Abstract
Variants of mitochondrial DNA (mtDNA) are commonly used as markers to track human evolution because of the high sequence divergence and exclusive maternal inheritance. It is assumed that the inheritance is clonal, i.e. that mtDNA is transmitted between generations without germline recombination. In contrast to this assumption, a number of studies have reported the presence of recombinant mtDNA molecules in cell lines and animal tissues, including humans. If germline recombination of mtDNA is frequent, it would strongly impact phylogenetic and population studies by altering estimates of coalescent time and branch lengths in phylogenetic trees. Unfortunately, this whole area is controversial and the experimental approaches have been widely criticized as they often depend on polymerase chain reaction (PCR) amplification of mtDNA and/or involve studies of transformed cell lines. In this study, we used an in vivo mouse model that has had germline heteroplasmy for a defined set of mtDNA mutations for more than 50 generations. To assess recombination, we adapted and validated a method based on cloning of single mtDNA molecules in the λ phage, without prior PCR amplification, followed by subsequent mutation analysis. We screened 2922 mtDNA molecules and found no germline recombination after transmission of mtDNA under genetically and evolutionary relevant conditions in mammals.
INTRODUCTION
Mutations in mitochondrial DNA (mtDNA) are the most frequently used genetic marker in phylogenetic and population studies in mammals. This popularity rests upon three pillars: the high sequence divergence of mtDNA, the exclusive maternal transmission and the assumed lack of germline recombination. This means that mtDNA is used as a molecular marker showing patterns of clonal inheritance, which reduces the complexity of the analyses as bi-parental inheritance does not have to be taken into account. The possible occurrence of mtDNA recombination in the mammalian maternal germline has far reaching implications, because even modest frequencies of germline recombination would substantially affect coalescent time estimates and branch lengths in phylogenetic trees (1). During the last two decades numerous reports have questioned the assumption that there is no mtDNA recombination based on analyses of human patient tissues (2,3), tissue culture cells (4,5) and tissues from interspecies animal hybrids (6,7). However, many of these studies have relied heavily on polymerase chain reaction (PCR) amplification with no or little control for recombination by jumping PCR, which may produce chimeric PCR products (8) that falsely are interpreted as evidence for in vivo recombination. Another common limitation of these reports is that they mostly cover events in somatic tissues or are based on analyses of transformed cell lines and therefore have little relevance for understanding mtDNA evolution of animals. To complicate matters further, it should be noted that bi-parental inheritance of mtDNA has been robustly documented in some organisms, like the mussel Mytilus galloprovincialis (9). Within mammals, heteroplasmy, i.e. the presence of two mtDNA variants within the same individual, is more common than previously thought (10,11) and paternal leakage of mtDNA has been reported under unusual conditions, such as interspecific mouse crosses (12,13). Hence, there are certainly documented circumstances where different types of mtDNA molecules coexist in an individual, but it is unclear to what extent recombination of mtDNA actually occurs in the mammalian germline. Unfortunately, technical limitations combined with the lack of appropriate in vivo experimental systems has left the research field in uncertainty, with claims both for and against recombination of mammalian mtDNA being questioned. In this report, we have used a maternal mouse line that has been heteroplasmic for a defined set of mtDNA mutations for more than 50 generations in combination with a protocol based on λ phage cloning for sequence analysis of single mtDNA molecules. We analyzed 2922 mtDNA molecules and found no recombinant molecules when mtDNA was transmitted in the maternal germline under genetically and evolutionarily relevant conditions. These findings thus demonstrate that recombination does not contribute to mtDNA variation in mammals.
MATERIALS AND METHODS
Mouse breeding
The NZB-BALB/c heteroplasmic mouse line was generated by cytoplast transfer, as previously described (14), and has been maintained by backcrossing of heteroplasmic females to inbred BALB/c males for more than 50 generations.
Preparation of mtDNA for λ phage cloning
Liver and kidney mtDNA was extracted and purified according to standard protocols. In brief, tissues were homogenized with 15 strokes of a teflon homogenizer at 1500 rpm in mitochondrial isolation buffer [MIB; 225 mM mannitol, 7.5 mM sucrose, 1 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1 mM ethylenediaminetetraacetic acid (EDTA) pH 8.0, 0.1% bovine serum albumin and 0.025% β-mercaptoethanol]. Cell debris was pelleted by low speed centrifugation (10 min at 600 g) and mitochondria were recovered from the supernatant by high-speed centrifugation (10 min at 5000g). Mitochondrial pellets were washed three times in MIB, using a loose-fit glass homogenizer to resuspend the pellets (Wheaton, USA). The final pellet was placed on a two-step sucrose gradient cushion (1.5 M/1M), followed by 1 h centrifugation at 77 000 g (SW32Ti, Sorvall). Mitochondria were recovered from the gradient, pelleted in mitochondrial gradient buffer (MGB; 10 mM HEPES pH 7.8, 10 mM EDTA pH8.0), re-suspended in mitochondrial lysis buffer (MLB; 75 mM NaCl, 50 mM EDTA pH 8.0, 20 mM HEPES pH 7.8) and lysed with 1% Sarkosyl, followed by phenol/chloroform extraction and isopropanol/NaCl precipitation. The concentration mtDNA was determined on a DyNAQuant 200 (Hoefer).
λ Phage cloning
Sucrose-gradient purified mtDNA (100–200 ng) was digested with the BglII endonuclease (NewEngland BioLabs). Appropriate cohesive ends were generated by Klenow DNA polymerase extension in the presence of 0.17 mM dGTP and dATP, followed by standard phenol/chloroform extraction and ethanol/salt precipitation supplemented with 1 µl Pellet paint (Novagen). The DNA pellet was dissolved in 1.8 µl ddH20, 1 µl l λ phage vector arms (Stratagene), 0.35 µl 10x T4 DNA ligase buffer, 0.3 µl T4 DNA ligase (NewEngland BioLabs)—with a DNA concentration of >0.2 µg/µl—and incubated for 20 h at +4°C. The ligation reaction was added to an in vitro packaging extract (Stratagene) and incubated for 90 min at room temperature. Packaged phage library titers were prepared in 500 µl of SM buffer (10 mM NaCl, 8 mM MgSO4, 50 mM TRIS–HCl pH 7.5) and 20 µl of chloroform. Individual λ clones were expanded in an infected host Escherichia coli strain, which had entered the lytic cycle for plaque formation. The host E. coli was grown according to conditions in the provided protocol and pelleted and dissolved in 10 mM MgSO4 to an OD600 of 0.5. Two hundred µl of E. coli cells were then incubated with an appropriate volume of phage (0.01–2 µl) at +37°C for 15 min. The phage—E. coli solution was then diluted with 3.5 ml of liquid NZYM top agar, spread on NZYM agar plates and incubated over night at +37°C. The following day individual plaques were picked using a 1-ml pipette tip and eluted into a suitable solution for further analysis. Re-plating experiments suggest that a single plaque contains approximately 50 000 copies of the cloned mtDNA.
PCR and restriction fragment length polymorphism analysis
One microlitre of eluted phage was used as PCR template per reaction. DNA was extracted from phage particles by a 10-min incubation at 95°C as the initial step in the PCR. PCR primers were designed to amplify appropriate parts of the mtDNA genome to be used in the restriction fragment length polymorphism (RFLP) screening for recombination (Supplementary Table S1). PCR products were cleaved with the appropriate endonucleases (Supplementary Table S2) and the digested PCR products separated by gel electrophoresis in 1.5% agarose gels.
PCR and sequencing of full-length mtDNA molecules
PCR primers, covering the entire mtDNA sequence (Supplementary Table S3) and containing M13 sequencing tags at their 5′-ends, were designed to amplify 27 overlapping PCR products. PCR products were purified with AMPure (Agencourt) and sequenced using BigDye, version 3.1 chemistry (Applied Biosystems). Cycle sequencing reactions were purified using X-terminator solution (Applied Biosystems) and run on a 3130xL capillary sequencer. Sequences were aligned to the C57BL/6J reference sequence (accession NC 005089.1) using the Seqscape V2.5 software (Applied Biosystems) and all polymorphisms were numbered according to the C57BL/6J reference sequence.
Quantification of heteroplasmy levels
Relative heteroplasmy levels for whole-tissue extracts, isolated mitochondria and phage titer were quantified by last-cycle fluorescent PCR–RFLP analysis. The relative heteroplasmy levels of the final clones were also quantified by PCR–RFLP and the cleaved products were separated on 1.5 % agarose gels. PCR and RFLP analyses were performed as previously described (15), using primers containing a 5′ M13-tag and an internal digestion control (forward: TGT AAA ACG ACG GCC AGT TCT CCG TGC TAC CTA AAC ACC; reverse: CAG GAA ACA GCT ATG ACC TAG GTT GGT GCC GGA TAT TGT G). For the last-cycle fluorescent analysis, the final cycle of a standard PCR was spiked with 5 μM M13F-hex primer. PCR products were purified using Agencourt Ampure (Beckman Coulter) and an aliquot was digested by NlaIV endonuclease (New England Biolabs). The digested product (1 μl) was separated by fragment analysis on a 3130xL DNA Analyzer and analyzed using the GeneMapper v.4.0 software (Applied Biosystems). Peak heights were used to determine relative heteroplasmy levels. Average values from three independent experiments were used for the quantification of mtDNA mutation levels.
RESULTS
PCR amplification induces abundant in vitro recombination artifacts
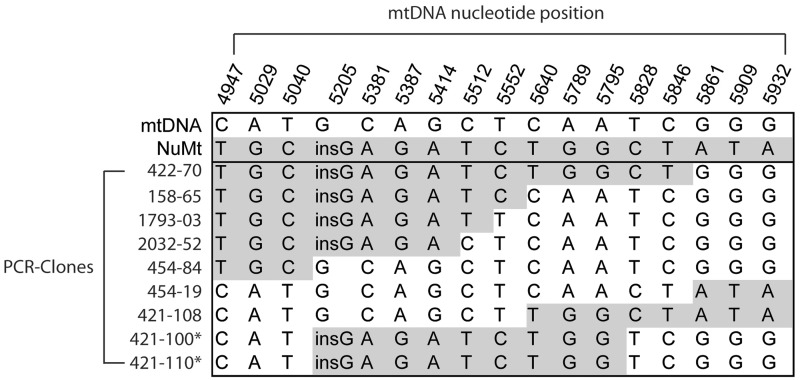
The amplification of DNA by PCR can lead to the creation of artificial chimeric molecules by polymerase jumping (8). We have previously applied a standard PCR-clone-sequence protocol to detect mtDNA mutations in C57BL/6N mice (16,17). From these available datasets, we extracted sequences that had been filtered due to similarity with nuclear mtDNA pseudogenes (NuMts). We identified NuMts in 41 of 3940 clones (∼1% of samples). Strikingly, nine of the clones (Figure 1) showed evidence of eight independent PCR-induced recombination events between NuMts and mtDNA (Figure 1). This high frequency was present despite the very low abundance of nuclear DNA relative to mtDNA in the samples. Cloning and subsequent sequencing of PCR products is therefore not suitable for identifying mtDNA recombination events because artificial in vitro generated chimeric molecules are very frequent and indistinguishable from true in vivo recombinants.
Figure 1.
Sequence of artificially created recombinant mtDNA molecules. The sequence patterns of mtDNA molecules analyzed by the PCR-clone-sequencing method are shown. The mtDNA reference sequence variants are depicted in white and the NuMts in grey. In total, 9 out of 41 of clones showed evidence of PCR-induced recombination between NuMts and mtDNA. Asterisk indicates two clones generated by the same in vitro event.
Cloning of mtDNA in the λ phage does not create artificially chimeric mtDNA molecules
The unsuitability of a PCR-clone-sequencing approach to study mtDNA recombination prompted us to develop an alternative method to capture large stretches of native mtDNA. The λ phage has a long tradition of being the vehicle of choice to clone a variety of genomic targets. Utilizing this vector, we developed a direct cloning protocol that enables us to trap single full-length mtDNA molecules from mitochondrial-enriched DNA preparations using as little as 10 ng of input DNA. By sequencing entire phage inserts we were able to show experimentally that λ phage cloning can successfully trap full-length mtDNA molecules. Next, we created an artificially heteroplasmic sample by mixing liver tissue from C57BL/6 and NZB mice, that differ by 90 positions in their mtDNA, to experimentally test whether chimeric molecules are artificially created by the λ cloning approach. A standard mtDNA extraction procedure was then performed on the mixture of liver tissue from the two mouse strains, followed by λ phage cloning. In order to screen a large number of mtDNA clones, we chose four strain-specific polymorphisms distributed across the mitochondrial genome (nucleotide positions 2814, 5463, 9985 and 15 657) for RFLP analysis (Figure 2a and b). Individual plaques were analyzed for recombinants carrying both NZB and C57BL/6 mtDNA variants. In total, we screened 100 molecules and detected no recombinant molecules.
Figure 2.
RFLP analyses of mtDNA in heteroplasmic mice. (a) RFLP patterns obtained from restriction enzyme digestion of λ phage cloned mtDNA molecules from liver showing pure BALB/c (lanes 1–4) or NZB (lanes 9–12) patterns. One cloned molecule (lanes 5–8) shows a mixed NZB/BALB/c cleavage pattern. The screened nucleotide variants (2814, 5463, 9985 and 15 657) are shown on top of each lane. (b) Schematic depiction of mouse mtDNA with the non-coding control region (yellow) and genes encoding proteins (green), ribosomal RNAs (rRNAs; blue) and transfer RNAs (tRNAs; brown). The four sites that were used for screening to distinguish between NZB and BALB/c mtDNA (nucleotide variants 2814, 5463, 9985 and 15 657) are indicated with arrows.
Cloning of mtDNA in the λ phage is unbiased
In order to ensure that the cloning method is not biased toward a subgroup of mtDNA molecules, we extracted mtDNA from a previously published mouse line with a heteroplasmic 3875delC mutation in the tRNAMet gene (15). We scored the 3875delC genotype in 830 cloned full-length mtDNA molecules from two heteroplasmic tRNAMet mice (418 and 412 for mice 1 and 2, respectively), by PCR–RFLP. Heteroplasmy levels in liver tissue, mtDNA extracts and the phage titers for the two respective animals were quantified by last-cycle fluorescent PCR–RFLP analysis. Subsequent comparison of heteroplasmy levels in the liver tissue, in purified mtDNA, in λ phage titers and the final phage clones revealed no statistically significant variation (Chi-square test), indicating that the method is unbiased toward which molecule is cloned (Table 1).
Table 1.
The λ phage protocol does not favor cloning of specific mtDNA genotypes
| Mouse 1 |
Mouse 2 |
|||
|---|---|---|---|---|
| Heterpolasmy (%) | P-value | Heterpolasmy (%) | P-value | |
| Liver tissue | 74.48 | 68.16 | ||
| mtDNA extract | 74.77 | 0.95 | 66.93 | 0.79 |
| Phage titer | 75.37 | 0.81 | 66.68 | 0.77 |
| Lambda clones | 75.12 | 0.88 | 71.26 | 0.51 |
Determination of the λ phage replication error rate
We experimentally tested the λ phage replication error rate by replica-plating a single λ clone containing mtDNA 60 times in 6 parallel lines, corresponding to 360 passages of a single λ-mtDNA clone. The entire mtDNA from the initial clone as well as all final six clones were fully sequenced. In comparison with the input sequence no new mutations were found, thus supporting an extremely low error rate during λ phage DNA replication, previously estimated to be 7.7 × 10-8 mutations/bp (18).
No evidence for germline recombination of mammalian mtDNA
After having adapted and validated the direct λ phage method to clone mtDNA from mouse tissue samples, we applied this technique to experimentally test whether recombination of mtDNA occurs in the mammalian germline. The NZB-BALBc mouse line was generated by cytoplast transfer between an NZB-derived cytoplasm and a BALBc embryo >20 years ago (14). Heteroplasmy levels in these mice segregate in a random manner, best explained by random genetic drift and the heteroplasmy levels in oocytes are dependent on the levels in the mother (14). Different estimates have found that the proportion of NZB mtDNA in oocytes ranges from 0.9% to 41% (14,19). As these mice have retained an intracellular heteroplasmy between NZB and BALB/c mtDNA for >20 years, they pose an ideal model to investigate mtDNA recombination and clonality in the maternal mammalian germline.
We used the above mentioned RFLP analysis to screen single mtDNA molecules cloned in the λ phage (Figure 2a and b). After screening a total of 2922 mtDNA molecules from liver and kidney samples from NZB-BALB/c animals (n = 2), we identified only a single aberrant molecule with a mixed BALB/c-NZB digestion pattern (Figure 2a). Sequencing this entire chimeric molecule revealed that the informative SNPs were almost exclusively NZB-derived with only one single nucleotide position representing a BALB/c SNP (15 657c>T). All adjacent polymorphisms were NZB (positions 15 603 and 15 917, Supplementary Table S4), suggesting that this variant represents homoplasy, i.e. a single mutational event, rather than a recombination event. In total, we screened 70 128bp of mtDNA by RFLP analysis and identified only a single sequence variant, which suggests that the mutation rate is ∼1.43 × 10-5 mutations/bp. This is in good concordance with the mutation load estimate obtained by PCR-clone-sequencing (Supplementary Table S5). In order to identify any additional spontaneous or homoplastic variants that might have fixed in the two genomes, we sequenced the entire mtDNA of nine BALBc and eight NZB-derived λ clones. All NZB mtDNA clones analyzed differed from previously reported NZB sequence (20) at two positions (2767g>C, 7778g>T), whereas both the NZB and BALB/c molecules differed by a single adenine insertion in the polyadenine-stretch of the tRNAArg gene. As we were unable to determine whether these changes were present in the initial founder animals, we cannot determine whether the NZB variants were acquired during the last 20 years of mouse breeding and clonally expanded, or whether these variants were already present in the original NZB strain.
DISCUSSION
The creation of novel recombinant sequence variants requires not only an enzymatic process for mtDNA recombination, but also the presence of different sequence variants of mtDNA. To some extent the exclusive maternal transmission of mtDNA provides a natural barrier against recombination. However, there are reports of low-level heteroplasmy in mammals, including humans (10,11), suggesting that this may in fact be the prevailing situation. Sperm could theoretically provide a source of divergent mtDNA and transmission of paternal mtDNA occurs in interspecific crosses (12,13). However, under normal physiological conditions, paternal mitochondria are actively targeted and destroyed in the oocyte by an incompletely understood mechanism, which is present throughout the animal kingdom (21–24) with a few exceptions (12,13). Recent high-resolution STED microscopy studies of primary cells and cell lines in tissue culture have revealed that mammalian mitochondrial nucleoids typically contain a single copy of mtDNA (25,26), further reducing the chance that two molecules will be within close proximity to favor recombination events. However, the nucleoid organization of mtDNA in the maternal germline has not yet been studied and may possibly be different from cells in culture.
Despite arguments against mtDNA recombination, there are nevertheless many reports suggesting the occurrence of recombination in the mammalian mtDNA. Analysis of recombination by PCR amplification is prone to artifacts and these results are therefore controversial (21). In this study, we sequenced cloned PCR products and could unequivocally demonstrate relatively high levels of polymerase jumping between mtDNA and nuclear pseudogene templates, although the latter were present at very low levels. We thus conclude that PCR amplification prior to cloning is an unreliable method when studying mtDNA recombination. These results lead us to establish a direct cloning approach by using the λ phage to trap individual mtDNA molecules prior to amplification. Control experiments showed that this λ phage cloning approach is unbiased and does not create artificial recombinants.
The NZB-BALB/c mouse line analyzed in this study has been heteroplasmic for two essentially neutral mtDNA variants, that differ at 90 nucleotide positions, for more than 50 generations. We applied a direct cloning approach followed by sequencing of individual mtDNA molecules and were unable to detect any mtDNA recombination events despite the extensive period of heteroplasmy. The mtDNA germline bottleneck effect allows only a small fraction of the mtDNA copies in the maternal germline to contribute to the mature oocyte and sequence variants can therefore be rapidly fixed within a population (27). One previous study in heteroplasmic mice, using a very limited number of generations, found no recombination in neutral mtDNA variants, whereas <1% recombinant molecules were found in a mouse line carrying deleted mtDNA (28). Importantly, it should be noted that the analysis of these mouse lines mainly covered somatic recombination events, but the results nevertheless suggest that recombination may be possible under scenarios of mitochondrial dysfunction. The results in our study clearly show that recombination is vanishingly rare both in somatic tissues and in the germline under conditions of neutral heteroplasmy. As low-level heteroplasmy under neutral conditions seems to be the common state in nature, recombination should not contribute to genetic variation and distribution of mtDNA variants in most natural animal populations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The European Research Council [ERC-2010-AdG 268897 to N.G.L.]; Swedish Research Council [521-2010-2766 to N.G.L.]; Jane and Aatos Erkko Foundation and the Academy of Finland (to B.J.B.); Åke Wiberg Foundation [367990950 to C.F.]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Schierup MH, Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–891. doi: 10.1093/genetics/156.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zsurka G, Hampel KG, Kudina T, Kornblum C, Kraytsberg Y, Elger CE, Khrapko K, Kunz WS. Inheritance of mitochondrial DNA recombinants in double-heteroplasmic families: potential implications for phylogenetic analysis. Am. J. Hum. Genet. 2007;80:298–305. doi: 10.1086/511282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kraytsberg Y, Schwartz M, Brown TA, Ebralidse K, Kunz WS, Clayton DA, Vissing J, Khrapko K. Recombination of human mitochondrial DNA. Science. 2004;304:981. doi: 10.1126/science.1096342. [DOI] [PubMed] [Google Scholar]
- 4.Fan W, Lin CS, Potluri P, Procaccio V, Wallace DC. mtDNA lineage analysis of mouse L-cell lines reveals the accumulation of multiple mtDNA mutants and intermolecular recombination. Genes Dev. 2012;26:384–394. doi: 10.1101/gad.175802.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Aurelio M, Gajewski CD, Lin MT, Mauck WM, Shao LZ, Lenaz G, Moraes CT, Manfredi G. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 2004;13:3171–3179. doi: 10.1093/hmg/ddh326. [DOI] [PubMed] [Google Scholar]
- 6.Ujvari B, Dowton M, Madsen T. Mitochondrial DNA recombination in a free-ranging Australian lizard. Biol. Lett. 2007;3:189–192. doi: 10.1098/rsbl.2006.0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo X, Liu S, Liu Y. Evidence for recombination of mitochondrial DNA in triploid crucian carp. Genetics. 2006;172:1745–1749. doi: 10.1534/genetics.105.049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pääbo S, Irwin DM, Wilson AC. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- 9.Zouros E, Oberhauser Ball A, Saavedra C, Freeman K. An unusual type of mitochondrial DNA inheritance in the blue mussle Mytilus. Proc. Natl Acad. Sci. USA. 1994;91:7463–7467. doi: 10.1073/pnas.91.16.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne BAI, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horváth R, Taylor RW, Samuels DC, Santibanez-Koref M, Chinnery PF. Universal heteroplasmy of human mitochondrial DNA. Hum. Mol. Genet. 2013;22:384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Schönberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am. J. Hum. Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyllensten U, Wharton D, Josefsson A, Wilson AC. Paternal inheritance of mitochondrial DNA in mice. Nature. 1991;352:255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- 13.Kaneda H, Hayashi JI, Takahama S, Taya C, Lindahl KF, Yonekawa H. Elimination of paternal mitochondrial DNA in intraspecific crosses during early embryogenesis. Proc. Natl Acad. Sci. USA. 1995;92:4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 15.Freyer C, Cree LM, Mourier A, Stewart JB, Koolmeister C, Milenkovic D, Wai T, Floros VI, Hagström E, Chatzidaki EE, et al. Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat. Genet. 2012;44:1282–1285. doi: 10.1038/ng.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanrooij PH, Uhler JP, Shi Y, Westerlund F, Falkenberg M, Gustafsson CM. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012;40:10334–10344. doi: 10.1093/nar/gks802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross JM, Stewart JB, Hagström E, Brené S, Mourier A, Freyer C, Lagouge M, Hoffer BJ, Olson L, Larsson N-G. Germline mtDNA mutations aggravate ageing and can impair brain development. Nature. 2013;501:412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- 20.Moreno-Loshuertos R, Acín-Pérez R, Fernández-Silva P, Movilla N, Pérez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enríquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 21.Elson JL, Lightowlers RN. Mitochondrial DNA clonality in the dock: can surveillance swing the case? Trends Genet. 2006;22:603–607. doi: 10.1016/j.tig.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Sato M, Sato K. Maternal inheritance of mitochondrial DNA: degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy. 2012;8:424–425. doi: 10.4161/auto.19243. [DOI] [PubMed] [Google Scholar]
- 23.Al Rawi S, Louvet-Valleé S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Allophagy: a macroautophagic process degrading spermatozoid-inherited organelles. Autophagy. 2012;8:421–423. doi: 10.4161/auto.19242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLuca SZ, O’Farrell PH. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell. 2012;13:660–668. doi: 10.1016/j.devcel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson N-G, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl Acad. Sci. USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kukat C, Larsson N-G. mtDNA makes a U-turn for the mitochondrial nucleoid. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.04.009. doi: 10.1016/j.tcb.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Hauswirth WW, Laipis PJ. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc. Natl Acad. Sci. USA. 1982;79:4686–4690. doi: 10.1073/pnas.79.15.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato A, Nakada K, Akimoto M, Ishikawa K, Ono T, Shitara H, Yonekawa H, Hayashi J-I. Rare creation of recombinant mtDNA haplotypes in mammalian tissues. Proc. Natl Acad. Sci. USA. 2005;102:6057–6062. doi: 10.1073/pnas.0408666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.