Abstract
γH2AX formation by phosphorylation of the histone variant H2AX is the key process in the repair of DNA lesions including those arising at fragile sites under replication stress. Here we demonstrate that H2AX is dynamically reorganized to preoccupy γH2AX hotspots on increased replication stress by activated cell proliferation and that H2AX is enriched in aphidicolin-induced replisome stalling sites in cycling cells. Interestingly, H2AX enrichment was particularly found in genomic regions that replicate in early S phase. High transcription activity, a hallmark of early replicating fragile sites, was a determinant of H2AX localization. Subtelomeric H2AX enrichment was also attributable to early replication and high gene density. In contrast, late replicating and infrequently transcribed regions, including common fragile sites and heterochromatin, lacked H2AX enrichment. In particular, heterochromatin was inaccessible to H2AX incorporation, maybe partly explaining the cause of mutation accumulation in cancer heterochromatin. Meanwhile, H2AX in actively dividing cells was intimately colocalized with INO80. INO80 silencing reduced H2AX levels, particularly at the INO80-enriched sites. Our findings suggest that active DNA replication is accompanied with the specific localization of H2AX and INO80 for efficient damage repair or replication-fork stabilization in actively transcribed regions.
INTRODUCTION
Replication stress can cause the stalling and collapse of DNA replication forks, leading to the phosphorylation of H2AX (1). The phosphorylated form of H2AX, or γH2AX, colocalizes with anti-replication protein A, which is coupled with single strand DNA at stalled replication forks (2,3). It has been suggested that γH2AX can mark stalled replisomes even before the formation of double strand breaks (DSBs) (2,3). γH2AX has long been used as a marker of DSBs, as it is formed in chromatin surrounding DSB sites and triggers a series of molecular events that activate DNA repair response (4–6). Taken together, γH2AX enrichment may indicate the loci of stalled or broken replisomes.
Oncogene-induced replication stress particularly affects the regions of genomic fragility such as common fragile sites (CFSs) (7–10). Subtelomeric regions in mammalian cells are also fragile sites that experience increased replisome stalling and DSB formation under replication stress (11,12). A recent genome-wide study discovered that a high proportion of DNA lesions induced by replication stress are found in transcriptionally active gene-rich regions that replicate early, termed early replicating fragile sites (ERFSs) (3). Conflicts between the DNA replication and transcription machineries may explain the frequent stalling of replisomes at ERFSs (3,13). Facultative heterochromatin marked by H3K9 methylation is prone to somatic mutations in a variety of clinical tumors (14). However, the underlying mechanism remains mysterious.
Although γH2AX has been of research interest and the genome-wide location of γH2AX has been profiled (15,16), it remains unclear how the substrate molecule H2AX is regulated during chromatin packaging. Our previous genome-wide profiling (17) showed that H2AX itself is enriched in specific regions in cancer cell lines (Jurkat and HL-60). However, the main focus of our previous work was to compare the differences of γH2AX distribution in irradiated cells and cancer cells, and thus there were technical and biological limitations in the characterization of endogenous H2AX localization. First, we compared normal cells and transformed cells from different donors and therefore could not conclude whether H2AX is dynamically relocalized in response to endogenous stress. In fact, we did not experimentally impose any cellular stress at all but just observed the two different states. Second, although we observed that H2AX in cancer cells was enriched in specific regions, we were not able to propose a proper hypothesis concerning the mechanisms that direct H2AX localization. In this work, we sought to observe the dynamic changes of H2AX positioning before and after the stimulation or treatment of cells from the same donor while ruling out oncogenic effects intermingled with increased replication stress. In addition, we attempted to perform an in-depth characterization of H2AX localization by leveraging public genome-wide data for replication timing, replication-associated DSB locations, nucleosome occupancy, histone modifications, H2AZ, Pol2 and cancer mutation locations.
MATERIALS AND METHODS
Separation of CD4+ T cells
Human peripheral blood mononuclear cells were isolated from whole blood by standard Ficoll-Paque (GE Healthcare, Uppsala, Sweden) density gradient centrifugation. CD4+ T cells were isolated from peripheral blood mononuclear cells by magnetic separation using CD4 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to manufacturer’s protocol. Purity of the separated CD4+ T cells was evaluated with LSR II flow cytometer (BD Biosciences, San Jose, CA, USA) after staining with anti-CD3-APC and anti-CD4-PE. The purity was >95%.
CFSE labeling and activation of the separated CD4+ T cells
To assess proliferation of CD4+ T cells, the separated CD4+ T cells were labeled with 5 μM CFSE (Invitrogen, Carlsbad, CA, USA), and CFSE-labeled CD4+ T cells were re-suspended in RPMI 1640 containing 10% fetal bovine serum, 2 mM l-glutamine and 20 U/mL IL-2 (Peprotech, Rocky Hill, NJ, USA). For T cell stimulation, soluble anti-CD3 (0.1 μg/ml; BD Biosciences) and anti-CD28 (1 μg/ml; BD Biosciences) were added, and the culture was maintained for 96 h. Fluorescence intensity of the CFSE-labeled CD4+ T cells was examined with LSR II flow cytometer. CFSElow cells were considered to proliferate during the culture period, and the percentage of CFSElow fraction was calculated using FlowJo software (TreeStar, San Carlos, CA, USA). The fraction of CFSElow proliferating cells was 20–30% of CD4+ T cells in the presence of low dose (20 U/ml) of IL-2 without anti-CD3/anti-CD28-stimulation. Anti-CD3/anti-CD28 stimulation increased the fraction of CFSElow cells up to >50% in CD4+ T cells.
Western blot analysis
CD4+ T cells treated with IL-2 alone or IL-2 plus anti-CD3 and anti-CD28 antibodies were harvested. The cells were washed twice with ice-cold phosphate-buffered saline and were lysed in radioimmunoprecipitation assay lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate and 0.5% sodium deoxycholate, supplemented with a protease inhibitor cocktail]. Equal amounts of protein from cell lysates were boiled in sodium dodecyl sulfate-loading buffer for 10 min and separated on Any kD™ Mini-PROTEAN® TGX™ Precast Gel (Bio-Rad) and transferred to PVDF membranes (Millipore). The membranes were incubated with the primary antibodies against H2AX (Abcam, ab11175), γH2AX (Abcam, ab2893), H2AZ (Abcam, ab4174), SRCAP (Abcam, ab99408), INO80 (Abcam, ab118787), Pol2 (Abcam, ab5408) and GAPDH (Cell Signaling, #2118) overnight at 4°C. Anti-GAPDH antibody was purchased from Cell Signalling, Inc. Bound primary antibodies were detected with corresponding horseradish peroxidase-conjugated secondary antibodies. Blots were developed with enhanced chemiluminescence.
Processing of chromatin immunoprecipitation sequencing data
Chromatin immunoprecipitation (ChIP) was performed using the EZ-Magna ChIP™ kit (Millipore) according to the manufacturer’s instructions using antibodies against H2AX (Abcam, ab11175), γH2AX (Abcam, ab2893), H2AZ (Abcam, ab4174), SRCAP (Abcam, ab99408), INO80 (Abcam, ab118787) and Pol2 (Abcam, ab5408). ChIP DNA fragments were sequenced on Illumina HiSeq 2000. Chromatin immunoprecipitation sequencing (ChIP-seq) reads were mapped to the hg19 (GRCh37) human reference genome. The sequencing tags were extended to the average size of library fragments (i.e. 200 bp), and the number of overlapping sequence reads was obtained in 200-bp sliding windows across the genome. The relative enrichment of tags was obtained by a log2 ratio of (target read count/200 bp)/(total read count/genome size) as previously described (17,18). For the whole-genome scatterplots, 100-kb windows were used to obtain the target read count followed by the same normalization method. For the subtelomeric patterns, 4-Mb sliding windows were used to obtain the normalized readcount in the region of a given distance from the chromosome end, which was then averaged over multiple chromosome ends. Resting H2AX data were generated in our previous work (17) and were made available in the Gene Expression Omnibus (GEO) database under the accession number GSE25577. Sequencing data for H2AZ and Pol2 in resting CD4+ T cells were downloaded from http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.aspx. Tag extension, counting and normalization were performed in the same manner as described earlier in the text. Pol2 ChIP-seq data in HeLa cells were available in the GEO database under the accession number GSE12783. H3K9me3 ChIP-seq data in CD4+ T cells were downloaded from http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.aspxTag extension, counting and normalization were performed in the same manner as described earlier in the text. The MACS software (19) was used to identify the heterochromatin domains composed of H3K9me3 in CD4+ T cells. A total of 10 955 H3K9me3 domains with the average size of ∼1 kb were obtained with the default parameters. For mapping to heterochromatin, the identified domains were divided into 50 bins (with the bin size proportional to the domain size) and ±50 kb flanking regions were also broken down into 50 bins. The normalized readcounts to be mapped to heterchromatin were obtained from each bin and averaged over the 10 955 domains. H3K9me3 ChIP-seq data in HeLa cells were obtained from the ‘Histone Modifications by ChIP-seq from ENCODE/Broad Institute’ track of the UCSC genome browser. Tag extension, counting and normalization were performed in the same manner as described earlier in the text. We used the algorithm of HOMER (20) to identify 87 135 INO80-enriched regions and to overlap the normalized H2AX nucleosome density over the INO80 peaks for comparison between the wild-type and INO80-depleted cells (described later in the text).
Processing of nucleosome data and identification of nucleosome core positioning
MNase-digestion-based sequencing data for resting and activated mononucleosomes in CD4+ T cells (21) were obtained at http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcellnucleosomes.aspx. The MNase-seq reads were extended to 150 bp, and the number of reads was obtained in 200-bp sliding windows. Normalization was performed as described earlier. To avoid possible bias in the density of nucleosomes themselves in heterochromatic regions such as H3K9me3 domains, subtelomeres and CFSs, we subtracted the normalized mononucleosome scores in the resting or activated cells from the normalized H2AX, γH2AX or H2AZ scores in the resting or activated cells, respectively. The Nucleosome Positioning from Sequencing package (22), designed for the identification of mononucleosome positions based on ChIP-seq data, was used to identify the positions of H2AX-, H2AZ- and γH2AX-contaning nucleosomes. Different numbers of H2AX nucleosomes in the wild-type HeLa cells and INO80-knockdown cells were obtained at different P-values of each nucleosome detection, ranging from P = 10−3 to P = 10−10, as estimated using Poisson approximation. At P = 10−3, a total of 1 020 728 H2AX nucleosomes were identified in the wild-type HeLa cells and 350 507 H2AX nucleosomes in INO80-knockdown cells. For a positional comparison of H2AX, H2AZ and total nucleosomes with γH2AX nucleosomes in the activated T cells, a partial overlapping of 5–20% of the 146 bp nucleosome core was allowed.
Replication timing data
Replication timing in HeLa cells was ascertained by the isolation and sequencing of newly replicated DNA from six cell cycle fractions: G1/G1b, S1, S2, S3, S4 and G2 (23). Mapped reads from each cell-cycle phase were downloaded in BED format from the ‘Replication Timing by Repli-seq from ENCODE/University of Washington’ track of the UCSC genome browser. The number of sequence tags mapping within a 1-kb sliding window was obtained and normalized as described earlier. For overlapping with H2AX, Pol2 and H3K9me3 in HeLa cells, the correlations coefficient of the normalized readcounts across 1-kb windows were calculated and then scaled to 0 mean and unit variance across the six phases for plotting and comparison between H2AX, Pol2 and H3K9me3. We used another set of replication timing data based on the hybridization of early and late replication intermediates to NimbleGen oligonucleotide arrays (24). Wavelet-smoothed signals of log2 early/late S-phase ratios were downloaded from the ‘Replication Timing by Repli-chip from ENCODE/FSU’ track of the UCSC genome browser. Log2 ratios more than 1 and less than −1 were used to define early-replicating and late-replicating genomic regions, and the normalized H2AX readcounts in HeLa cells were mapped to each set of genomic regions.
Data for DSB locations, cancer mutations and CFSs
Genome-wide locations of DSBs induced by aphidicolin (aphidicolin-sensitive breakome) and those induced by neocarzinostatin (neocarzinostatin-sensitive breakome) were downloaded from http://breakome.eu (25). Normalized H2AX levels in 200-bp sliding windows were mapped to the inside of the obtained break regions (50 bins) and their ±500-kb flanking regions (50 bins). Because the number of the break regions was not large enough, smoothing with cubic spline (R function) was used to obtain a fitted line across the 100 bins. The compiled list of mutation locations in cancer was obtained from a previous study (14). A total of 88 905 mutations were available after liftover to the hg19 (GRCh37) reference genome. The number of the mutations was counted in a 10-kb sliding window and normalized as described earlier, which was mapped to the inside of the H3K9me3 domains (50 bins) and their ±500-kb flanking regions (50 bins) and then averaged over the 10 955 H3K9me3 domains. The compiled list of previously reported CFSs was obtained from a previous study (26) and provided in Supplementary Table S1. A total of 39 CFSs with the average size of 2.2 Mb were obtained. For mapping, each CFS was divided into 50 bins (with the bin size proportional to the size of the CFS) and ±500 kb flanking regions were divided into 50 bins. The normalized readcounts of H2AX and γH2AX were averaged over the 39 sites for each bin after adjusting for the potential bias of nucleosome density.
INO80 knockdown
Stable knockdown cells for INO80 were prepared as described previously (27). Briefly, the vector expressing INO80 small interfering RNA was constructed by inserting 5′ -GAT CCC CCT TGG TCT CCA TTT CAT ATT CAA GAG ATA TGA AAT GGA GAC CAA GTT TTT A-3′ and 5′-AGC TTA AAC TTG GTC TCC ATT TCA TAT TCT CTT GAA ATA TGA AAT GGA GAC CAA GGG G-3′ into the BglII–HindIII sites of pSuperior.puro vector (Oligoengine). HeLa-S3 cells were transfected using Lipofectamine 2000 (Invitrogen) with a vector expressing INO80 small interfering RNA or an empty vector by calcium phosphate and were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and puromycin as a selection marker at a concentration of 300 ng/ml.
RESULTS AND DISCUSSION
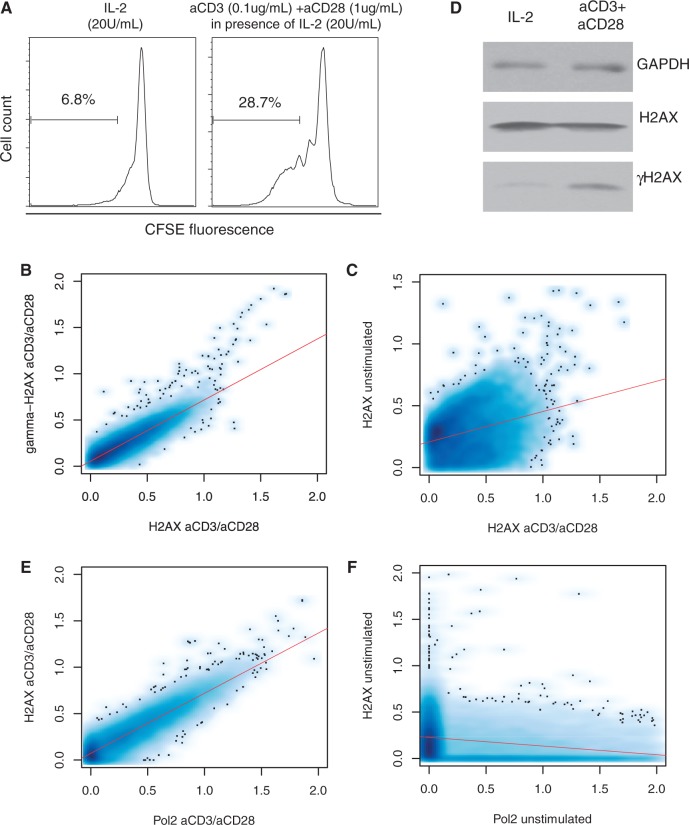
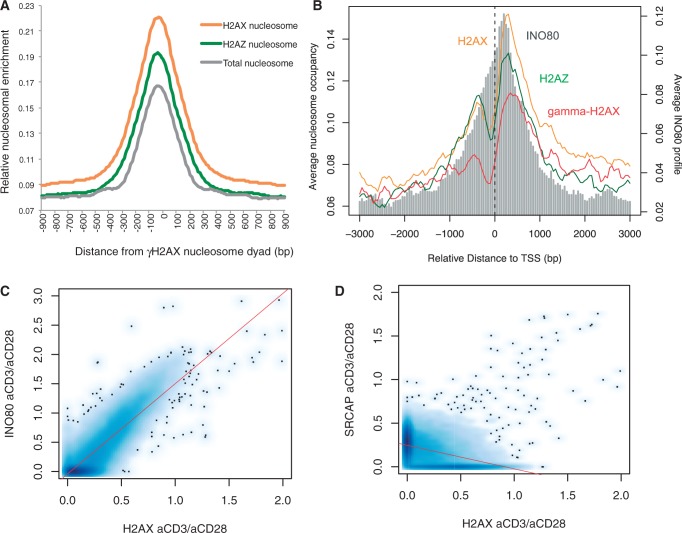
To understand the pure effect of active DNA replication independently of oncogene activation, we isolated resting CD4+ T cells, stimulated them with anti-CD3 and anti-CD28 antibodies (Figure 1A) and profiled H2AX and γH2AX patterns across the genome. In the activated cells, H2AX largely coincided with γH2AX (Figure 1B) apparently as a consequence of genome-wide repositioning from the resting state (Figure 1C). At the mononucleosome level, only 10–15% of γH2AX-containing nucleosomes overlapped with H2AX nucleosomes, reflecting the exclusive nature of the antibodies against the two histone forms. However, H2AX nucleosomes were enriched in the region centered on the γH2AX nucleosome dyad (Figure 2A). Therefore, the localization of H2AX to γH2AX hotspots observed in cancerous T cells (17) can be attributed to active cell proliferation (28). Interestingly, the H2AX patterns, when γH2AX levels were low under weaker activation by IL-2 alone (Figure 1D and Supplementary Figure S1), were similar to the anti-CD3/anti-CD28-activated H2AX patterns (Supplementary Figure S2), implying that the H2AX reorganization may begin immediately following cell proliferation cues before replication stress culminates.
Figure 1.
Genome-wide reorganization of H2AX on cell activation. (A) To assess proliferation of CD4+ T cells, the separated CD4+ T cells were labeled with CFSE. CFSE-labeled CD4+ T cells were re-suspended in 20 U/ml of IL-2. For T cell stimulation, soluble anti-CD3 and anti-CD28 antibodies were added and the culture was maintained for 96 h. (B) Genome-wide correlation between H2AX and γH2AX in the activated T cells. (C) Genome-wide correlation between H2AX in the activated T cells and H2AX in the resting T cells. (D) Western blot of H2AX and γH2AX in IL-2-stimulated cells and cells activated with anti-CD3 and anti-CD28 antibodies. (E) Genome-wide correlation between H2AX and Pol2 in the activated cells. (F) Genome-wide correlation between H2AX and Pol2 in the resting cells.
Figure 2.
Colocalization of H2AX with γH2AX and INO80. (A) Enrichment of H2AX-containing, H2AZ-containing and overall mononucleosomes in the region centered on the midpoint of the γH2AX nucleosome core. Relative enrichment was obtained by normalizing, using the total number of H2AX, H2AZ and overall mononucleosomes in the genome, respectively. (B) Normalized H2AX, γH2AX, H2AZ and INO80 levels were plotted according to the distance to the transcription start site of the RefSeq genes. (C) Genome-wide co-localization of H2AX and INO80 in the activated cells. (D) Genome-wide correlation between H2AX and SRCAP in the activated cells.
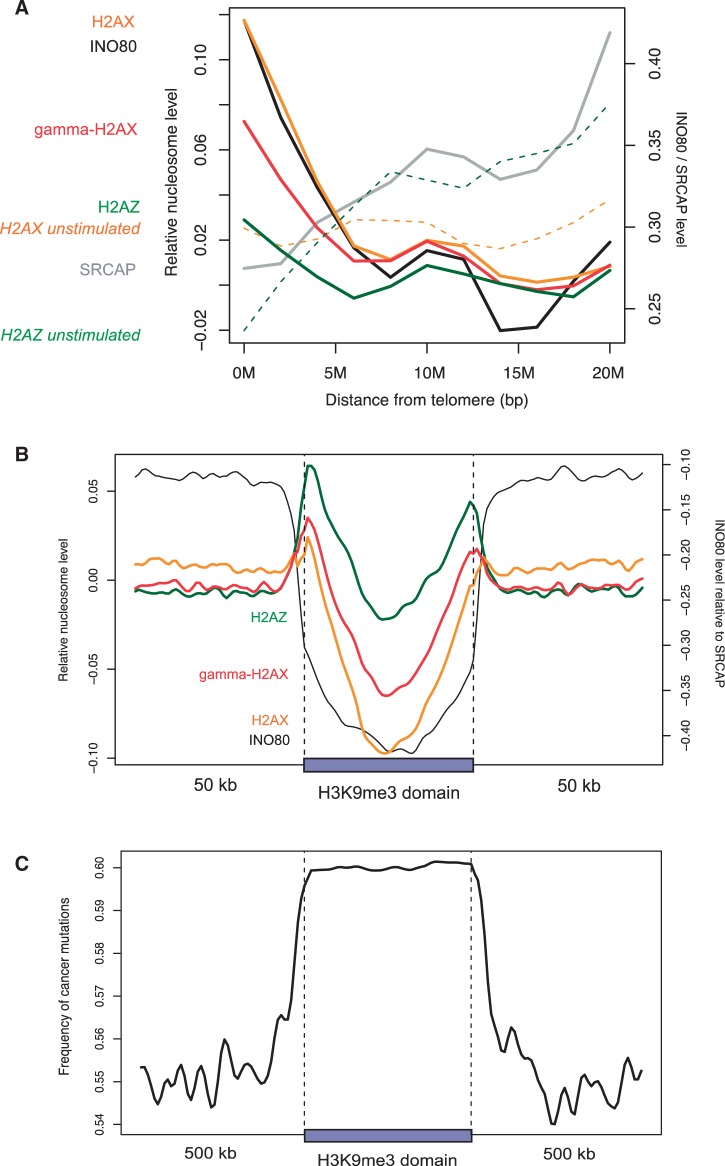
We examined H2AX enrichment at various fragile sites in the active cells. First, 39 CFSs compiled from published reports (26) did not show enrichment compared with the 1-Mb flanking regions (Supplementary Table S1 and Supplementary Figure S3). Furthermore, stimulated H2AX levels were even lower than resting H2AX levels (Supplementary Figure S3). Second, transcription activity, which is coupled with ERFSs (3), strongly correlated with H2AX localization in the active cells (Figure 1E), particularly at the transcription start site (Figure 2B and Supplementary Figure S 4). However, gene density per se was not directly associated with H2AX localization (Supplementary Figure 5). There was no correlation between H2AX and RNA polymerase II (Pol2) in the resting cells (Figure 1F). Third, subtelomeric regions were enriched for H2AX in the active state but not in the resting state (Figure 3A). Subtelomeres have not only high gene density but also high Pol2 density (Supplementary Figure S6). Finally, the heterochromatin domains identified based on H3K9me3 in CD4+ T cells were clearly devoid of H2AX (Figure 3B). It is known that heterochromatin regions and CFSs are rarely transcribed and tend to replicate in late S phase, whereas euchromatin is frequently transcribed and usually replicates in early S phase.
Figure 3.
Subtelomeric and heterochromatin patterns. (A and B) Normalized levels of H2AX and related molecules as a function of the distance to (A) chromosome ends and (B) H3K9me3 domains. To adjust for possible bias in the density of nucleosomes themselves, mononucleosome sequencing data in active T cells (21) were used to estimate the incorporation rate of H2AX, γH2AX and H2AZ. (C) The number of single nucleotide variations occurred in cancer (14) was counted in 10-kb windows and averaged across H3K9me3 domains.
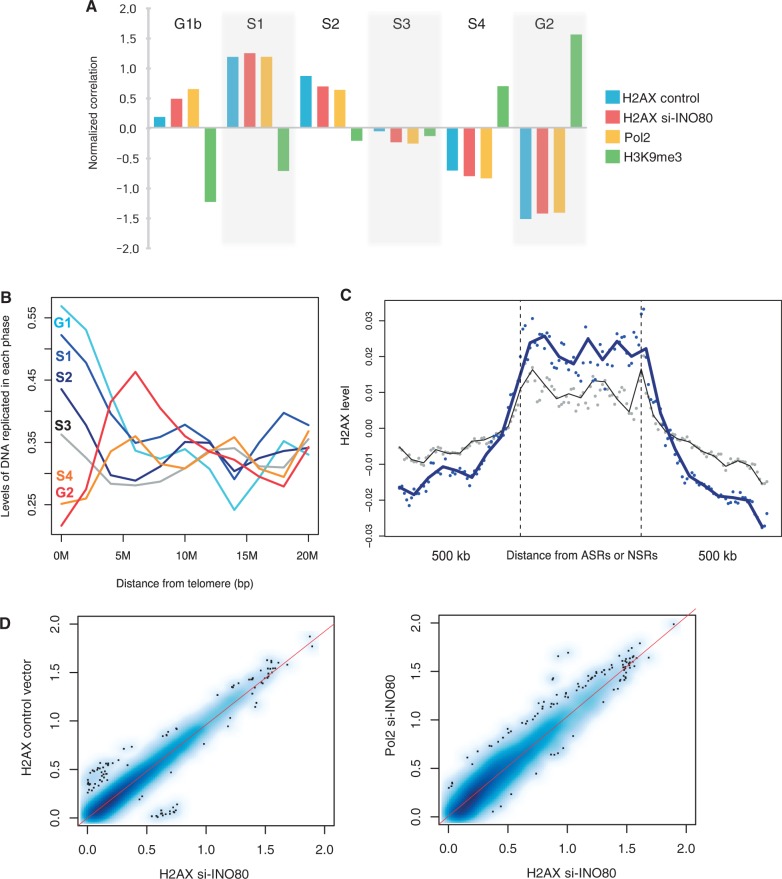
The above findings led us to characterize H2AX distribution in terms of replication timing. We obtained genome-wide H2AX positions in HeLa cells and mapped them with newly replicated DNA sequences from six cell cycle fractions (23). A substantial amount of γH2AX was observed in HeLa cells in the absence of any exogenous stimuli (25), indicating a high level of endogenous stress. The H2AX levels peaked in the S1 and S2 phases and then declined progressively (Figure 4A). The association of H2AX with early S phase was recapitulated by Pol2 in the same cells (Figure 4A). H3K9me3 in HeLa cells was correlated with late-replicating regions (Figure 4A). Subtelomeres were enriched for early replicating DNA (Figure 4B) and lacking H3K9me3 modification (Supplementary Figure S7). The H2AX enrichment in early replicating sites was confirmed by independent microarray data for the relative enrichment of early versus late S-phase nascent DNA (24) (Supplementary Figure S8).
Figure 4.
Replication timing and INO80 knockdown in HeLa cells. (A) The overlapping of H2AX, Pol2 and H3K9me3 in HeLa cells with genomic regions replicated in each phase of the cell cycle. The correlations of normalized readcounts in 1-kb windows were scaled to 0 mean and unit variance across the six phases. (B) The distribution of DNA fragments that replicate in each phase of the cell cycle as a function of the distance to chromosome ends. A 4-Mb sliding window was used to obtain the 200-bp normalized readcount from each cell cycle fraction, which was then averaged over multiple chromosome ends. (C) Normalized levels of H2AX in HeLa cells as a function of the distance to ASRs (blue dots) and neocarzinostatin-sensitive regions (gray dots). Spline smoothing was performed to obtain the fitting curves (dark blue and black curves). (D) Genome-wide correlations between H2AX before and after INO80 silencing (left) and between H2AX and Pol2 in the INO80-knockdown cells (right).
Although replication stress may be a major source of genome instability (29), DSBs may arise in cultured cells also because of physiological apoptosis or damage by reactive oxygen species (30). To observe the pure effect of replication stress, DNA polymerase inhibitors such as aphidicolin can be used to induce replication-fork stalling without cell cycle arrest (9). By directly detecting DSBs across the genome, >2000 aphidicolin-sensitive regions (ASRs) were identified in HeLa cells (25). For comparison, HeLa cells were treated with neocarzinostatin, a DSB-inducing drug, as a source of exogenous damage (25). We measured H2AX levels in ASRs and neocarzinostatin-sensitive regions along with their flanking regions. H2AX was enriched, particularly in ASRs (Figure 4C), an indication that specific H2AX positioning is related with replication stress.
To understand the underlying mechanism of specific H2AX localization, we investigated the role of INO80 and SRCAP. The INO80 and SRCAP complexes have crucial functions in DNA repair, checkpoint regulation and DNA replication (31). In particular, INO80 deletion increases sensitivity to induced DSBs in yeast (32). In fact, INO80 recruitment to induced DSBs is mediated by interactions with γH2AX, implicating its role in DNA repair (33). Moreover, INO80 is implicated in replication stress. For example, INO80 is recruited to replication forks, as cells enter S phase and promote efficient replication progression by stabilizing stalled replisomes (34,35). INO80 silencing causes delayed S-phase progression in humans (27). Furthermore, the chromatin remodeling complex is recruited to γH2AX and function in collaboration with SRCAP for histone-variant exchange by replacing γH2AX and H2AZ with free H2AX such that the substrates are dynamically restored for phosphorylation and DNA repair responses (36,37).
We mapped the genomic positions of INO80, SRCAP and H2AZ in the activated T cells. Across the genome, H2AX levels were proportional to INO80 density (Figure 2C) but not to SRCAP density (Figure 2D). INO80 was colocalized with Pol2 (Supplementary Figure S9) and concentrated on the transcription start site along with H2AX (Figure 2B). The subtelomeric enrichment of H2AX was precisely mirrored by the INO80 patterns, whereas SRCAP showed no relevance (Figure 3A). The increase in H2AZ levels after cell activation was not as significant as the increase in H2AX and INO80 levels (Figure 3A). Moreover, INO80 was strongly depleted from heterochromatin unlike H2AZ and SRCAP (Figure 3B).
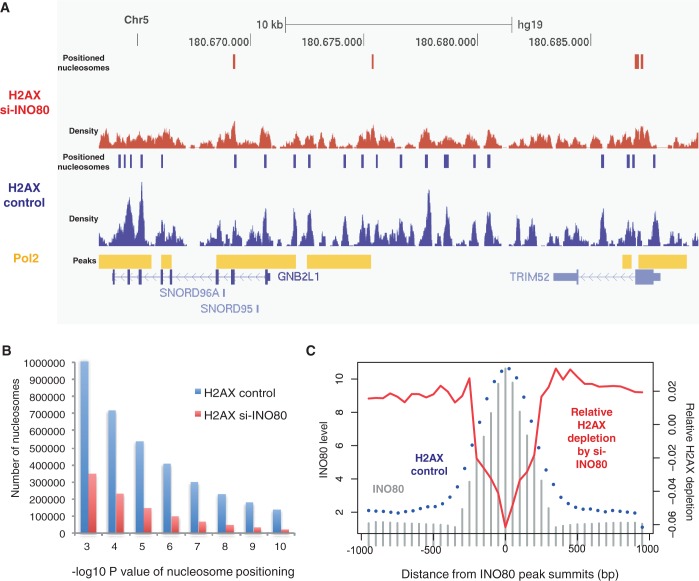
The intimate colocalization of INO80 with H2AX in cycling cells promoted us to investigate the changes in H2AX positioning on the silencing of INO80. Although the overall H2AX patterns, including the colocalization with Pol2, did not change considerably (Figure 4D), the number of H2AX nucleosomes reduced dramatically (from ∼1 to 0.35 million). As illustrated in a genomic region with high Pol2 density (Figure 5A), the overall H2AX distribution was preserved after INO80 knockdown, but H2AX density, and thus the number of positioned H2AX nucleosomes (Figure 5B), was reduced. We tested whether H2AX depletion is specifically due to the role of INO80 in H2AX deposition or because of secondary cellular effects caused by the absence of INO80. For this, we first identified INO80-enriched regions in actively dividing cells and then overlaid the normalized H2AX nucleosome density in the wild-type and INO80-knockdown cells. We obtained the magnitude of relative H2AX depletion by subtracting the wild-type profile from the INO80-knockdown profile and then overlapped the differences over the INO80 peaks. As shown in Figure 5C, while positioned H2AX nucleosomes (blue dots) nicely coincide with INO80 (gray bars) in active cells, the degree of relative H2AX depletion by INO80 silencing (red curve) is largest at the center of the INO80-enriched regions. This implies that H2AX depletion may be specifically owing to the role of INO80 in H2AX regulation. It appears that the dynamic cycle of H2A variant exchange catalyzed by INO80 and SWR1 (the SRCAP homolog) in yeast is conserved in human but without the involvement of SRCAP and H2AZ. It is tempting to suggest that INO80 may play a role in replenishing the substrate H2AX specifically at γH2AX hotspots by recognizing γH2AX and replacing it with H2AX.
Figure 5.
Changes in H2AX nucleosome density on INO80 knockdown. (A) The vertical bars indicate nucleosome positions before and after INO80 depletion, as predicted based on the sequencing tag density shown below. The Pol2 peaks are shown above the annotated transcripts. (B) The number of H2AX mononucleosome molecules (y axis) in the wild-type (blue) and INO80-knockdown (red) cells according to the P-value of each nucleosome detection (x axis) estimated using Poisson approximation. (C) INO80 density in the active cells (gray bars), H2AX nucleosomes occupancy in the wild-type cells (blue dots) and relative H2AX depletion on INO80 silencing (red curve) shown according to the distance from the INO80 peak summits.
Based on the depletion of H2AX and INO80 from heterochromatin, we suggest that deficient DNA repair owing to impaired H2AX deposition can explain the high mutation rates of heterochromatin in cancer genomes (14). We confirmed the enrichment of cancer mutations in the H3K9me3 domains that we identified (Figure 3C). The depletion was not only observed in the dividing cells (Figure 3B) but also in the resting cells (Supplementary Figure S10), an indication that there is inherent difficulty in H2AX incorporation (as measured by the H2AX level relative to overall nucleosome density) into heterochromatin and that the DNA repair deficiencies are manifested during active DNA replication. A recent study (38) demonstrated that cancer mutations are frequently found in late replicating regions, further supporting our hypothesis on the role of H2AX localization in DNA repair.
In this work, we observed that the activation of cell proliferation induced genome-wide H2AX reorganization. Among fragile sites, putative ERFSs and subtelomeres were enriched for H2AX and γH2AX, whereas CFSs and heterochromatin lacked H2AX and γH2AX enrichment. We propose that active DNA replication is accompanied with the specific localization of H2AX and INO80 to promote efficient DSB repair or replisome stabilization in transcriptionally active regions while permitting genomic instability in silent regions. Although a role for INO80 in the genome-wide reorganization of H2AX positioning has been suggested, the mechanism underlying the specific H2AX localization remains to be elucidated. We suspect the role of the transcription machineries in recruiting H2AX to the regions in which frequent replication-transcription collisions arise. However, this hypothesis remains to be tested.
ACCESSION NUMBER
All the raw data generated in this work are deposited in GEO under accession number GSE44309 (reviewer link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=frglxqwskqeqyjk&acc=GSE44309).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KAIST Future Systems Healthcare Project from the Ministry ofScience, ICT and Future Planning and by a grant from the National Research Foundation (NRF) of Korea [2012R1A1A1019094]. K.K. is a recipient of the Global PhD Fellowship of NRF. Computing facilities were supported by the NRF [2009-0086964] and the CHUNG Moon Soul Center of KAIST. Funding for open access charge: NRF [2012R1A1A1019094].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 2.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun H-W, McKinnon P, Wright G, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 5.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 8.Gorgoulis VG, Vassiliou L-VF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 9.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 10.Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu XR, Papavassiliou AG, et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 11.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zschenker O, Kulkarni A, Miller D, Reynolds GE, Granger-Locatelli M, Pottier G, Sabatier L, Murnane JP. Increased sensitivity of subtelomeric regions to DNA double-strand breaks in a human cancer cell line. DNA Repair. 2009;8:886–900. doi: 10.1016/j.dnarep.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortusewicz O, Herr P, Helleday T. Early replication fragile sites: where replication–transcription collisions cause genetic instability. EMBO J. 2013;32:493–495. doi: 10.1038/emboj.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 15.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szilard RK, Jacques P-É, Laramée L, Cheng B, Galicia S, Bataille AR, Yeung M, Mendez M, Bergeron M, Robert F, et al. Systematic identification of fragile sites via genome-wide location analysis of γ-H2AX. Nat. Struct. Mol. Biol. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo J, Kim SC, Lee HS, Kim JK, Shon HJ, Salleh NL, Desai KV, Lee JH, Kang ES, Kim JS, et al. Genome-wide profiles of H2AX and γ-H2AX differentiate endogenous and exogenous DNA damage hotspots in human cells. Nucleic Acids Res. 2012;40:5965–5974. doi: 10.1093/nar/gks287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi JK. Contrasting chromatin organization of CpG islands and exons in the human genome. Genome Biol. 2010;11:R70. doi: 10.1186/gb-2010-11-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, Nusbaum C, Myers R, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schones DE, Cui K, Cuddapah S, Roh T-Y, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, et al. Nucleotide-resolution dnA double-strand break mapping by next-generation sequencing. Nat. Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 27.Hur S-K, Park E-J, Han J-E, Kim Y-A, Kim J-D, Kang D, Kwon J. Roles of human INO80 chromatin remodeling enzyme in DNA replication and chromosome segregation suppress genome instability. Cell. Mol. Life Sci. 2010;67:2283–2296. doi: 10.1007/s00018-010-0337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Micco RD, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 29.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 30.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. γH2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Attikum HV, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 33.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 34.Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat. Struct. Mol. Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 35.Falbo KB, Alabert C, Katou Y, Wu S, Han J, Wehr T, Xiao J, He X, Zhang Z, Shi Y, et al. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat. Struct. Mol. Biol. 2009;16:1167–1172. doi: 10.1038/nsmb.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme Is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.