Abstract
DNA mimic proteins are unique factors that control the DNA binding activity of target proteins by directly occupying their DNA binding sites. The extremely divergent amino acid sequences of the DNA mimics make these proteins hard to predict, and although they are likely to be ubiquitous, to date, only a few have been reported and functionally analyzed. Here we used a bioinformatic approach to look for potential DNA mimic proteins among previously reported protein structures. From ∼14 candidates, we selected the Staphylococcus conserved hypothetical protein SSP0047, and used proteomic and structural approaches to show that it is a novel DNA mimic protein. In Staphylococcus aureus, we found that this protein acts as a uracil-DNA glycosylase inhibitor, and therefore named it S. aureus uracil-DNA glycosylase inhibitor (SAUGI). We also determined and analyzed the complex structure of SAUGI and S. aureus uracil-DNA glycosylase (SAUDG). Subsequent BIAcore studies further showed that SAUGI has a high binding affinity to both S. aureus and human UDG. The two uracil-DNA glycosylase inhibitors (UGI and p56) previously known to science were both found in Bacillus phages, and this is the first report of a bacterial DNA mimic that may regulate SAUDG’s functional roles in DNA repair and host defense.
INTRODUCTION
Research in the past decade has revealed several examples of regulatory proteins that mimic DNA. These proteins use negatively charged amino acids to imitate the charge distribution of DNA and thus prevent DNA from binding to its original target protein by direct competition (1,2). DNA mimic proteins can be found in virus, bacteria and eukaryotic cells, and they are involved in many important control mechanisms, including DNA repair, restriction, transcriptional control and DNA packaging (2). All of these observations suggest that DNA mimic proteins are essential to living cells, and the discovery of new mimics is potentially important in many areas of research. However, only a few DNA mimic proteins (<20) have so far been reported. The reason is that these proteins are hard to identify because their amino acid sequences and protein structures are extremely divergent (2).
We have been developing bioinformatic approaches to identify new DNA mimics, and here we found several candidates. One of these, the Staphylococcus conserved protein SSP0047, was selected for further study. In this report, we show that SSP0047 (or SAUGI; for Staphylococcus aureus uracil-DNA glycosylase inhibitor) acts as a uracil-DNA glycosylase inhibitor that breaks the uracil-removing activity of S. aureus uracil-DNA glycosylase (SAUDG). We also determined the structure of the SAUGI/SAUDG complex, and used surface plasmon resonance (BIAcore) to show that SAUGI has a high binding affinity to UDG.
Functionally, UDGs remove the uracils in DNA that result from the spontaneous deamination of cytosine or the incorporation of dUTP during replication (3,4). To date, only two uracil-DNA glycosylase inhibitors (UGI and p56) have been identified. One of these, phage PBS2 UGI, forms a tight and physiologically irreversible complex with a variety of UDG proteins in 1:1 molar stoichiometry (4–8). The other protein, p56, was identified in the Bacillus phage ϕ 29. Although its dimeric structure is different from the monomeric UGI, p56 has been shown to inhibit UDG’s activity as well (9–12). SAUGI is therefore the third uracil-DNA glycosylase inhibitor that has been identified, and the first in a species other than bacterial phage.
MATERIALS AND METHODS
Bioinformatic search for possible DNA mimic candidates in the protein structure database
To function as a DNA mimic, a protein must have two critical properties: a DNA-like arrangement of negative charges on its surface and an appropriate structural conformation (2,13). Here, we used these two properties to search for potential DNA mimic proteins in the Protein Data Bank (PDB). First, we used the 12 known DNA mimic proteins listed in Supplementary Table S1 (5–8,14–24) as starting queries to search on the DALI server (25) for proteins with loosely similar structures (Z-score >4.0 and root-mean-square deviation (RMSD) <3.5 Å; the RMSD is a measure of average deviation in distance between the aligned α-carbons in structural superimposition, while the Z-Score is a measure of alignment quality, with values above eight indicating good structural superimposition). Next, the list of candidate proteins was further reduced by applying additional constraints that were deduced from all 12 of the published DNA mimic proteins (Supplementary Table S1): (i) a protein size of <200 amino acids; (ii) a total of at least 10 aspartic acid and/or glutamic acid residues on the protein surface; and (iii) a negative charge on at least 10% of the surface residues. Finally, 14 proteins were considered potential DNA mimic proteins based on the similarity of negative charge distributions to the original query proteins (Supplementary Figure S1 and Supplementary Table S2).
Preparation and purification of recombinant SAUGI and SAUDG
For N-terminal His10-tagged SAUGI, the full-length SAUGI gene (NCBI sequence ID: AAL26663.1, amino-acid residues 1–112) with the stop codon was ligated into pET16b expression vector (Novagen). For C-terminal His6-tagged SAUDG, the full-length SAUDG gene (NCBI sequence ID: YP_040034.1, amino-acid residues 1–218) and human UDG gene (NCBI sequence ID and PDB: 1SSP_E, amino-acid residues 1–223) without the stop codon were ligated into pET21b expression vector (Novagen). All vectors were transformed into Escherichia coli BL21 (DE3), and after the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the recombinant proteins were expressed for 16 h at 16°C. Soluble proteins were purified by immobilized metal-ion chromatography with a Ni-NTA column, followed by gel filtration using Superdex 75 (GE Healthcare).
For tag-free SAUGI, a pET21b expression vector containing the full-length SAUGI gene with the stop codon was transformed into E. coli BL21 (DE3). After addition of 1 mM IPTG, the recombinant SAUGI protein without any tag was expressed for 16 h at 16°C. Soluble SAUGI was purified by Q anion exchange chromatography (GE Healthcare) with a gradient of 0–0.75 M NaC1 in 20 mM Tris, pH 7.4, buffer. SAUGI’s purity was further improved using gel filtration on a Superdex 75 column.
His-pulldown assays to identify proteins that interact with SAUGI
For the first His-pulldown assay, the soluble proteins extracted from S. aureus (200 µg) were mixed with purified N-terminal His10-tagged SAUGI (1 mg). The reaction mixture was adjusted to a final volume of 1 ml in a binding buffer of 1× phosphate buffered saline with 20 mM imidazole, and incubated for 16 h at 4°C. Incubation then continued with Ni-NTA beads (50 μl) for another 1 h. After incubation, the beads were washed with 5 ml of the same binding buffer. Proteins bound to the beads were then eluted by the addition of buffer containing 500 mM imidazole and resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. Protein bands of interest were excised from the gels and digested in-gel by trypsin. LC-nanoESI-MS/MS and the search program MASCOT were used to identify the proteins. In MASCOT, the selected variable modifications were methionine oxidation and cysteine carboxyamidomethylation. Significant hits (as defined by Mascot probability analysis) were regarded as positive identification. In another His-pulldown assay to confirm the interaction between SAUGI and SAUDG, 3 μM purified C-terminal His6-tagged SAUDG was mixed with 1 μM untagged SAUGI. Subsequent steps in this assay were the same as described above.
Gel filtration to determine the native molecular weights of target proteins
A Superdex 75 10/300 gel filtration column (GE Healthcare) was used to estimate the approximate molecular weights (MWs) of SAUGI and SAUDG, as well as the SAUGI/SAUDG complex. The buffer used in this assay was 30 mM Tris–HCl, pH 7.4, 100 mM NaCl, 5% glycerol, 1 mM EDTA and 1 mM Dithiothreitol (DTT). Gel filtration standard proteins [ovalbumin (43 kDa), carbonic anhydrase (29 kDa), RNase A (13.7 kDa) and aprotinin (6.5 kDa)] were used to create a calibration curve for protein MWs on the same gel filtration column. For each protein, the logarithm of MW was plotted against Kav, which was calculated as follows: Kav = (Ve–Vo)/(Vt–Vo), where Ve is the elution volume, Vo is the column void volume using blue dextran 2000 and Vt is the total column bed volume (24 ml for the Superdex 75 10/300 gel filtration column).
Crystallization, data collection and structure determination of SAUDG and the SAUDG/SAUGI complex
For SAUDG crystallization, 2 µl of purified C-terminal His6-tagged SAUDG solution (16 mg/ml; 20 mM Tris, pH 8.0, and 100 mM NaCl) were mixed with 2 µl of a reservoir containing 0.1 M Hepes sodium, pH 7.5, 10% 2-propanol and 18% PEG4000 as a precipitant. For crystallization of the SAUDG/SAUGI complex, 1 mg of SAUGI and 2.5 mg of C-terminal His6-tagged SAUDG were mixed in 100 µl of complex buffer (30 mM Tris–HCl, pH 7.4, 5% glycerol, 100 mM NaCl, 1 mM EDTA and 1 mM DTT) at room temperature (RT) for 1 h. A Superdex 75 gel filtration column was then used to separate the complex from the uncomplexed monomers. Subsequently, 2 µl of the complex solution (20 mg/ml) were mixed with 2 µl of a reservoir containing 0.1 M sodium Hepes, pH 7.2, and 15 % PEG20000 as a precipitant. Equilibration with the reservoir was achieved by the sitting drop method. Before flash cooling, ethylene glycol at a final concentration of 15% was added as a cryoprotectant for both the SAUDG and SAUDG/SAUGI complex crystals.
Native X-ray diffraction data from the SAUDG and SAUDG/SAUGI complex crystals were collected on beamline 13B at the National Synchrotron Radiation Research Center in Hsinchu, Taiwan. The data were processed using HKL2000 (26). Molecular replacement calculations were performed by the program Molrep (27). For SAUDG, the crystal structure of Vibrio cholerae uracil-DNA glycosylase (PDB number: 2JHQ) was used as a search model, and for the SAUDG/SAUGI complex, a search model was constructed by replacing the UGI in the crystal structure of E. coli UDG/UGI (PDB number: 2UGI) with the Staphylococcus saprophyticus hypothetical protein SSP0047 (PDB number: 2KCD). Both initial maps were produced after preliminary refinement using Refmac (28), and the programs Buccaneer (29), Coot (30) and Refmac (28) were used in the subsequent model building and refinement. Model bias was removed by simulated annealing and checking against composite maps calculated by CNS (31) using 20 different models. Statistics for the data collection and refinement are shown in Table 1. The PyMOL program (32) was used for the structural analyses and also for figure production.
Table 1.
Data collection and refinement statistics of SAUDG and SAUGI/SAUDG crystals
| Data collection | SAUDG | SAUGI/SAUDG |
|---|---|---|
| Wave length (Å) | 1.00000 | 1.00000 |
| Space group | P21 | P212121 |
| Unit cell a, b, c (Å) | 56.4, 79.5, 62.6 | 51.6, 86.4, 88.5 |
| β (°) | 112.1 | |
| Resolution (Å) | 50–1.48 (1.53–1.48) | 25–2.2 (2.28–2.20) |
| Unique reflections | 82 029 (7976) | 20 216 (1636) |
| Redundancy | 4.3 (4.1) | 9.1 (6.8) |
| Completeness (%) | 96.6 (94.7) | 97.6 (80.7) |
| I/σ(I) | 26.5 (2.6) | 27.7 (6.8) |
| Rmerge (%) | 6.1 (66.3) | 7.0 (21.2) |
| Refinement | ||
| Rwork (%) | 16.5 | 17.1 |
| Rfree (%) | 19.7 | 22.1 |
| Bond RMSD (Å) | 0.011 | 0.017 |
| Angle RMSD (°) | 1.24 | 1.6 |
| Mean B value/no of atom | 19.4/4019 | 14.0/2970 |
| Ramachandran plot (%) | ||
| Most favored (%) | 96.73 | 95.34 |
| Allowed (%) | 2.80 | 3.73 |
| Outliers (%) | 0.47 | 0.93 |
Determination of the binding affinities between UGIs and UDGs
The binding affinities of SAUGI and UGI to SAUDG and human UDG were determined by surface plasmon resonance using a BIAcore T200 (GE Healthcare) according to the protocols provided by the manufacturer. Briefly, the anti-His antibody was immobilized on the CM5 chip surface using the standard amine coupling method. The C-terminal His6-tagged UDG was then immobilized on the CM5 chip surface by binding to the anti-His antibody. Typically 20–30 response units were immobilized on the individual flow cells of the sensor chip. Next, SAUGI or UGI in 10 mM Hepes, 150 mM NaCl and 0.05% v/v Surfactant P20 was injected for 1 min at a flow rate of 30 µl/min. Association (ka) and dissociation (kd) kinetic constants were calculated by BIAevaluation 3.1 software using a simple 1:1 Langmuir model. The equilibrium dissociation constant KD values were calculated by the equation KD = kd/ka.
Determination of SAUGI’s ability to inhibit SAUDG activity
The methods used here to measure the uracil removing activity of SAUDG were modified from procedures described in previous reports (9,33). For the single-stranded DNA (ssDNA) activity of SAUDG, a 40-mer Hex-labeled oligodeoxynucleotide containing a single uracil (ssDNA19U; 5′-Hex-GTAAAACGACGGCCAGTGUATTCGAGCTCGGTACCCGGGG-3′) was used as the DNA substrate. For SAUDG’s double-stranded DNA (dsDNA) activity, 40-mer dsDNA fragments were prepared by mixing equal volumes and concentrations of ssDNA19U and its no-uracil complementary oligonucleotide (5′-FAM-CCCCGGGTACCGAGCTCGAAGGCACTGGCCGTCGTTTTAC-3′), incubating the reaction mixtures at 95°C for 5 min, and then lowering the temperature to RT for another 30 min. For each reaction, different amounts of purified recombinant SAUGI were mixed with C-terminal His6-tagged SAUDG. The reaction mixtures were adjusted to a volume of 9 μl in reaction buffer (for the ssDNA reaction: 20 mM Tris–HCl, pH 8.0, 1 mM DTT, 1 mM EDTA; for the dsDNA reaction: 50 mM potassium acetate, 20 mM Tris–acetate, pH 7.9, 10 mM magnesium acetate and 1 mM DTT) and preincubated at 37°C for 15 min. Subsequently, 1 μl of reaction buffer containing the respective ssDNA and dsDNA substrates was added to a final concentration of 0.5 μM, and the incubation continued for 15 min. Lastly, to cleave the apyrimidinic sites in the ssDNA, NaOH was added to a final concentration of 0.2 M and allowed to react at 90°C for 30 min, while apurinic-apyrimidinic (AP) endonuclease I (4 units, New England Biolabs) was added and allowed to react at 37°C for 30 min to cleave the apyrimidinic sites in the dsDNA. The dsDNA reaction was terminated by raising the temperature to 95°C for 5 min. The ssDNA and dsDNA reaction products were then mixed with 2× sample loading buffer (95% formamide, 20 mM EDTA, 1 mg/ml bromophenol blue) and run on a 20% polyacrylamide gel (0.5× Tris-borate-EDTA buffer, 8 M urea). The results were visualized using ultraviolet light.
RESULTS
Bioinformatic search for DNA mimic candidates from the PDB
In total, 14 potential DNA mimic proteins were identified based on the properties of the known DNA mimic proteins. The structures and negative charge distributions of these proteins are all similar to their respective query proteins (Supplementary Figure S1 and Supplementary Table S2). Among these candidates, the Staphylococcus hypothetical protein SSP0047 (PDB number: 2KCD) is similar to the uracil-DNA glycosylase inhibitor from phage PBS2 (UGI; PDB number: 1UGI) with a Z-score of 5.8 and RMSD of 2.9 Å. Although there are several other candidates with higher Z-scores and/or lower RMSD scores, we selected SSP0047 for further study because the negative charge distribution on its surface bears the highest similarity to its original query protein (Supplementary Figure S1). Interestingly, although the protein fold and charge distributions of SSP0047 and UGI are similar, their amino acid sequences show only a weak homology (Supplementary Figure S2). The function of this protein had not been identified before, but its nuclear magnetic resonance (NMR) structure was determined by Northeast Structural Genomics Consortium (34) and released in 2009. A comparison of the distances between the carboxyl groups on the side chains of SSP0047 and the phosphate groups on B-form DNA suggests that this protein is a DNA mimic (Supplementary Figure S3). Taken together, these data suggest that SSP0047 may act as a UGI-like uracil-DNA glycosylase inhibitor. Since the SSP0047 protein is conserved in different Staphylococcus spp, in the following experiments we decided to focus on the SSP0047 homolog from the important human pathogen S. aureus. The protein is subsequently referred to as SAUGI.
The interaction between SAUGI and SAUDG
A His pulldown assay was used to identify possible interacting partners of SAUGI. In this assay, SAUGI was used as bait and proteins extracted from S. aureus were used as prey. The result showed many S. aureus proteins being pulled down by SAUGI (Supplementary Figure S4). Six protein bands were selected and identified using mass spectrometry. Interestingly, within these bands there were at least 8 DNA binding proteins, including DNA topoisomerase IV A subunit (GyrA/ParC), catabolite control protein A (CcpA), phosphosugar binding transcriptional regulator (RpiR family), accessory gene regulator A (agrA), two-component system response regulator (WalR/VicR), GTP-sensing transcriptional pleiotropic repressor (CodY), DNA damage-inducible repressor (LexA) and SAUDG (Supplementary Table S3). Since the protein folding of SAUGI is similar to that of UGI (Supplementary Figure S2), we hypothesized that SAUDG is likely a good target for SAUGI. We therefore further investigated the interaction between these two proteins.
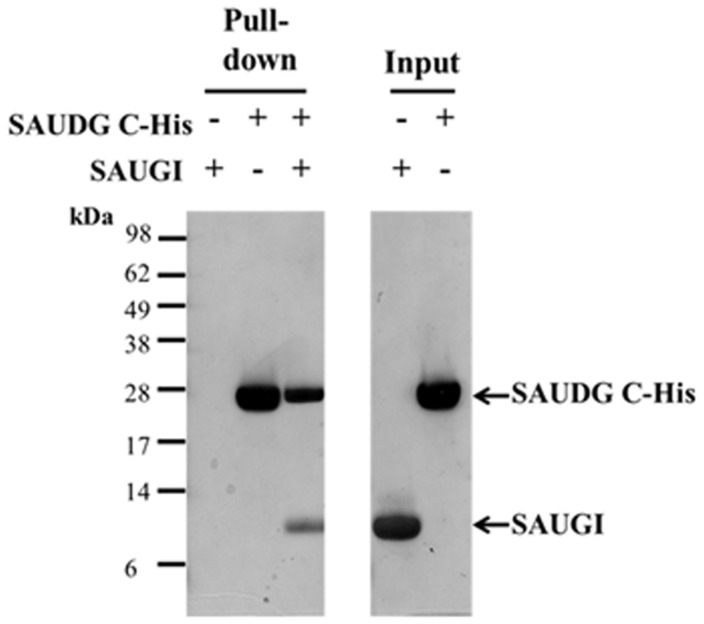
First, the interaction between SAUGI and SAUDG was reconfirmed by another His pulldown assay, which showed that SAUGI without any tag could be pulled down by C-terminal His6-tagged SAUDG (Figure 1). Gel filtration chromatography gave calculated MWs for SAUDG and the SAUGI/SAUDG complex that were close to the respective theoretical MWs of the SAUDG monomer and the monomer-to-monomer complex (Supplementary Figure S5A and Table 2). The calculated MW of SAUGI was ∼50% greater than its predicted monomer weight, but this was probably due to nonspecific interactions between SAUGI and the column material (Supplementary Figure S5A and Table 2). Analytical ultracentrifugation results for SAUGI and SAUDG were both close to the theoretical monomer values, while the calculated MW for the complex was within 10% of its theoretical MW (Supplementary Figure S5B and Table 2). Taken together, these results suggest that SAUGI forms a monomer-to-monomer complex with SAUDG that corresponds to the previously reported UGI/UDG complex.
Figure 1.
The SAUGI-SAUDG interaction confirmed by His-pulldown. In this assay, SAUGI without any tag was used as prey and C-terminal His6-tagged SAUDG was used as bait. The result showed that SAUGI was pulled down by C-terminal His6-tagged SAUDG.
Table 2.
The results of gel filtration and analytical ultracentrifugation (AUC)
| Gel filtration |
Analytical ultracentrifugation |
||||
|---|---|---|---|---|---|
| Theoretical MWa (kDa) | Calculated MW (kDa) | Calculated MWb (kDa) | Sc predicted by HYDROPRO | Observed S from AUC | |
| SAUGI monomer | 14.4 | 21.2 | 14.6 | 1.68 | 1.79 |
| SAUDG monomer | 26.0 | 25.3 | 26.5 | 2.64 | 2.68 |
| SAUGI/SAUDG complex | 40.4 | 36.7 | 36.6 | 3.40 | 3.31 |
aThe theoretical MW is based on the amino acid sequence.
bThe program SEDFIT (http://www.analyticalultracentrifugation.com) was used to calculate the MWs by using sedimentation coefficients from each protein peak.
cHYDROPRO (35) predicted the S values based on the atomic coordinates reported in this study.
The crystal structures of SAUDG and SAUGI/SAUDG
SAUDG has a conserved fold that is similar to the reported UDG structures from other species. Structural alignment shows that the Leu184 of SAUDG corresponds to the protruding residue Leu191 of E. coli UDG, Phe191 of Bacillus subtilis UDG and Leu272 of human UDG (Supplementary Figure S6A). The side chains of these residues insert into DNA’s minor groove to replace the flipped-out uracil, and they are crucial for the formation of a stable complex between UDG and its respective substrate (36,37). Additionally, three loops [4-Pro, Gly-Ser and Leu272 loops; (37)] that accomplish compression of the phosphates in the human UDG/DNA complex are also conserved in these UDG structures (Supplementary Figure S6A). Structural alignment of SAUDG with the human UDG/DNA complex further suggested the probable location of DNA binding region in SAUDG (Supplementary Figure S6B).
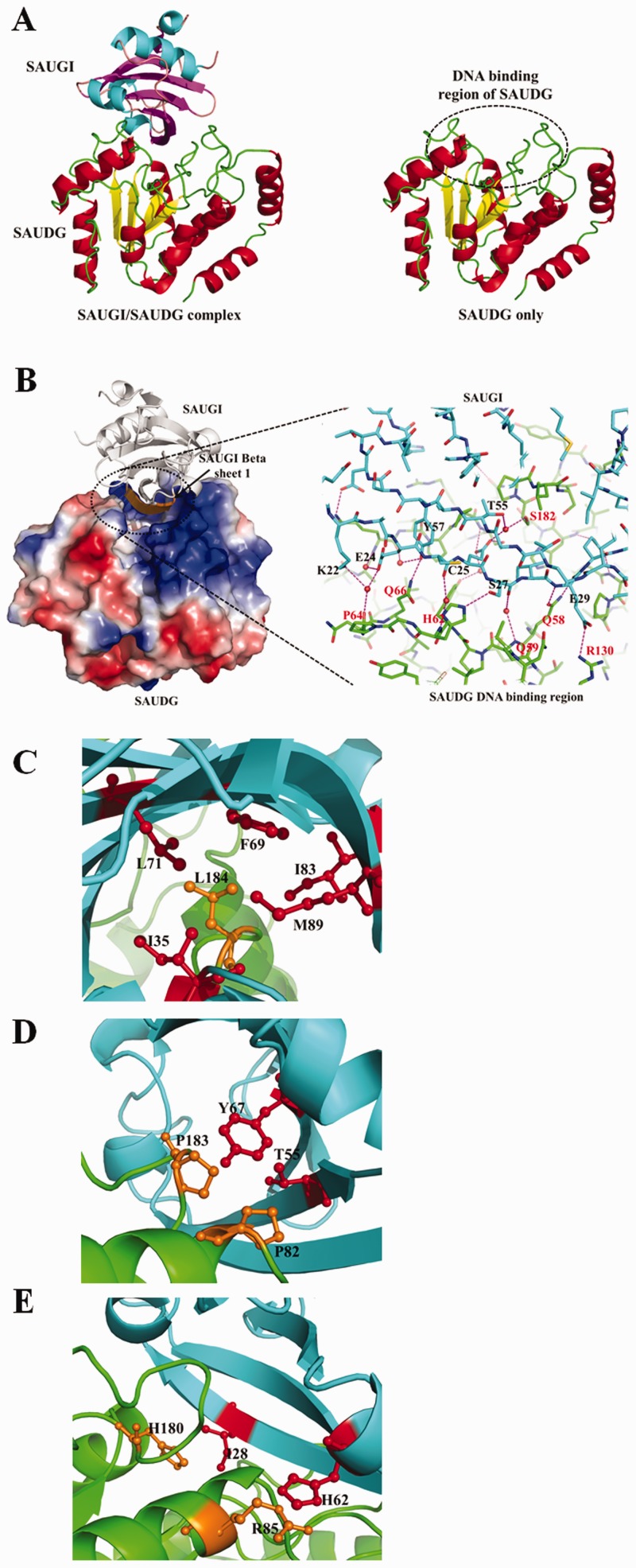
The complex crystal structure of SAUGI/SAUDG is shown in Figure 2A. SAUGI binds directly to the DNA binding region of SAUDG. In the SAUGI/SAUDG interface, at least 20 water molecules were observed. These presumably form a network of hydrogen bonds between the proteins, and so far 12 direct hydrogen bonds and 8 water-mediated bonds have been found (Figure 2B and Supplementary Table S4). In addition, the side chain of the protruding residue Leu184 in SAUDG is surrounded by the side chains of Ile35, Phe69, Leu71, Ile83 and Met89 in SAUGI, and this provides hydrophobic interactions that further stabilize the complex (Figure 2C). Adjacent to this hydrophobic region, SAUDG also has Pro82 and Pro183 posed against the side chains of Thr55 and Tyr67 in SAUGI, which contributes further to the nonpolar interactions (Figure 2D). Near the center of the interface, there are potential van der Waals interactions between the side chain of His180 in SAUDG and the facing side chain of Ile28 in SAUGI (Figure 2E). Although it lies outside the main interface region, the side chain of Arg85 in SAUDG is also stacked against that of His62 in SAUGI (Figure 2E). All of these interactions contribute to the tight binding between SAUGI and SAUDG.
Figure 2.
Crystal structures of the SAUGI/SAUDG complex and SAUDG. (A) Ribbon diagrams of the SAUGI/SAUDG complex and SAUDG. The α-helices and β-strands are, respectively, colored red and yellow in SAUDG and cyan and magenta in SAUGI. The secondary structural elements and the amino acid sequences of SAUGI and SAUDG can be found in Supplementary Figure S7. (B) The SAUGI/SAUDG interface. In the magnified inset, selected residues of SAUGI and SAUDG are labeled in black and red, respectively. The pink dashed lines represent hydrogen bonds. Four of the residues on the SAUGI strand β-1, Glu24, Cys25, Ser27 and Glu29, are hydrogen-bonded to SAUDG, which suggests the importance of strand β-1 for SAUGI’s binding affinity. (C–E) Other important interactions that contribute to the binding between SAUGI (cyan) and SAUDG (green). The SAUGI side chains are colored red, while those of SAUDG are orange.
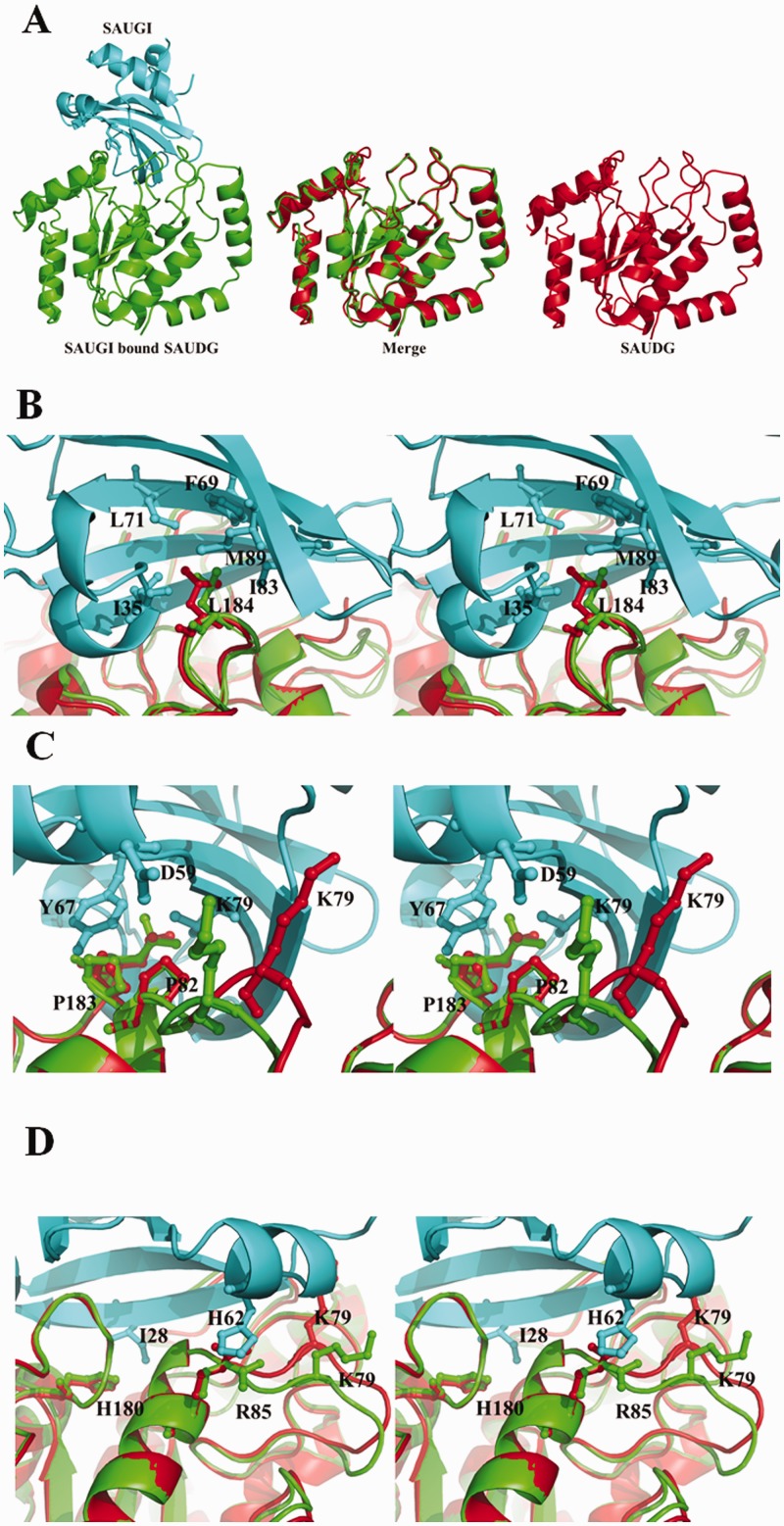
Conformational changes in SAUDG and SAUGI on complex formation
The binding between SAUGI and SAUDG did not induce any large conformational changes in SAUDG (Figure 3A; the RMSD fit is 0.345 Å). The most significant conformational change was found in the loop region formed by residues 75–80 (Figure 3B), which corresponds to the phosphate-compressing 4-Pro loop in the human UDG/DNA complex. The Lys79 side chain on this loop moves forward and forms a hydrogen bond with the Asp59 side chain of SAUGI (Supplementary Table S4). The side chain orientations of Leu184 and Arg85 on SAUDG were also affected by their interactions with the SAUGI residues (Figure 3C and D).
Figure 3.
A comparison of SAUGI-bound SAUDG and unbound SAUDG. (A) The SAUGI-bound SAUDG and the unbound SAUDG are, respectively, colored green and red. SAUGI is colored cyan. The merged image in the center shows the superimposed structures of the bound and unbound SAUDG. Conformational differences between the bound and unbound SAUDG are seen in the loop region of residues 75–80 (B) and in the side chain orientations of Leu184 (C) and Arg85 (D). Panels (B–D) are in stereo. The 2Fo-Fc maps of these regions in SAUGI-bound SAUDG and unbound SAUDG are shown in Supplementary Figure S8.
For SAUGI, an RMSD fit of 1.22 Å was calculated between the unbound SSP0047 (as determined by NMR) and SAUDG-bound SAUGI (as determined by radiographic crystallography). Structural differences were seen in two loop regions (Supplementary Figure S9; residues 41–49; 73–75). However, it is not clear whether these differences were due to the binding to SAUDG, or to the different determination methods (i.e. NMR versus radiography).
The binding affinity of SAUGI and SAUDG
The binding affinities between various uracil-DNA glycosylase inhibitors and UDGs were measured using surface plasmon resonance (BIAcore) (Supplementary Figure S10 and Table 3). The results showed that SAUGI and SAUDG have rapid association and slow disassociation rates. This turns out a high equilibrium dissociation constant (KD = 1 nM) and implies SAUGI is a strong inhibitor of SAUDG. Interestingly, SAUGI also binds to human UDG with a high equilibrium dissociation constant (KD = 2.5 nM), a relatively slow association rate and the same slow disassociation rate. Moreover, for bacterial SAUDG, the binding affinity of UGI is about twice that of SAUGI, while for human UDG, the binding affinity of UGI is 12–13 times greater than SAUGI’s.
Table 3.
The binding affinities of uracil-DNA glycosylase inhibitors and UDGs
| UGIs to UDGs | ka (1/Ms) | kd (1/s) | KD (nM) |
|---|---|---|---|
| SAUGI to SAUDG | 2 472 386 ± 16 363 | 0.00296 ± 0.00010 | 1.198 ± 0.029 |
| Phage UGI to SAUDG | 8 904 442 ± 419 216 | 0.00610 ± 0.00068 | 0.685 ± 0.080 |
| SAUGI to human UDG | 495 685 ± 22 047 | 0.00124 ± 0.00001 | 2.509 ± 0.088 |
| Phage UGI to human UDG | 6 315 673 ± 187 973 | 0.00119 ± 0.00010 | 0.189 ± 0.021 |
SAUGI inhibits the uracil-removing activity of SAUDG
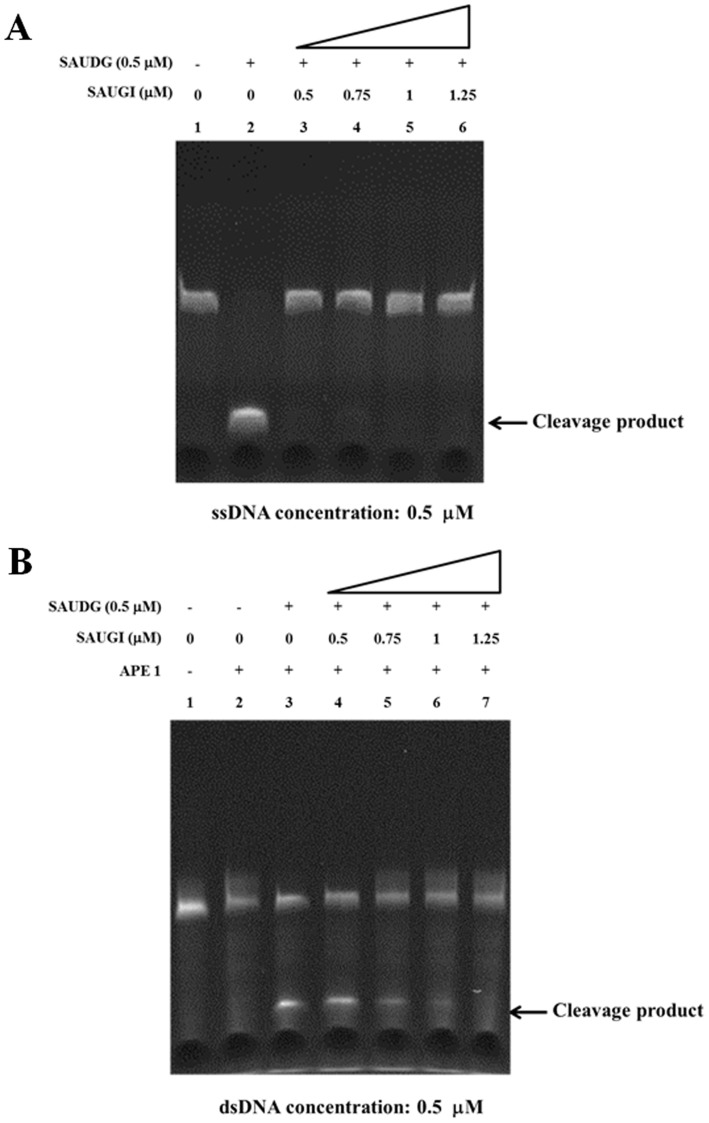
To further confirm that SAUGI acts as a uracil-DNA glycosylase inhibitor, the SAUDG activity was determined by an in vitro glycosylase cleavage assay in the presence or absence of SAUGI. In the absence of SAUGI, SAUDG removed the uracil from both ssDNA and dsDNA, as shown by the cleavage at the resulting apyrimidinic site when treated with heat and alkali (ssDNA) or with AP endonuclease (dsDNA) (Figure 4A lane 2 and Figure 4B lane 3). In contrast, the amounts of cleaved product were progressively reduced by the addition of increasing quantities of SAUGI (Figure 4A and B). SAUDG activity was completely inhibited when the molar ratio of SAUGI to SAUDG reached 1:1 for the ssDNA assay and 2:1 for the dsDNA assay. The slight difference in the inhibiting effect of SAUGI on the ssDNA and dsDNA uracil-removing activity by SAUDG may be due to the different reaction buffers used.
Figure 4.
Inhibition of SAUDG’s uracil-removing activity by SAUGI. (A) ssDNA. (B) dsDNA. Addition of the DNA mimic SAUGI reduced the specific uracil-removing activity of SAUDG in a dosage-dependent manner.
DISCUSSION
The two previously known uracil-DNA glycosylase inhibitors are well-studied DNA mimic proteins that specifically inhibit the activity of UDGs. One of these mimics, phage PBS2 UGI, contains a DNA-like surface that binds to the DNA binding region of UDG and thus blocks its uracil-removing activity (5–8,38). This serves to protect the special genomic DNA of PBS2, which is unusual in that it incorporates uracil residues instead of thymine residues (39). The other uracil-DNA glycosylase inhibitor, p56, was identified from Bacillus phage ϕ 29 (9–12). Interestingly, despite its similar function, the dimeric structure of p56 is different from that of the monomeric UGI (10). This p56 has later been shown to form a complex with both Bacillus UDG and herpes simplex virus (HSV) UDG (11,12). Unlike phage PBS2, the genomic DNA of ϕ 29 does not contain uracil residues, but Serrano-Heras (2006) hypothesized that p56 might knock out the host-encoded UDG base-excision repair system because this UDG activity could still be harmful to phage replication if uracil residues occur in the replicative intermediates (40). This hypothesis was recently confirmed (41).
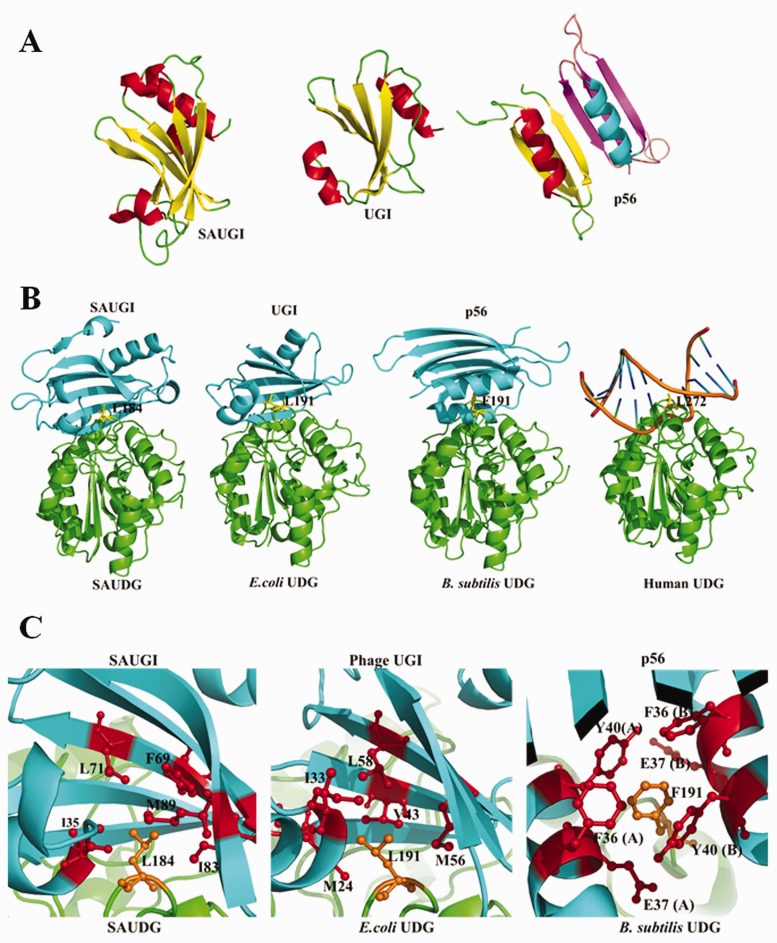
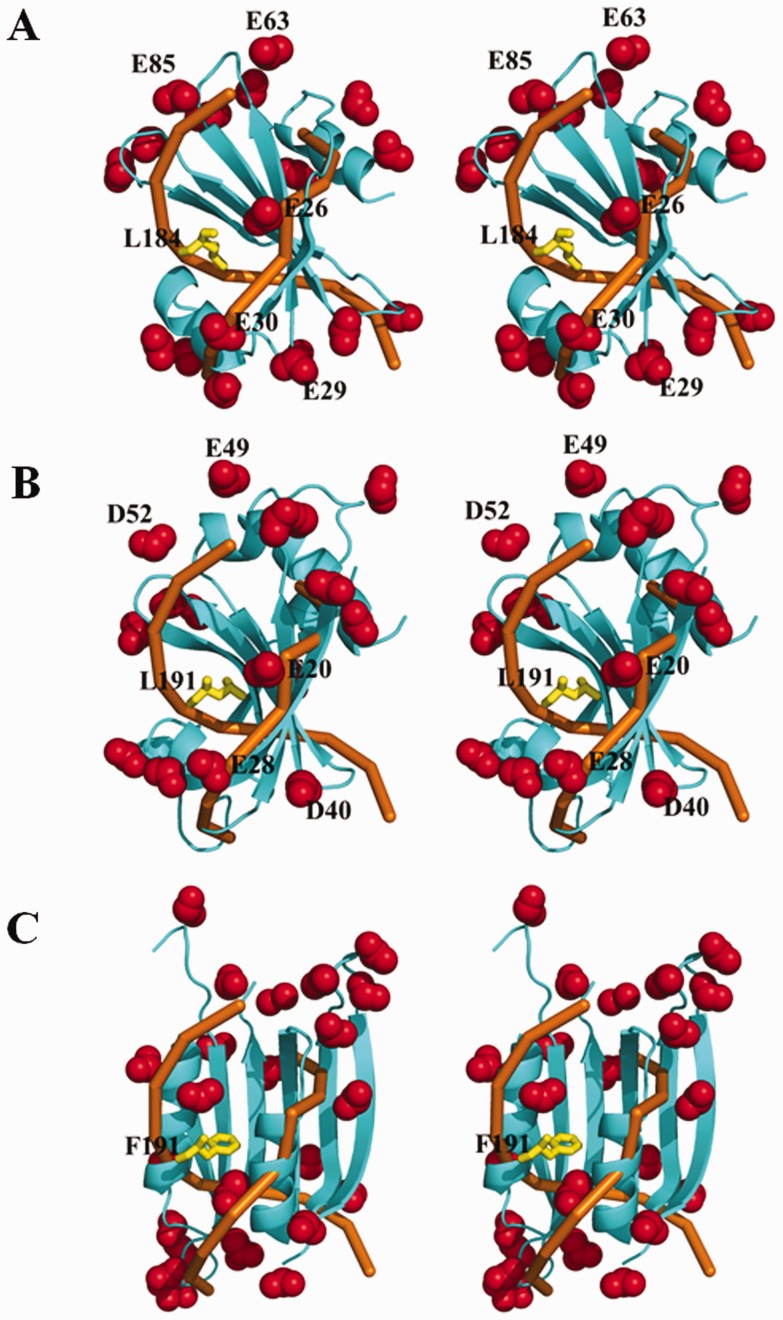
In terms of the protein folding and charge distribution, SAUGI is a DNA mimic that resembles UGI rather than p56 (Figure 5A). Figure 5B and C further shows how SAUGI, UGI and p56 all share a conserved UDG binding strategy, which involves targeting their respective UDG’s protruding residue (i.e. SAUDG Leu184, E. coli UDG Leu191 and B. subtilis UDG Phe191; see Supplementary Figure S6A) by means of a hydrophobic pocket. Interestingly, unlike the conformational changes seen in the loops and β-zippers of human UDG when it binds to DNA (37), only minor conformational changes are seen when these uracil-DNA glycosylase inhibitors bind to their targets (Supplementary Figure S11). All of these uracil-DNA glycosylase inhibitors therefore bind to their targets in conformations that are close to the unbound conformations. As shown in Figure 6, while many of the carboxyl groups on the side chains of SAUGI and UGI share similar locations, most of the carboxyl groups on the side chains of p56 occupy different locations. The matching carboxyl groups on SAUGI and UGI surround the hydrophobic pockets that are used to bind to SAUDG Leu184 and E. coli UDG Leu191, respectively, and they may be functionally important for these two uracil-DNA glycosylase inhibitors.
Figure 5.
A structural comparison of SAUGI, UGI, p56 and their respective complexes with UDG. (A) The known uracil-DNA glycosylase inhibitors. The α-helices and β-strands are, respectively, colored red and yellow in SAUGI, UGI and one molecule of the p56 dimer, and cyan and magenta in the other p56 molecule. (B) A comparison of the UDG complexes with SAUGI, UGI, p56 and DNA. The uracil-DNA glycosylase inhibitors and the UDGs are, respectively, colored cyan and green. The protruding residues from the UDGs are shown as yellow sticks. (C) Details of the hydrophobic binding pockets in the interacting regions of the three uracil-DNA glycosylase inhibitors. Residues of the protein inhibitors and UDGs are shown as red and orange sticks, respectively.
Figure 6.
The side-chain carboxyl groups of three uracil-DNA glycosylase inhibitors that match the human UDG-bound DNA phosphate backbone. The carboxyl groups on (A) SAUGI, (B) UGI and (C) p56 are shown as red spheres, while the protruding UDG residues are shown as yellow sticks. The UDG-bound DNA phosphate backbone is colored orange. A good correspondence is seen between the several carboxyl groups on SAUGI and UGI (i.e. Glu 26/Glu 20, Glu29/Asp 40, Glu30/Glu 28, Glu 85/Asp 52 and Glu 63/Glu49). All three figures are in stereo.
At this time, the biological meaning of SAUGI remains unclear. In particular, why would Staphylococcus produce a protein that inhibits its own DNA repair system? One possible explanation is as follows. It has recently been shown that the replication of DNA viruses can be blocked by increasing the amount of dUTP in the viral genome (42). Many viruses, such as poxviruses, HSV and retroviruses, encode dUTPase and/or UDG to prevent genomic uracilation (43), while HIV-1 virus also avoids uracilation of its genome by recruiting human UDG (44,45). Meanwhile, among the 27 bacteriophages that infect S. aureus, genomic information shows that 13 of them encode dUTPase (46). This suggests that these Staphylococcus phages are sensitive to genomic uracilation. By inhibiting UDG activity and thus preventing the removal of spontaneously deaminated cytosine from phage DNA, SAUGI may act as an anti-phage factor in Staphylococcus. However, this hypothesis still needs to be verified, and other roles for SAUGI are also possible. For example, SAUGI may simply act as a regulator that modulates the SAUDG activity in Staphylococcus. As a first step in exploring this possibility, it would be interesting to observe the effects of up- or downregulation of SAUGI expression in Staphylococcus.
While the high binding affinity of UGI to human UDG might be cytotoxic to human cells, we note that SAUGI’s relatively low binding affinity to human UDG (Supplementary Figure S10 and Table 3) might allow it to directly suppress UDG repair activity in S. aureus and other human pathogens. In addition, SAUGI-based drugs might be used to decrease the replication of viruses such as HSV by inhibiting their UDG activity. Lastly, the His-pulldown assay showed that SAUGI interacted with other DNA binding proteins in addition to SAUDG (Supplementary Figure S4 and Supplementary Table S3), and we have further confirmed the interaction between SAUGI and the SALEX transcription factor (LexA) using a bis (sulphosuccinimidyl) suberate (BS3) cross-linking assay (Supplementary Figure S12). These results suggest that SAUGI may also act to regulate other DNA binding proteins.
In conclusion, this is the first case of a DNA mimic protein identified by using a bioinformatic approach, and it also represents the first uracil-DNA glycosylase inhibitor from a non-phage species.
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org. The PDB ID codes of SAUDG and SAUGI/SAUDG complex are 3WDF and 3WDG, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [47–48].
FUNDING
This work was supported financially by Academia Sinica, Taipei Medical University and National Science Council [022371, TMU102-AE1-B12 and NSC102-2319-B-001-003]. Funding for open access charge: Academia Sinica and National Science Council [022371 and NSC102-2319-B-001-003].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the staff of beamline BL13B1 at the Radiation Research Center (NSRRC) in Hsinchu, Taiwan, for their help in X-ray crystal data collection. They also thank the Core Facilities for Proteomics Research located at the Institute of Biological Chemistry, Academia Sinica, for their help in Proteomic mass spectrometry analyses.
REFERENCES
- 1.Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair. 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Dryden DTF. DNA mimicry by proteins and the control of enzymatic activity on DNA. Trends Biotechnol. 2006;4:378–382. doi: 10.1016/j.tibtech.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Hagen L, Peña-Diaz J, Kavli B, Otterlei M, Slupphaug G, Krokan HE. Genomic uracil and human disease. Exp. Cell Res. 2006;312:2666–2672. doi: 10.1016/j.yexcr.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Parikh SS, Putnam CD, Tainer JA. Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res. 2000;460:183–199. doi: 10.1016/s0921-8777(00)00026-4. [DOI] [PubMed] [Google Scholar]
- 5.Mol CD, Arvai AS, Sanderson RJ, Slupphaug G, Kavli B, Krokan HE, Mosbaugh DW, Tainer JA. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 6.Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 7.Kaushal PS, Talawar RK, Krishna PD, Varshney U, Vijayan M. Unique features of the structure and interactions of mycobacterial uracil-DNA glycosylase: structure of a complex of the Mycobacterium tuberculosis enzyme in comparison with those from other sources. Acta Crystallogr. D Biol. Crystallogr. 2008;64:551–560. doi: 10.1107/S090744490800512X. [DOI] [PubMed] [Google Scholar]
- 8.Géoui T, Buisson M, Tarbouriech N, Burmeister WP. New insights on the role of the gamma-herpesvirus uracil-DNA glycosylase leucine loop revealed by the structure of the Epstein-Barr virus enzyme in complex with an inhibitor protein. J. Mol. Biol. 2007;366:117–131. doi: 10.1016/j.jmb.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Serrano-Heras G, Ruiz-Masó JA, del Solar G, Espinosa M, Bravo A, Salas M. Protein p56 from the Bacillus subtilis phage phi29 inhibits DNA-binding ability of uracil-DNA glycosylase. Nucleic Acids Res. 2007;35:5393–5401. doi: 10.1093/nar/gkm584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asensio JL, Pérez-Lago L, Lázaro JM, González C, Serrano-Heras G, Salas M. Novel dimeric structure of phage ϕ 29-encoded protein p56: insights into uracil-DNA glycosylase inhibition. Nucleic Acids Res. 2011;39:9779–9788. doi: 10.1093/nar/gkr667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baños-Sanz JI, Mojardín L, Sanz-Aparicio J, Lázaro JM, Villar L, Serrano-Heras G, González B, Salas M. Crystal structure and functional insights into uracil-DNA glycosylase inhibition by phage ϕ 29 DNA mimic protein p56. Nucleic Acids Res. 2013;41:6761–73. doi: 10.1093/nar/gkt395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole AR, Ofer S, Ryzhenkova K, Baltulionis G, Hornyak P, Savva R. Architecturally diverse proteins converge on an analogous mechanism to inactivate uracil-DNA glycosylase. Nucleic Acids Res. 2013;41:8760–8775. doi: 10.1093/nar/gkt633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez BE, Voloshin ON, Camerini-Otero RD, Bax A. Solution structure of DinI provides insight into its mode of RecA inactivation. Protein Sci. 2000;9:2161–2169. doi: 10.1110/ps.9.11.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons LM, Yeh DC, Orban J. Solution structure of the highly acidic protein HI1450 from Haemophilus influenzae, a putative double-stranded DNA mimic. Proteins. 2004;54:375–383. doi: 10.1002/prot.10607. [DOI] [PubMed] [Google Scholar]
- 16.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DT. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 17.Liu D, Ishima R, Tong KI, Bagby S, Kokubo T, Muhandiram DR, Kay LE, Nakatani Y, Ikura M. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 18.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl Acad. Sci. USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 20.León E, Navarro-Avilés G, Santiveri CM, Flores-Flores C, Rico M, González C, Murillo FJ, Elías-Arnanz M, Jiménez MA, Padmanabhan S. A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix. Nucleic Acids Res. 2010;38:5226–5241. doi: 10.1093/nar/gkq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Court R, Cook N, Saikrishnan K, Wigley D. The crystal structure of lambda-Gam protein suggests a model for RecBCD inhibition. J. Mol. Biol. 2007;371:25–33. doi: 10.1016/j.jmb.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 22.McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, et al. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 2009;37:4887–4897. doi: 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang HC, Wang HC, Ko TP, Lee YM, Leu JH, Ho CH, Huang WP, Lo CF, Wang AHJ. White spot syndrome virus protein ICP11: a histone-binding DNA mimic that disrupts nucleosome assembly. Proc. Natl Acad. Sci. USA. 2008;105:20758–20763. doi: 10.1073/pnas.0811233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HC, Ko TP, Wu ML, Ku SC, Wu HJ, Wang AHJ. Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor. Nucleic Acids Res. 2012;40:5718–5730. doi: 10.1093/nar/gks177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. Protein-structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 28.Vagin AA, Steiner RS, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organisation of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2284–2295. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 29.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.The PyMOL Molecular Graphics System, Version 1.5.0.4. 2010. Schrödinger, LLC, Portland, OR. http://www.pymol.org/citing (11 October 2013, date last accessed)
- 33.Bulgar AD, Weeks LD, Miao Y, Yang S, Xu Y, Guo C, Markowitz S, Oleinick N, Gerson SL, Liu L. Removal of uracil by uracil DNA glycosylase limits pemetrexed cytotoxicity: overriding the limit with methoxyamine to inhibit base excision repair. Cell Death Dis. 2012;3:e252. doi: 10.1038/cddis.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramelot TA, Ding K, Chen CX, Jiang M, Ciccosanti C, Xiao R, Liu J, Baran MC, Swapna G, Acton TB, et al. Solution NMR structure of SSP0047 from Staphylococcus saprophyticus. Northeast Structural Genomics Consortium Target SyR6. Northeast Structural Genomics Consortium. 2009 http://www.nesg.org/ (11 October 2013, date last accessed) [Google Scholar]
- 35.Ortega A, Amorós D, García de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys J. 2011;101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Handa P, Roy S, Varshney U. The role of leucine 191 of Escherichia coli uracil DNA glycosylase in the formation of a highly stable complex with the substrate mimic, ugi, and in uracil excision from the synthetic substrates. J. Biol. Chem. 2001;276:17324–17331. doi: 10.1074/jbc.M011166200. [DOI] [PubMed] [Google Scholar]
- 37.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Mosbaugh DW. Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J. Biol. Chem. 1989;264:1163–1171. [PubMed] [Google Scholar]
- 39.Takahashi I, Marmur J. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature. 1963;197:794–795. doi: 10.1038/197794a0. [DOI] [PubMed] [Google Scholar]
- 40.Serrano-Heras G, Salas M, Bravo A. A uracil-DNA glycosylase inhibitor encoded by a non-uracil containing viral DNA. J. Biol. Chem. 2006;281:7068–7074. doi: 10.1074/jbc.M511152200. [DOI] [PubMed] [Google Scholar]
- 41.Serrano-Heras G, Bravo A, Salas M. Phage phi29 protein p56 prevents viral DNA replication impairment caused by uracil excision activity of uracil-DNA glycosylase. Proc. Natl Acad. Sci. USA. 2008;105:19044–19049. doi: 10.1073/pnas.0808797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sire J, Quérat G, Esnault C, Priet S. Uracil within DNA: an actor of antiviral immunity. Retrovirology. 2008;5:45. doi: 10.1186/1742-4690-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen R, Wang H, Mansky LM. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 2002;83:2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- 44.Guenzel CA, Hérate C, Le Rouzic E, Maidou-Peindara P, Sadler HA, Rouyez MC, Mansky LM, Benichou S. Recruitment of the nuclear form of uracil DNA glycosylase into virus particles participates in the full infectivity of HIV-1. J. Virol. 2012;86:2533–2544. doi: 10.1128/JVI.05163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weil AF, Ghosh D, Zhou Y, Seiple L, McMahon MA, Spivak AM, Siliciano RF, Stivers JT. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl Acad. Sci. USA. 2013;110:448–457. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl Acad. Sci. USA. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollastri G, Baldi P, Fariselli P, Casadio R. Prediction of coordination number and relative solvent accessibility in proteins. Proteins. 2002;47:142–153. doi: 10.1002/prot.10069. [DOI] [PubMed] [Google Scholar]
- 48.Wang HC, Wu ML, Ko TP, Wang AHJ. Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein. Nucleic Acids Res. 2013;41:5127–5138. doi: 10.1093/nar/gkt201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.