaky scanning and reinitiation at downstream AUGs (20,23,41,42). Whether or not peptide-dependent regulation occurs in our constructs is not known, but based on the observations of others we feel that it is unlikely given the degree of inhibition exhibited by each uORF. It has also been suggested that translation of mRNAs containing uAUGs may occur via a cap-independent mechanism in order to circumvent regulatory uAUGs/uORFs. However, our findings do not support an IRES-mediated mechanism operating on the constructs used in this study since initiation at the main AUG reduced concomitantly with each additional uORF. We therefore conclude that ribosomes arriving at the main AUG have done so via cap-dependent linear scanning. Our data has relevance to the estimated 37–57% of mRNAs that contain uORFs with weak AUG contexts (13) by showing that efficient initiation of the main AUG of these transcripts can be mediated by leaky scanning and reinitiation mechanisms.
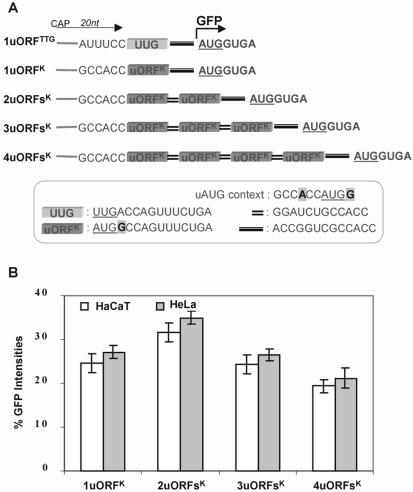
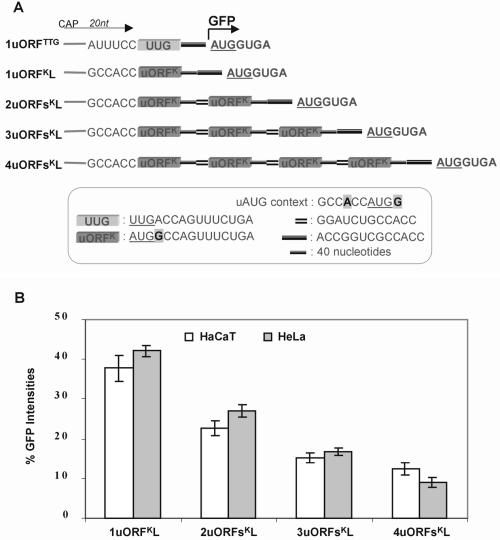
We next produced a second set of constructs that were similar to the previous set except that the uORFs contained strong uAUGs set in the most optimal context (Fig. 5A). These constructs were designed to investigate the efficiency of the reinitiation mechanism while minimizing the contribution from the leaky scanning mechanism (see Fig. 1). Since the contexts for the main AUG and uAUGs are identical, these constructs offer a direct indication of ribosome initiation at each uAUG. The results revealed that initiation at the main AUG was indeed significantly inhibited compared with the corresponding constructs with weak uAUGs (compare Figs 4B and 5B), indicating that leaky scanning contributes significantly to initiation at the main AUG in the first set. Notably, we did not observe complete inhibition of initiation at the main AUG even in a construct with four uORFs, which surprisingly exhibited initiation at 20% of control levels (Fig. 5B). These results demonstrate that a significant number of ribosomes were able to recharge and reinitiate at a downstream AUG after scanning and initiating at four uAUGs with optimal context. Although a number of reports have investigated the influence of intercistronic length and uORF size on reinitiation efficiency, no study has to date explored the consequence of an increasing number of uORFs or compared weak with strong uAUG contexts. The best characterized 5′-UTR with multiple uORFs is the yeast GCN4 transcript, which contains four small uORFs each encoding two or three amino acids and with uAUGs residing in relatively optimal contexts (11). In these aspects the 5′-UTR of the GCN4 transcript closely resembles the constructs shown above, although reinitiation at downstream ORFs has been shown to be dependent on long intercistronic regions which are thought to be necessary to allow ribosomes the time needed to acquire competency (43,44). This differs from the uORF used in our study, which demonstrated a greater degree of reinitiation capacity without requiring long intercistronic spacers in order to resume scanning.
Figure 5.
Flow cytometry analysis of constructs containing multiple uORFs with the Kozak consensus sequence flanking uAUGs and a 12 nt spacer between ORFs in transiently transfected mammalian cells. (A) Schematic showing the 5′-UTR structure of constructs containing differing numbers of uORFs with ‘strong’ uAUGs. Construct design was as for Figure 4A except that the Kozak sequence flanked uAUGs. The insert box shows the context of uAUGs, the sequences of the uORFs, the intercistronic spacer between uORFs and the spacer between the ultimate uORF and the main ORF. (B) Graphical representation of GFP intensities determined by flow cytometry of constructs shown in (A). All values are relative to construct 1uORFTTG.
Interestingly, the inhibition of the main AUG imposed by one uORF was greater than that produced by two uORFs (compare constructs 1uORFK and 2uORFsK in Fig. 5B). An explanation for this apparent anomaly may be found in the length of the spacer between the uORF and the main AUG, since the ability of ribosomes to reinitiate at downstream AUGs is impaired by short-length spacers (15–18). The lower levels of reinitiation at the main AUG in construct 1uORFK could be due to the relative short spacer of 13 nt, whereas the higher reinitiation levels observed in construct 2uORFsK may be due to the second uORF acting as a 27 nt spacer giving a total length of 40 nt. This explanation also suggests the possibility that some ribosomes reaching the fifth AUG in construct 4uORFsK may have escaped reinitiation at some uAUGs due to the short spacer between uORFs.
In order to ensure maximum reinitiation, a third set of constructs was designed with longer spacers (Fig. 6A). These constructs had the same leader and uORF sequences as the constructs shown in Figure 5, but the intercistronic spacer was lengthened from 12 to 52 nt as a 52 nt spacer had been shown by others to allow maximum recovery of ribosome reinitiation (15,16). It was therefore expected that expression levels of the main ORF in these constructs would be a measure of the true capacity of the reinitiation mechanism, since the majority of ribosomes reaching the main AUG codon would do so after translating each of the uORFs. The data shows that initiation at the main AUG was lower in these constructs than in the two previous sets of constructs (compare Figs 4 and 5 with 6B). We found that ∼40% of ribosomes were able to initiate twice, which is consistent with a previous report (15), and ∼25% were able to initiate three times. Even in a construct with four uORFs (with the most optimal AUG context) separated by relatively long intercistronic spacers, initiation at the main AUG was as high as 10% of total initiation capacity (Fig. 6B). These results indicate that nearly 10% of ribosomes entering from the 5′-cap site are capable of reinitiating at least five times. In addition, we assessed GFP expression by mutant constructs in which the start codon of the GFP reporter was changed to UUG but which were otherwise identical to constructs 5uORFs, 4uORFsK and 4uORFsKL. The mutant constructs produced near background reporter levels (data not shown), indicating that initiation of the main ORF of the constructs used here was principally through the AUG start codon and not through non-AUG initiator codons or by other mechanisms. While it is unlikely that a natural mRNA would contain multiple uORFs each with strong uAUGs, our data show that initiation at the main AUG in such a transcript would still occur by the reinitiation mechanism.
Figure 6.
Constructs containing multiple uORFs with the Kozak consensus sequence flanking uAUGs and a 52 nt spacer between ORFs and their analysis in transiently transfected mammalian cells. (A) Schematic showing the 5′-UTR structure of constructs containing differing numbers of uORFs with ‘strong’ uAUGs and a longer intercistronic spacer. Construct design was as for Figure 5A except with longer spacers between uORFs. Each construct contains a 53 nt spacer between the main AUG of the GFP reporter and the preceding uORF. The insert box shows the context of uAUGs, the sequences of the uORFs, the intercistronic spacer between uORFs and the spacer between the ultimate uORF and the main ORF. (B) Graphical representation of GFP intensities determined by flow cytometry of constructs shown in (A). All values are relative to construct 1uORFTTG.
Several recent bioinformatic studies have identified an increasing number of mRNAs containing multiple uAUGs/uORFs (5,7–9) and, therefore, it has been predicted that the number of mRNAs using the reinitiation mechanism for translation of the main ORF is expected to increase. Notwithstanding that the 5′-UTRs analyzed in the present study were artificial and on average shorter than native 5′-UTRs with uAUGs/uORFs, the results show that ribosomes are able to reach a downstream AUG via context-dependent leaky scanning and/or reinitiation mechanisms. The present study has clearly demonstrated translation of a downstream main ORF, albeit not at high levels, even in mRNAs with multiple ‘strong’ uAUGs and uORFs. The level of reporter expression seen in these constructs implies that translation of authentic mRNAs with uAUGs/uORFs would still result in biologically significant levels of their main ORF product. This is especially so when it is considered that these messengers usually encode regulatory proteins that are normally present at low concentrations. The prevalence of mRNAs with uAUGs/uORFs may also be indicative of functions for these features beyond merely limiting expression of the main ORF. For at least two transcripts there is evidence that uAUGs/uORFs act as important regulatory sites for the control of protein levels in response to cellular and environmental signals, as has been observed for GCN4 by uORF4 in response to starvation conditions in yeast (43) and human CD36 by uORF1 in response to high glucose levels (25). Whether these examples will serve as general paradigms for other transcripts remains to be determined, as does the complete repertoire of uAUGs/uORFs function. Although this study was conducted on mRNAs containing uORFs and uAUGs, the results are also relevant to the initiation of internal AUGs located downstream of the major ORF, which play a role in generating protein diversity (12). The ability of ribosomes to leaky scan through several uAUGs as found in the present study raises the prospect that initiation at an internal AUG may be far more common than is currently envisioned. In conclusion, the ability of ribosomes to initiate at a downstream AUG in mRNAs with uAUGs/uORFs has been systematically investigated and the presence of uAUGs/uORFs does not necessarily preclude initiation at the main start codon by leaky scanning and reinitiation mechanisms.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Norbert Fusenig for the HaCaT cells and Tom Hadwen and Tina Christy for their technical support. We are grateful to Drs Derek Kennedy and George Muscat for critical reading of the manuscript. This work was supported by a grant from the NHMRC (Aust) and Uniseed Pty Ltd.
REFERENCES
- 1.Hellen C.U. and Sarnow,P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev., 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 2.Hershey J. and Merrick,W. (2000) The pathway and mechanism of initiation of protein synthesis. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 38–88. [Google Scholar]
- 3.Jackson R.J. and Kaminski,A. (1995) Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA, 1, 985–1000. [PMC free article] [PubMed] [Google Scholar]
- 4.Cazzola M. and Skoda,R. (2000) Translational pathophysiology: a novel molecular mechanism of human disease. Blood, 95, 3280–3288. [PubMed] [Google Scholar]
- 5.Davuluri R.V., Suzuki,Y., Sugano,S. and Zhang,M.Q. (2000) CART classification of human 5′ UTR sequences. Genome Res., 10, 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kochetov A.V., Ischenko,I.V., Vorobiev,D.G., Kel,A.E., Babenko,V.N., Kisselev,L.L. and Kolchanov,N.A. (1998) Eukaryotic mRNAs encoding abundant and scarce proteins are statistically dissimilar in many structural features. FEBS Lett., 440, 351–355. [DOI] [PubMed] [Google Scholar]
- 7.Pesole G., Gissi,C., Grillo,G., Licciulli,F., Liuni,S. and Saccone,C. (2000) Analysis of oligonucleotide AUG start codon context in eukaryotic mRNAs. Gene, 261, 85–91. [DOI] [PubMed] [Google Scholar]
- 8.Rogozin I.B., Kochetov,A.V., Kondrashov,F.A., Koonin,E.V. and Milanesi,L. (2001) Presence of AUG triplets in 5′ untranslated regions of eukaryotic cDNAs correlates with a “weak” context of the start codon. Bioinformatics, 17, 890–900. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y., Ishihara,D., Sasaki,M., Nakagawa,H., Hata,H., Tsunoda,T., Watanabe,M., Komatsu,T., Ota,T., Isogai,T., Suyama,A. and Sugano,S. (2000) Statistical analysis of the 5′ untranslated region of human mRNA using “oligo-capped” cDNA libraries. Genomics, 64, 286–297. [DOI] [PubMed] [Google Scholar]
- 10.Gray N.K. and Wickens,M. (1998) Control of translation initiation in animals. Annu. Rev. Cell. Dev. Biol., 14, 399–458. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch A.G. (1997) Translational regulation of yeast GCN4. J. Biol. Chem., 272, 21661–21664. [DOI] [PubMed] [Google Scholar]
- 12.Kozak M. (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene, 299, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijer H.A. and Thomas,A.A.M. (2002) Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J., 367, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris D.R. and Geballe,A.P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol., 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Child S.J., Miller,M.K. and Geballe,A.P. (1999) Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem., 274, 24335–24341. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. (1987) Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol., 7, 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lincoln A.J., Monczak,Y., Williams,S.C. and Johnson,P.F. (1998) Inhibition of CCAAT/enhancer-binding protein alpha and beta translation by upstream open reading frames. J. Biol. Chem., 273, 9552–9560. [DOI] [PubMed] [Google Scholar]
- 18.Luukkonen B.G., Tan,W. and Schwartz,S. (1995) Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol., 69, 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alderete J., Jarrahian,S. and Geballe,A. (1999) Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J. Virol., 73, 8330–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaba A., Wang,Z., Krishnamoorthy,T., Hinnebusch,A.G. and Sachs,M. (2001) Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J., 20, 6453–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Gaba,A. and Sachs,M.S. (1999) A highly conserved mechanism of regulated ribosome stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J. Biol. Chem., 274, 37565–37574. [DOI] [PubMed] [Google Scholar]
- 22.Law G.L., Raney,A., Heusner,C. and Morris,D.R. (2001) Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem., 276, 38036–38043. [DOI] [PubMed] [Google Scholar]
- 23.Meijer H.A. and Thomas,A.A. (2003) Ribosomes stalling on uORF1 in the Xenopus Cx41 5′ UTR inhibit downstream translation initiation. Nucleic Acids Res., 31, 3174–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin X., Turcott,E., Englehardt,S., Mize,G.J. and Morris,D.R. (2003) The two upstream open reading frames of oncogene mdm2 have different translational regulatory properties. J. Biol. Chem., 278, 25716–25721. [DOI] [PubMed] [Google Scholar]
- 25.Griffin E., Re,A., Hamel,N., Fu,C., Bush,H., McCaffrey,T. and Asch,A.S. (2001) A link between diabetes and atherosclerosis: glucose regulates expression of CD36 at the level of translation. Nature Med., 7, 840–846. [DOI] [PubMed] [Google Scholar]
- 26.Calkhoven C.F., Muller,C. and Leutz,A. (2000) Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev., 14, 1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 27.Calkhoven C.F., Muller,C., Martin,R., Krosl,G., Pietsch,H., Hoang,T. and Leutz,A. (2003) Translational control of SCL-isoform expression in hematopoietic lineage choice. Genes Dev., 17, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarrazin S., Starck,J., Gonnet,C., Doubeikovski,A., Melet,F. and Morle,F. (2000) Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5′ upstream open reading frames in Fli-1 mRNA. Mol. Cell. Biol., 20, 2959–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong W., Hsieh,C.C., Kurtz,A.J., Rabek,J.P. and Papaconstantinou,J. (2001) Regulation of CCAAT/enhancer-binding protein-beta isoform synthesis by alternative translational initiation at multiple AUG start sites. Nucleic Acids Res., 29, 3087–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemmings-Mieszczak M., Hohn,T. and Preiss,T. (2000) Termination and peptide release at the upstream open reading frame are required for downstream translation on synthetic shunt-competent mRNA leaders. Mol. Cell. Biol., 20, 6212–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pooggin M.M., Futterer,J., Skryabin,K.G. and Hohn,T. (2001) Ribosome shunt is essential for infectivity of cauliflower mosaic virus. Proc. Natl Acad. Sci. USA, 98, 886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yaman I., Fernandez,J., Liu,H., Caprara,M., Komar,A.A., Koromilas,A.E., Zhou,L., Snider,M.D., Scheuner,D., Karfman,R.J. and Hatzoglou,M. (2003) The Zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell, 113, 519–531. [DOI] [PubMed] [Google Scholar]
- 33.Wang X-Q. and Rothnagel,J.R. (2001) Post-transcriptional regulation of the GLI1 oncogene by the expression of alternative 5′ untranslated regions. J. Biol. Chem., 276, 1311–1316. [DOI] [PubMed] [Google Scholar]
- 34.Boukamp P., Petrussevska,R.T., Breitkreutz,D., Hornung,J., Markham,A. and Fusenig,N.E. (1988) Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol., 106, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gey G.O., Coffman,W.D. and Kubicek,M.T. (1952) Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res., 12, 264–265. [Google Scholar]
- 36.Kozak M. (1984) Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature, 308, 241–246. [DOI] [PubMed] [Google Scholar]
- 37.Kozak M. (1986) Point mutations define a sequence flanking the AUG initiation codon that modulates translation by eukaryotic ribosomes. Cell, 44, 283–292. [DOI] [PubMed] [Google Scholar]
- 38.Kozak M. (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem., 226, 19867–19870. [PubMed] [Google Scholar]
- 39.Kozak M. (2001) New ways of initiating translation in eukaryotes? Mol. Cell. Biol., 21, 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. (1989) Circumstances and mechanism of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol. Cell. Biol., 9, 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao J. and Geballe,A.P. (1995) Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J. Virol., 69, 1030–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mize G.J., Ruan,H., Low,J.J. and Morris,D.R. (1998) The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J. Biol. Chem., 273, 32500–32505. [DOI] [PubMed] [Google Scholar]
- 43.Abastado J.-P., Miller,P.F., Jackson,B.M. and Hinnebusch,A.G. (1991) Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol., 11, 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant C.M., Miller,P.F. and Hinnebusch,A.G. (1994) Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron. Mol. Cell. Biol., 14, 2616–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]