Abstract
Splicing of precursor messenger RNA is a hallmark of eukaryotic cells, which is carried out by the spliceosome, a multi-megadalton ribonucleoprotein machinery. The splicing reaction removes non-coding regions (introns) and ligates coding regions (exons). The spliceosome is a highly dynamic ribonucleoprotein complex that undergoes dramatic structural changes during its assembly, the catalysis and its disassembly. The transitions between the different steps during the splicing cycle are promoted by eight conserved DExD/H box ATPases. The DEAH-box protein Prp43 is responsible for the disassembly of the intron-lariat spliceosome and its helicase activity is activated by the G-patch protein Ntr1. Here, we investigate the activation of Prp43 by Ntr1 in the presence and absence of RNA substrate by functional assays and structural proteomics. Residues 51–110 of Ntr1 were identified to be the minimal fragment that induces full activation. We found protein–protein cross-links that indicate that Prp43 interacts with the G-patch motif of Ntr1 through its C-terminal domains. Additionally, we report on functionally important RNA binding residues in both proteins and propose a model for the activation of the helicase.
INTRODUCTION
During eukaryotic gene expression, defined parts of the chromosomal DNA are transcribed to precursor messenger RNAs (pre-mRNAs), which are processed before being exported to the nucleus. The processing includes the addition of a 5′-cap as well as a 3′-polyA tail (1,2) and splicing. Splicing of pre-mRNA consists of the removal of non-coding sequences (introns) and the ligation of expressed sequences (exons), which result in a mature mRNA (3). Splicing of pre-mRNAs requires two consecutive transesterification reactions and is catalyzed by the spliceosome, a multi-megadalton and highly dynamic ribonucleoprotein (RNP) complex (3,4). The spliceosome is formed by five uridine-rich small nuclear RNAs (U snRNAs) and numerous proteins; a subset of these is stably associated with the U snRNAs, making up the respective small nuclear RNPs (U snRNPs). The U1, U2, U4/U6 and U5 snRNPs are part of the major (or U2-dependent) spliceosome. In some genes of metazoans and plants, an additional intron class termed U12-type introns has been found; these are spliced by the minor spliceosome, which comprises the U11, U12, U4atac/U6atac and U5 snRNPs (5,6).
The spliceosome is a single-turnover enzymatic machine, which assembles on its substrate, catalyzes the splicing reaction and dissociates afterwards, with multiple changes in its protein composition during the different steps. Thus, at different stages of the splicing cycle, the spliceosome varies greatly in protein composition, with the number of implicated proteins averaging between 110 and 180 in metazoans (7).
The dynamics of the spliceosome are controlled by eight conserved DExD/H-box ATPases, which promote the events leading to the assembly of the spliceosome, the catalysis of the splicing reaction and the dissociation of the intron-lariat spliceosome (8,9). The term “DExD/H” is based on a conserved amino acid sequence in the helicase motif 2. The DEAD-box protein Sub2 (UAP56 in humans) recruits the U2 snRNP to the spliceosome (10), which is then partially remodelled by the DEAD-box protein Prp5 (11). Prp28, an ATPase that also contains the DEAD-motif, is responsible for the release of the U1 snRNP (12). The Ski2-like ATPase Brr2 plays a role in the release of the U4 snRNP during spliceosomal assembly (13).
During the catalysis of the splicing reaction, the DEAH-box ATPases Prp2 and Prp16 are required for the release of the splicing proteins SF3a/SF3b and Yju2/Cwc25, respectively (14–16). Prp22, which is also a DEAH-box protein, is needed during the second transesterification step and also catalyzes the release of the mature mRNA (17,18). Finally, the DEAH-box ATPase Prp43 is involved in the disassembly of the post-spliceosome, leading to the release of the intron lariat, the U6 snRNA, a 20–25 S U2 snRNP and an 18 S U5 snRNP (19,20). It has also been shown that Prp43, in addition to its function during splicing, is involved in the biogenesis of the small and the large ribosomal subunit (21,22)
Additionally, the spliceosomal DExD/H-box ATPases Prp5, Prp16, Prp22, Prp28 and Prp43 have also been suggested to be involved in ensuring the fidelity of splicing and acting as proofreaders (23–25).
Like all members of the superfamily two helicases, the spliceosomal DExD/H helicases contain a central helicase core that is formed by two RecA-like domains (26,27) and is responsible for nucleotide binding and the interaction with the RNA (28). In addition, there are additional domains present in these enzymes that have been shown to regulate their activity or to act as platforms for interaction partners, as their action during the splicing cycle has to be precisely regulated (9,29).
Two of the spliceosomal DEAH-box ATPases, Prp2 and Prp43, have been shown to interact during the splicing cycle with proteins that contain a G-patch motif, namely, Prp2 with Spp2 and Prp43, with Ntr1 (30,31). The G-patch motif encompasses ∼50 residues, among which six are highly conserved glycines; it is predominantly found in proteins associated with nucleic acid interactions (32).
Although no effect of Spp2 on the enzymatic activities of Prp2 has been reported so far, the G-patch motif containing part of Ntr1 strongly stimulates the helicase activity of Prp43, and hence is regarded as an activator of Prp43 (33). Recently, the crystal structure of Prp43 from Saccharomyces cerevisiae (yPrp43) in complex with adenosine diphosphate (ADP) revealed that it shares some features with the processive DNA-helicase Hel308 (34–36). In yPrp43, the two RecA-like domains are followed by a winged helix, a ratchet and an OB-fold domain; an N-terminal extension connects the first RecA-like domain with the C-terminal (NTE) OB-fold domain. The three C-terminal domains of yPrp43 are also present in human Prp22 and seem to be a conserved feature of all four spliceosomal DEAH-box proteins (37). Although this crystal structure is the first reported for a DEAH-box protein and provides insights into the domain organization of this family, it explains neither the binding site of the G-patch motif Ntr1 nor the structural changes that transform yPrp43 into its active form. Furthermore, no 3D structure of a G-patch motif is known to date.
Understanding the detailed dynamics that underlie helicase activation requires extensive structural and biochemical studies. Here, we show that the G-patch motif of Ntr1 is intrinsically unstructured but can adopt secondary-structure elements on binding to Prp43. In addition, we identified the minimal fragment needed for activation. We then set out to investigate structural changes associated with the binding of Ntr1 to Prp43 by applying a series of structural probing techniques in combination with mass spectrometry. These are protein–protein (38), protein–RNA cross-linking (39) and hydroxyl-radical footprinting (40), all of which have recently proven to be useful tools in the investigation of multiprotein and RNP complexes. We were able to define the sites of the interaction between Prp43 and Ntr1 at a molecular level and, furthermore, results of hydroxyl-radical probing led us to suggest changes in the conformation of Prp43 on Ntr1 binding. In addition, our protein–RNA cross-linking data demonstrate that (i) Ntr1 is in direct contact with the RNA through its G-patch motif and (ii) on binding to Ntr1 in the presence of RNA, the ratchet domain of Prp43 becomes more exposed and available for cross-linking as compared with Prp43 bound to RNA alone.
MATERIALS AND METHODS
Molecular cloning
Prp43 was amplified from genomic yeast DNA (S. cerevisiae S288c) using the oligonucleotides Prp43-for and Prp43-rev (Supplementary Table S1 for the sequences of all oligonucleotides) and inserted into a pENTRY-IBA10 vector following the IBA StarGate manual (version December 2008) from IBA GmbH. Subsequently, the open reading frame (ORF) of Prp43 was transferred into a pASG-IBA3 plasmid, adding a C-terminal strep affinity sequence to the protein.
Ntr1(1–120) was amplified from genomic yeast DNA using the oligonucleotides Ntr1-for and Ntr1(120)-rev and inserted into a pENTRY-IBA10 vector following the IBA StarGate manual. Subsequently, the ORF of Ntr1(1–120) was transferred into a pPSG-IBA5 plasmid, adding an N-terminal strep affinity sequence to the protein.
Ntr1(51–110) was amplified from genomic yeast DNA using the oligonucleotides Ntr1(51)-for and Ntr1(110)-rev and inserted into a pENTRY-IBA10 vector following the IBA StarGate manual. Subsequently, the ORF of Ntr1(51–110) was transferred into a pASG-IBA25 plasmid, adding an N-terminal glutathione S-transferase (GST) affinity sequence to the protein.
For the cloning of expression plasmids for the fusion proteins, the sequences of Prp43 and Ntr1-fragments were amplified from genomic yeast DNA. (Parts of) the sequences of Prp43 and Ntr1 were amplified for the following vectors: pETM-13-Prp43-Ntr1(51–110) (oligonucleotides Prp43-IF-for, Prp43-IF-51-rev, Ntr1-51-FL-for and Ntr1-110-rev), pETM-13-Prp43(1–745)-Ntr1(51–110) (oligonucleotides Prp43-IF-for, Prp43-745-IF-51-rev, Ntr1-51-745-for and Ntr1-110-rev), pETM13-Prp43-Ntr1(51–102) (oligonucleotides Prp43-IF-for, Prp43-IF-51-rev, Ntr1-51-FL-for and Ntr1-102-rev), pETM13-Prp43-Ntr1(51–99) (oligonucleotides Prp43-IF-for, Prp43-IF-51-rev, Ntr1-51-FL-for and Ntr1-99-rev), pETM13-Prp43-Ntr1(59–110) (oligonucleotides Prp43-IF-for, Prp43-IF-59-rev, Ntr1-59-FL-for and Ntr1-110-rev) and pETM13-Prp43-Ntr1(62–110) (oligonucleotides Prp43-IF-for, Prp43-IF-62-rev, Ntr1-62-FL-for and Ntr1-110-rev). The respective fragments were transferred into a NcoI/SalI (Fermentas)-digested pETM-13 expression vector using the In-Fusion technique (Clontech) and following the user manual (version October 2011), and a C-terminal Strep affinity sequence was included in the sequence of the reverse oligonucleotides.
Site-directed mutagenesis of the plasmid pETM-13_Prp43-Ntr1(51–110) was performed by plasmid amplification via polymerase chain reaction using the primer pairs Prp43_K418E-for/Prp43_K418E-rev, Prp43_Y595A-for/Prp43_Y595A-rev, Prp43_Y610A_S614A-for/Prp43_Y610A_S614A-rev, Prp43_Y630A-for/Prp43_Y630A-rev and Ntr1_K67E-for/Ntr1_K67E-rev, respectively.
Protein expression and purification
The plasmid pASG-IBA3-Prp43 was transformed into Rosetta2 (DE3) cells (Novagen). Expression cultures of ten 2-L flasks containing 500 ml 2YT media were grown at 37°C at 220 rpm until an OD600 of 0.6 was reached. The expression of Prp43-Strep was induced by adding 200 µg anhydrotetracyclin per litre of expression culture, and the cell cultures were incubated at 16°C and at 220 rpm overnight.
Cells were harvested by centrifugation at 4000 × g for 15 min at 4°C. The cells were resuspended in buffer A (400 mM NaCl, 50 mM Tris/HCl pH 7.5, 2 mM MgCl2) and were disrupted using a microfluidizer (Microfluidics). The soluble fraction was separated from the unsoluble fraction by centrifugation at 30 000 × g for 45 min at 4°C.
Prp43-Strep was bound to a StrepTactin HP Sepharose column (GE Healthcare) and eluted by buffer A supplemented with 2 mM d-desthiobiotin. Prp43-Strep was purified to homogeneity using a Superdex 200 (26/60) column (GE Healthcare) in buffer S (100 mM NaCl, 10 mM Tris pH 7.5, 2 mM MgCl2).
The Prp43-Ntr1 fusion proteins were expressed, harvested and purified in the same way as described for Prp43, with the only difference that the protein expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM to the expression culture instead of anhydrotetracyclin.
Strep-Ntr1(1–120) and GST-Ntr1(51–110) were expressed and harvested as described for Prp43, with the differences that the expression of Ntr1(1–120) was induced by adding IPTG to a final concentration of 1 mM to the expression culture and that the expression cultures were incubated after induction at 30°C at 220 rpm for 5 h for both Ntr1 truncations.
Strep-Ntr1(1–120) was isolated from the supernatant using a StrepTactin HP Sepharose column in buffer A. The elution fractions were desalted into buffer B (50 mM NaCl, 10 mM Tris/HCl, pH 7.5, 2 mM MgCl2) using a HiPrep Desalting 26/10 column (GE Healthcare) and bound to a Source 30 Q column (GE Healthcare), from which the pure protein was eluted by a gradient of buffer C (600 mM NaCl, 100 mM Tris/HCl pH 7.5, 2 mM MgCl2).
GST-Ntr1(51–110) was isolated from the soluble fraction using a GSH Sepharose 15-ml column (GE Healthcare), from which it was eluted by buffer A supplemented with 30 mM reduced glutathione. The GST affinity sequence was removed by incubation with 0.01 mg PreScission Protease (GE Healthcare) per 1 mg protein. Ntr1(51–110) was separated from the GST by performing a size exclusion chromatography using a Superdex 75 (26/60) column (GE Healthcare) in buffer S. For all protein samples, the A260/A280 ratio was determined after the last purification step to analyse any potential nucleic acid contamination. During establishing of the purification protocols, it was observed that an additional final Source 30 Q column purification step neither changed the A260/A280 nor the ATPase activities, showing that no nucleic acid was present in the samples.
Complex formation
The complexes were formed by incubation of Prp43 with a 5-fold molecular excess with the Ntr1-fragment and further purified by a S200 size exclusion chromatography in buffer S. For the protein–protein cross-linking and the hydroxyl radical experiments, the complexes were transferred into buffer M (100 mM NaCl, 10 mM sodium phosphate buffer, pH 7.4, 2 mM MgCl2) using a HiPrep Desalting 26/10 column (GE Healthcare).
Determination of the ATPase activity
The ATPase assay was performed as described previously (37), with the difference that 5 µM of A20 RNA was used and that the measurements were performed in buffer S.
Determination of the helicase activity
The unwinding assay was performed by fluorescence spectroscopy as described previously (41). For the strand annealing, 4.5 µM of the two RNA strands (5′-GCGCCUACGGAGCUGGUGGCGUAGGCGCAAAAAAAAAAAAAAAAAAAA-3′ and 5′-(Cy5)-GCGCCUACGCCACCAGCUCCGUAGGCGC-(BBQ)-3′, IBA GmbH) was supplemented with 1 µl 100 mM Tris, pH 8.0, incubated at 80°C for 5 min and cooled down to 20°C with a step size of 3°C/min. Subsequently, the sample was diluted to a 1 µM double-stranded RNA (dsRNA) solution.
The 150 µl reaction mixtures contained 20 nM protein, 50 nM dsRNA, 10 mM Tris, pH 7.5, 100 mM sodium chloride and 2 mM MgCl2. The samples were incubated at 20°C in the fluorescence spectrometer Fluoromax III (Horiba Jobin Yvon) for 15 min, and the measurements were started by adding ATP to a final concentration of 1 mM. The excitation wavelength was set to 643 nm, and the fluorescence at 667 nm was measured every 2 s during 10 min. For the determination of the maximal reaction velocity (vmax), the initial slope v0 (ΔF/min) of the fluorescence was determined three times independently for each sample. The amount of unwound dsRNA was calculated with respect to the signal at 667 nm for the annealed and completely unwound dsRNA, respectively.
Determination of dissociation constants
For the determination of dissociation constants, 1 µM Prp43 was incubated with the Ntr1-fragment in concentrations between 0.1 and 12 µM. Each sample was incubated for 2 h, and subsequently the ATP hydrolysis rate at 100 mM ATP at 20°C was determined. For each concentration, three independent measurements were performed. The ATPase activity was plotted against the concentration of the Ntr1-fragment and fitted to a hyperbola using SigmaPlot 11.0 (Systat Software). The Kd was directly deduced from the equation of the hyperbola fit.
Far-UV circular dichroism spectroscopy
The protein sample was desalted in buffer CD (100 mM sodium phosphate, pH 7.5) and diluted to 0.1 mg/ml. Three hundred microlitres of the sample was pipetted into a cuvet with a path length of 1 mm and transferred into a Chirascan CD spectrometer (AppliedPhotophysics). The spectra were recorded in far-ultraviolet (UV) range from 195–260 nm at 20°C with a time-per-point value of 1 s and a path length of 1 nm. Every measurement was performed for five times independently. For the final absorption curve, the values of the negative control, which contained exclusively the sample buffer, were subtracted from the values of the protein sample. Trifluorethanol (TFE) was added in concentrations between 10 and 40% (v/v) to the samples.
Secondary structure prediction
The IUPRED server was used to predict disordered regions in proteins by the calculation of unfavourable estimated energies (42). The ANCHOR server combines the disorder tendency predicted by IUPRED with the sensitivity to structural environment, and thus identifies protein regions that might form defined secondary structure elements when binding to a globular protein (43).
Hydroxyl radical probing and digestion of peptides
Proteins were probed by chemically generated hydroxyl radicals, as described in (44). Seventy microlitres of a 50-µM protein complex solution was added to a reaction tube containing 10 µl of 13 mM Fe-ethylenediaminetetraacetic acid (Sigma-Aldrich; Merck, respectively) solution and 10 µl of 0.2 M sodium ascorbate (Sigma-Aldrich) solution, both in 50 mM Na2HPO4 buffer, pH 6.5. For control purposes, a 17 µl aliquot was transferred to 40 µl quenching buffer (2 M Tris, pH 7.2, Prolabo). Finally, 10 µl of 3% v/v H202 (Sigma-Aldrich) was added to the Fenton mixture to start the reaction. At 10, 60, 150 and 300 s, 17 µl aliquots were removed and transferred to new microcentrifuge tubes containing 40 µl quenching buffer.
Protein pellets were dissolved and denatured in a 50 µl acetonitrile:buffer (100 mM ammonium bicarbonate) mixture (80:20), by intense vortexing, followed by a 10-min incubation in a thermomixer at 25°C and 500 rpm. Fifty microlitres of a 2-mM dithiothreitol (DTT) solution in 80% acetonitrile (ACN) was added. The sample was incubated for 30 min at 25°C and 500 rpm. To alkylate cysteine residues, 50 µl of a 12-mM iodoacetamide solution in 80% ACN was added and incubated for further 30 min at 25°C and 500 rpm. Samples were then predigested with Lys-C (Roche Diagnostics GmbH), added in a 1/20 w/w enzyme to protein ratio and incubated for 3 h at 25°C and 500 rpm. Trypsin (Promega) was added in a 1/20 w/w enzyme to protein ratio, and the sample incubated overnight under the same conditions. Samples were dried in a vacuum concentrator and stored at −20°C until posterior use (45).
UV-induced cross-linking of γ-[32P]-ATP-labelled RNA and Prp43/Ntr1 complex
The single-stranded RNA oligonucleotide (5′-GACGGGCUGUUGGUGAUGGAC-3′, Roche Kulmbach GmbH) was radiolabelled with γ-[32P]-ATP (6000 Ci/mmol) and 1 µL T4 polynucleotide kinase following standard procedures. The resulting labelled RNA was incubated for 30 min on ice with ∼50-fold molar excess of Prp43/Ntr1(1-120). The resulting protein–RNA complex divided into 6.8 µl aliquots and irradiated at 254 nm for 0, 1, 2, 5, 10 and 12 min, using an in-house constructed cross-linking device, including 4 8 W lamps (F8T5BL Sankyo Denki). The cross-linked samples were analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and the resulting gel was coomassie-stained and bands containing radiolabelled RNA were visualized with a STORM phosphoimager (GE Healthcare).
UV-induced protein–RNA cross-linking and enrichment of cross-linked peptides
UV cross-linking and enrichment were performed as described in (46). Briefly, 1 nmol of the single-stranded RNA oligonucleotide 5′-GACGGGCUGUUGGUGAUGGAC-3′ was incubated in a 1:1 molar ratio with either Prp43/Ntr1 or Prp43 on ice for 30 min. The resulting mixture was taken to 100 µl in 10 mM Tris, pH 7.5, 100 mM NaCl, 2 mM MgCl2 buffer, transferred to black polypropylene microplates (Greiner Bio-One) and irradiated at 254 nm for 10 min. Samples were ethanol precipitated, denatured in 4 M urea, 50 mM Tris HCl, pH 7.9, and digested for 2 h at 52°C with 1 µg RNase A and 1 U RNase T1 (Ambion, Applied Biosystems). RNA digestion was followed by overnight trypsin proteolysis at 37°C. Samples were desalted on a C18 (Dr Maisch GmbH) column. Enrichment of cross-linked peptides was performed on a TiO2 (GL Sciences) column. Both columns were prepared in-house, and the enrichment was achieved essentially as in the procedure described in (46); after evacuation to dryness, the sample was dissolved in 200 mg/ml 2,5-dihydroxy benzoic acid (Sigma–Aldrich) in 80% ACN containing 5% trifluoroacetic acid; it was then loaded on the TiO2 column, washed with 200 mg/mL dihydroxy benzoic acid in 80% ACN, 5% trifluoroacetic acid and eluted with 0.3 M ammonia.
Protein–protein cross-linking
The optimal cross-linker to protein ratio was determined by using 2.5 µg aliquots of the Prp43/Ntr1 complex and a series of cross-linker molar excesses of 5, 10, 25, 50, 100 and 200 as well as a control. Samples were allowed to react with freshly prepared BS3 for 30 min at room temperature. The chosen cross-linker to protein ratio was 30, which led to a high yield, but to a homogeneous sharp band as visualized on SDS-PAGE. The cross-linked samples were analysed by SDS page on a 4–12% Bis-Tris gel (Invitrogen) with MES as running buffer and stained with coomassie blue.
For MS analysis, both Prp43/Ntr1(1–120) and Prp43/Ntr1(51–110) were cross-linked with freshly prepared BS3 and BS2G (Pierce, Thermo Scientific) in a 20:1 and 30:1 cross-linker to protein ratio, allowed to react and separated as described earlier. Cross-linked bands were excised from the gel and combined in a single reaction cup according to cross-linking reagent. In-gel digestion and extraction of peptides was achieved as described elsewhere (47). The solution of extracted peptides was concentrated on a vacuum evaporator to 5 µl and diluted to a final volume of 10 µl to reach a sample solvent composition of 5% v/v ACN and 1% v/v FA. Samples were submitted for immediate analysis to the mass spectrometer.
Nano-liquid chromatography and mass spectrometric methods
For liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses, hydroxyl radical footprinting and UV-induced cross-linking samples were dissolved in 30 and 10 µl sample solvent (5% v/v ACN, 1% v/v FA), respectively. Five microlitres was injected onto a nano-liquid chromatography system (Agilent 1100 series, Agilent Technologies) including an ∼2-cm-long, 150-μm inner diameter C18 trapping column in-line with an ∼15-cm-long, 75-μm inner diameter C18 analytical column (both packed in-house, C18 AQ 120 Å 5 μm, Dr Maisch GmbH). Peptides were loaded on the trapping column at a flow rate of 10 μL/min in buffer A (0.1% FA in H2O, v/v) and subsequently eluted and separated on the analytical column with a gradient of 7–38% buffer B (95% acetonitrile, 0.1% FA in H2O, v/v) with an elution time of 33 min (0.87%/min) and a flow rate of 300 nl/min.
Online electrospray mass spectrometry was performed with an LTQ-Orbitrap Velos instrument (Thermo Scientific), operated in data-dependant mode using a TOP10 method. MS scans were recorded in the range of 350–1600 m/z. The 10 most intense ions were selected for subsequent MS/MS. Both precursor ions and well as fragment ions were scanned in the Orbitrap. Fragment ions were generated by HCD activation (higher energy collision dissociation, normalized collision energy = 40), and recorded from m/z = 100. As precursor ions as well as fragment ions were scanned in the Orbitrap, and the resulting spectra were measured with high accuracy (<5 ppm), both in the MS and MS-MS levels.
Both protein–protein and protein–RNA cross-linking samples were submitted to LC-MS analysis immediately after their preparation. Nano-LC chromatography was performed as described earlier, except for the gradient, which extended over 97 min. Mass spectrometric analysis of protein–protein cross-linked peptides was performed in the same instrument, with a TOP8 method in data-dependent acquisition mode. MS1 ions were recorded in the range of 300–1800 m/z at 60 000 resolution. Fragmentation was generated by collision induced activation and only precursor ions of charge state ≥3 were selected for fragmentation. Fragment ions were acquired in the Orbitrap at 7500 resolution. Dynamic exclusion was set at 30 s with a repeat count of 1.
Data analysis
Hydroxyl radical footprinting
Data analysis was performed using MaxQuant 1.1.1.14 (48). Database search was performed against Prp43 and Ntr1(1–120) sequences, with Andromeda considering 14 variable modifications: hydroxylation (+16 Da) in Hys, Val, Met, Leu, Tyr, Arg, Pro, Ile, Phe; dioxidation (+32 Da) of Met, Tyr, Phe; deguanidination (−43 Da) in Arg and ring-opening (−22) in His (49). By making use of extracted ion chromatograms, obtained with the label-free algorithm from MaxQuant, the ratios of oxidized to non-oxidized peptides in which a specific residue is involved were determined, and thus the dose–response plots for modified peptides were obtained (50).
UV-induced protein–RNA cross-linking
Protein–RNA cross-linking experiments were analysed with OpenMS (51,52) and OMSSA (53) as search engine. Data analysis workflows were assembled especially for our laboratory by Timo Sachsenberg (Prof. Oliver Kohlbacher, Applied Bioinformatics Group, Eberhard Karls University, Tübingen), based on (46) and a manuscript in preparation. MS.raw files were converted into the.mzML format with msconvert (54). High-scoring candidate spectra were manually annotated to completion.
Protein–protein cross-linking
Data analysis was performed with MassMatrix as described in their publications and manuals (55,56). Thermo Scientific raw files were converted to the mzxmL data format with MMConverter and submitted to database search with the following parameters: peptide length between 6 and 40 amino acid long, 10 ppm MS1 tolerance and 0.02 Da MS2 tolerance. Oxidation in methionine was set as a variable modification, whereas carbamidomethylation of cysteine was set as a fixed modification. Spectra were searched against a FASTA file composed of the sequences of Prp43 and Ntr1(1–120) and a reversed decoy database. The resulting matches were validated manually.
RESULTS
The G-patch motif of Ntr1 is intrinsically unstructured
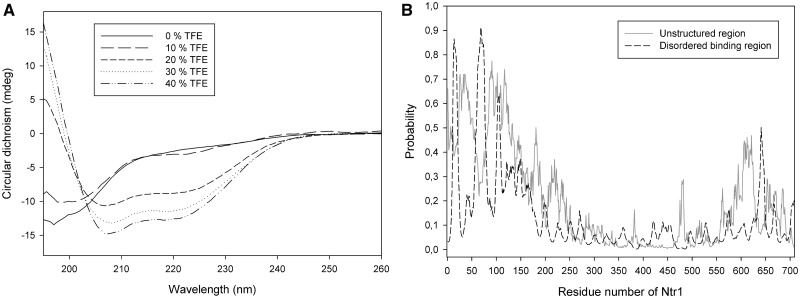
Far-UV circular dichroism spectroscopy (CD) was used to analyse the content of secondary structure elements of truncated Ntr1(51–110). The CD-spectrum of this Ntr1-fragment indicated the absence of any secondary structure elements (Figure 1A). The capacity of evolving secondary structure elements was investigated by determination of far-UV CD spectra in presence of the small molecule TFE at different concentrations. TFE is known to support the formation of alpha-helices (57). For concentrations of ≥20% (v/v) of TFE, the CD spectrum of Ntr1(51–110) changes significantly and gives rise to two local minima at 208 and 222 nm, indicating the presence of alpha-helices (Figure 1A).
Figure 1.
Secondary structure analysis of Ntr1. (A) CD spectra of Ntr1(51–110) at different concentrations (0–40% v/v) of TFE. The circular dichroism (y-axis) is shown with respect to the wavelength (x-axis). (B) Prediction of the presence of disordered binding regions (dashed line) and unstructured regions (continuous line) for full-length Ntr1. The probability to be unstructured or a disordered binding region, respectively (y-axis), is shown with respect to the residue number (x-axis).
Hence, the CD spectra suggest that Ntr1(51–110) is natively unstructured, and that it has the capacity to form alpha-helices. The signal for beta-strands in a CD spectrum is detected by a decrease in signal between 200 and 210 nm and is equal to the signal for an alpha-helix in this range, and the presence of a beta-strand in yNtr1(51–110) cannot be proved in the measured CD spectra.
To learn whether the described potential secondary structure elements could be evolved on binding of Ntr1(51–110) to Prp43, an additional bioinformatical approach using ANCHOR and IUPRED (42,43) was chosen. Although the predicted disorder probability is high for the 150 N-terminal residues, it is on a low level for the residues 151–600 and increases again for the last 100 C-terminal residues (Figure 1B). Three regions are predicted to be disordered binding regions: residues 11–20, 60–77 and 102–107; of these, the second and third are part of the minimal required activating fragment Ntr1(51–110). Residues 60–77 overlap with a region predicted to form alpha-helices—interestingly, this region is predicted to be unstructured at a probability of 0.3, which is lower compared with the residues 1–150, but still quite high compared with the region between 150 and 600.
Combining the experimental data collected by far-UV CD spectroscopy with the bioinformatical predictions, the following structural model for the G-patch motif of Ntr1 can be outlined: the residues forming the G-patch of Ntr1 are intrinsically unstructured, but they have characteristics indicating a tendency to form secondary structure elements, namely, alpha-helices and beta-strands. The formation of these secondary structure elements could be induced on binding to the globular interaction partner, the spliceosomal helicase Prp43. Regarding full-length Ntr1, the G-patch containing N-terminal 150 residues and the C-terminal 100 residues are predicted to be unstructured, whereas the central part of the protein (residues 150–600) seems to form a structured region.
Ntr1(51–110) is the minimal functional fragment required for the activation of Prp43
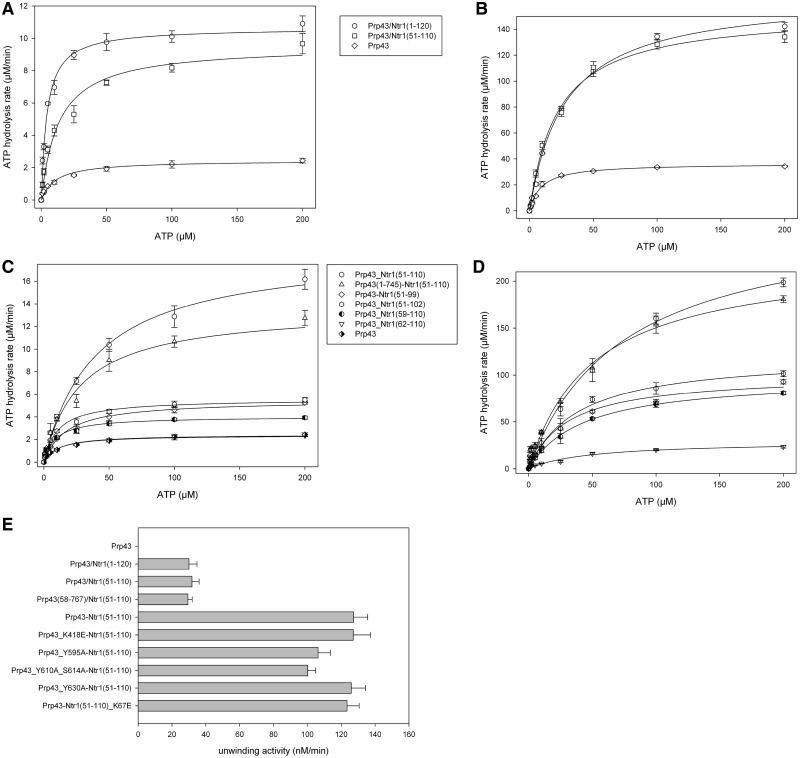
Prp43 and Ntr1(1–120), an Ntr1-fragment that has been reported as sufficient for the stimulation of the helicase activity of Prp43 (31), were expressed and purified separately. The complex of Prp43 and Ntr1(1–120) was formed by mixing both proteins and further purified via a size exclusion chromatography (Supplementary Figure S2A and B). By expressing, purifying and characterizing different N-terminal fragments of Ntr1, it turned out that the 60 amino acids containing peptide Ntr1(51–110) were still able to bind to Prp43 (Supplementary Figure S2C and D) and were sufficient to stimulate the ATPase and helicase activity of Prp43. Furthermore, it could be shown that both Ntr1 truncations were able to stimulate the ATPase activity of Prp43, even in absence of RNA (Figure 2A, B and E and Tables 1 and 2). Although the maximal ATP hydrolysis rate of Prp43 was 2.43 ± 0.12 µM/min in absence and 36.47 ± 0.51 µM/min in presence of RNA, it was stimulated to a similar extent in complex with Ntr1(1–120) (11.18 ± 0.34 µM/min and 166.48 ± 2.38 µM/min, respectively) and Ntr1(51–110) (9.54 ± 0.37 µM/min and 153.40 ± 3.08 µM/min, respectively).
Figure 2.
ATPase and helicase activity of different Prp43/Ntr1 complexes and fusion proteins. (A and B) ATPase activity of Prp43, Prp43/Ntr1(51–110) and Prp43/Ntr1(1–120) in absence (A) and presence of A20-RNA (B). The ATP hydrolysis rate (y-axis) is shown with respect to the ATP concentration (x-axis). (C and D) ATPase activity for different Prp43-Ntr1 fusion proteins in absence (A) and presence (B) of A20-RNA. (E) Helicase activity of Prp43, Prp43/Ntr1 complexes and the Prp43-Ntr1(51–110) fusion protein. The maximal unwinding velocity is shown.
Table 1.
ATPase activities of Prp43 and the complexes Prp43/Ntr1(1–120) and Ntr1(51–110)
| Protein | In absence of RNA |
In presence of A20-RNA |
||
|---|---|---|---|---|
| KM (µM) | vmax (µM/min) | KM (µM) | vmax (µM/min) | |
| Prp43 | 11.07 ± 2.07 | 2.43 ± 0.12 | 9.23 ± 0.53 | 36.47 ± 0.51 |
| Prp43/Ntr1(1–120) | 16.37 ± 1.81 | 11.18 ± 0.34 | 27.95 ± 1.25 | 166.48 ± 2.38 |
| Prp43/Ntr1(51–110) | 13.42 ± 1.93 | 9.54 ± 0.37 | 22.48 ± 1.50 | 153.40 ± 3.08 |
Table 2.
Unwinding activities for Prp43, Prp43/Ntr1 complexes and Prp43-Ntr1(51–110) fusion proteins
| Protein | vmax (nM/min) |
|---|---|
| Prp43 | n.d. |
| Prp43/Ntr1(1–120) | 30.0 ± 4.8 |
| Prp43/Ntr1(51–110) | 31.8 ± 4.2 |
| Prp43(58–767)/Ntr1(51–110) | 29.4 ± 2.4 |
| Prp43-Ntr1(51–110) | 127.2 ± 8.4 |
| Prp43_K418E-Ntr1(51–110) | 127.1 ± 10.3 |
| Prp43-Y595A-Ntr1(51–110) | 106.3 ± 7.3 |
| Prp43_Y610A_S614A-Ntr1(51–110) | 100.2 ± 4.7 |
| Prp43_Y630A-Ntr1(51–110) | 125.9 ± 8.4 |
| Prp43-Ntr1(51–110)_K67E | 123.4 ± 7.2 |
As the expression and purification of soluble shorter fragments of Ntr1 was not feasible, they were fused directly to the C-terminus of Prp43. Interestingly, the fusion of Prp43 and Ntr1(51–110) [referred to as Prp43-Ntr1(51–110)] behaves like the functional complex of both proteins, as it exhibits a maximal ATP hydrolysis rate, which is about twice as high compared with the complex of both proteins (18.79 ± 0.66 µM/min). In the size exclusion chromatography, the fusion protein Prp43-Ntr1(51-110) eluted as a monomer, indicating an intramolecularinteraction of the G-patch motif and the helicase molecule of the the fusion protein. (Supplementary Figure S3). Comparison of the ATPase activity revealed similar ATPase activities of the fusion proteins Prp43-Ntr1(1–120), Prp43-Ntr1(51–120) and Prp43-Ntr1(51–110). Furthermore, the ATPase activity of the fusion protein Prp43-Ntr1(51–110) turned out to be about twice as high as of the prepared complex Prp43/Ntr1(51–110), in the absence as well as in the presence of A20-RNA. By further decreasing the size of the Ntr1-fragment fused to Prp43, the ATPase activity decreased dramatically in both cases (Figure 2C and D, Table 3). To exclude that this effect was due to a too short linker, the ATPase activity of the fusion protein Prp43(1–745)-Ntr1(51–110) was determined, which resulted in an activity significantly higher than for the fusion proteins with smaller Ntr1 fragments. The fusion protein Prp43(1–745)-Ntr1(51–110), which lacks the 22 flexible C-terminal residues of Prp43, showed comparable activities, even when a shorter linker was used.
Table 3.
ATPase activities of different Prp43-Ntr1 fusion proteins
| Protein | In absence of RNA |
In presence of A20-RNA |
||
|---|---|---|---|---|
| KM (µM) | vmax (µM/min) | KM (µM) | vmax (µM/min) | |
| Prp43-Ntr1(51–110) | 39.53 ± 3.92 | 18.79 ± 0.66 | 73.43 ± 12.17 | 271.90 ± 19.45 |
| Prp43(1–745)-Ntr1(51–110) | 28.33 ± 4.61 | 13.59 ± 0.87 | 50.18 ± 9.50 | 225.92 ± 16.29 |
| Prp43-Ntr1(51–102) | 17.50 ± 2.43 | 5.50 ± 0.22 | 38.74 ± 5.29 | 121.33 ± 5.82 |
| Prp43-Ntr1(51–99) | 10.68 ± 1.38 | 5.60 ± 0.18 | 30.68 ± 6.16 | 100.56 ± 6.61 |
| Prp43-Ntr1(59–110) | 8.77 ± 1.09 | 4.02 ± 0.12 | 43.06 ± 2.36 | 98.10 ± 1.95 |
| Prp43-Ntr1(62–110) | 10.62 ± 2.18 | 2.38 ± 0.12 | 48.07 ± 11.49 | 29.35 ± 2.64 |
Using the stimulatory effect on the ATPase activity of Prp43 on Ntr1 binding, an approximate determination of the binding constants of Ntr1(1–120) and Ntr1(51–110) to Prp43 under steady-state turnover conditions was possible. Therefore, the ATPase activity of Prp43 was measured in the presence of different concentrations of the truncated Ntr1 variant (Supplementary Figure S4A and B). The dissociation constant of the Prp43/Ntr1(1–120) complex (1.49 ± 0.05 µM) turned out to be slightly lower than that of the Prp43/Ntr1(51–110) complex (2.24 ± 0.11 µM). This indicates that residues within the first 50 residues of Ntr1 are important for binding to Prp43, but not for the activation of the helicase.
Taken together, Ntr1(51–110) is sufficient for the full activation of Prp43. Furthermore, both Ntr1(1–120) and Ntr1(51–110) stimulate not only the helicase, but also the ATPase activity of Prp43.
Protein–protein cross-linking indicates that Ntr1 interacts with the C-terminal domains of Prp43
In the absence of high-resolution 3D data derived from crystallization and/or nuclear magnetic resonance experiments, mass-spectrometric (MS) approaches can provide information about the spatial rearrangement of protein–protein and protein–ligand complexes. We set out to gain structural information on how Ntr1 and Prp43 interact by using cross-linking and subsequent analysis of the cross-linked protein regions by mass spectrometry. We studied the binary complex by chemical cross-linking with BS3 and BS2G, which link primary amines, in this case lysine residues, with an 11.4 Å and a 7.4 Å spacer arm, respectively. The cross-linked complexes were separated by SDS-PAGE and cross-linked peptides were identified by MS after in-gel digestion.
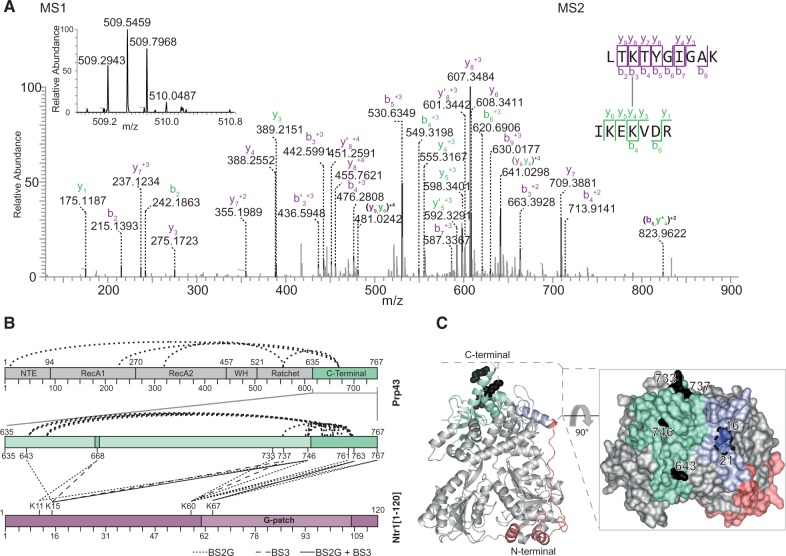
In total, 42 intramolecular and 16 interprotein cross-links were identified (Figure 3B and Supplementary Table S3). The intramolecular cross-links located within the crystal structure of Prp43 were well within the maximum distance that the cross-linkers can span, thus providing a positive control for the method (Supplementary Figure S5). It is important to point out that cross-linked residues do not directly contact each other, but provide information about proximal locations.
Figure 3.
Protein–protein cross-linking map and localization on the C-terminal domain of Prp43. (A) MS1 and MS2 spectra of cross-linked K60 in LTKTYGIGAK [Ntr1(1–120)] to K746 in IKEKVDR (Prp43). MS1 spectrum shows the isotopic envelope of the quadruply charged cross-linked peptide pair. The MS2 spectrum, zoomed in the region of 0–900 m/z for clarity, depicts the identified peptide fragments. (B) Cross-linking map depicts both intramolecular and intermolecular cross-links identified in Prp43-Ntr1(1–120). Dark green areas indicate regions not solved in the crystal structure. Both BS3 and BS2G cross-links are represented, with a full line for a cross-link identified in both, and differently dashed lines for those cross-links identified only with one particular cross-linker. Both cross-linkers, despite not fully overlapping, aim at the same cross-linked regions: the very C-terminal region of Prp43 and the beginning of the G-patch motif of Ntr1. (C) Localization of Prp43 cross-linked residues, and rotated surface representation. Residues cross-linked to Ntr1 are depicted in black, and the residues in the N-terminal domain determined to be protected in the footprinting studies are in blue.
Notably, all of the interprotein cross-links between Prp43 and Ntr1(1–20) are located within the C-terminal OB-fold domain of Prp43 (635–767, Figure 3B), whereas the corresponding cross-linked sites within Ntr1(1–120) are more widely distributed throughout the protein’s sequence and also include K67 within the G-patch motif (Figure 3B and Supplementary Tables S3 and S4). Many of the cross-links at the C-terminal region of Prp43 involve lysine residues located close to the C-terminus of Prp43 (residues 746–767), a region that is not seen in the crystal structure and is thought to be very flexible (shaded in Figure 3C).
Interestingly, K668, which lies in a loop connecting two beta-strands of the OB-fold domain of Prp43 and is not defined in the crystal structure, appears to be an important cross-linking hub. This particular position is cross-linked to residues K11 and K15 of Ntr1 (Figure 3B), indicating that this residue is located close to the Ntr1 interaction site and is flexible. The cross-linked residues within the minimal interaction domain of Ntr1 (1–120) also exhibit a particular feature: K67 in the G-patch motif cross-links exclusively to residues K763 and K767 within the region closest to the C-terminus of Prp43. Importantly, K60 of Ntr1, which lies close to the G-patch motif, cross-links to a wider area in the C-terminal part of Prp43 (733–767, Figure 3B), encompassing residues that are present and others that are not defined in the crystal structure of Prp43. Residues K11 and K15, located outside the G-patch region, cross-link to the 643–668 region and to others as far away as 733–746 (Figure 3B). These cross-links indicate that the residues 1–50 of Ntr1, although they are not involved in the activation, might also bind to Prp43, thereby leading to the observed higher binding affinity of Ntr1(1–120) to Prp43 compared with Ntr1(51–110).
These cross-linking results on Ntr1(1–120) in complex with Prp43 could be confirmed by cross-linking experiments on the minimal functional fragment of Ntr1 (51–110). Not unexpectedly, a similar cross-linking pattern was obtained: K60 and K67 of Ntr1 were cross-linked to the region of Prp43 closest to the C-terminus (Supplementary Figure S6 and Supplementary Table S4). Most of the observed cross-links originate from K60 of Ntr1, joining it to lysine residues between amino acids 733–763 of Prp43. In addition, we identified a cross-link between K67 in Ntr1 and K766 of Prp43, as well as an additional contact between K78 in the G-patch motif of Ntr1 and K4 in the NTE of Prp43. It is of note that K4 is also part of a flexible part of the helicase: it is one of the 19 amino acid residues encompassing the N-terminal tail, which has been shown to be in proximity to the C-terminal domain of Prp43 (PDB-ID 2xau).
A representation of the cross-linked residues on the 3D structure of Prp43 (Figure 3C, Supplementary Figure S8) reveals that they are located on a small surface region of the helicase on the C-terminal OB-fold domain. Most of the residues involved belong to the last 35 residues that are flexible and not required in vivo (58). As mentioned earlier, the identified residues indicate proximal contacts and not direct interactions, and they strongly indicate that the G-patch motif interacts with the three C-terminal domains of Prp43, namely, the Winged Helix (462–525), the Ratchet (521–634) and the OB-fold domain (635–767).
Binding of Ntr1 induces domain movements in Prp43
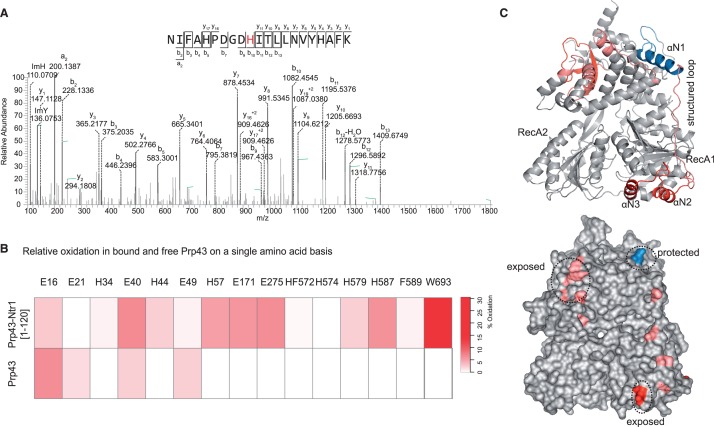
Having identified the C-terminal domains of Prp43 as a major interaction site of Ntr1, we next asked whether the binding of Ntr1 to Prp43 might induce changes in the conformation of Prp43 that could lead to the enhanced RNA unwinding activity (33). Monitoring solvent accessibility changes of residues of Prp43 that are induced by Ntr1 binding can help identify changes in the overall conformation. Hydroxyl radical probing of proteins yields information on relative solvent accessibility, as OH radicals can covalently modify side chains of amino acid residues (40). Therefore, we probed Prp43 alone and the complex Prp43/Ntr1(1–120) with hydroxyl radicals generated by the Fenton reaction in a time-dependent manner. Proteins were hydrolysed with the endoproteinase trypsin, and the modified peptides and amino acids were identified by mass spectrometry. By quantitatively comparing the rates of oxidation of a particular peptide in two states, i.e. bound and unbound, information about the relative solvent accessibility of the two states can be obtained. Particular residues at which the modification occurred can be identified and quantified by tandem mass spectrometry (Supplementary Figure S1A).
In total, 44 peptides in Prp43 were identified and could be quantified, of which 8 peptides showed differences in their levels of oxidation when Ntr1(1–120) was bound to Prp43 (Supplementary Figure S7). These differences in modification levels concern different regions on Prp43, and several residues of the N-terminal extension (NTE) of Prp43 showed differences on binding of Ntr1(1–120). The residue E16 was less strongly oxidized (modified to 6.1 ± 1.9% in Prp43, and only to 2.7 ± 0.9% in the complex), which could have been due either to direct binding and thus protection by Ntr1(1–120), or to a movement ‘into’ the C-terminal domain and thus away from the surface of the protein. Additionally, residues E40, H44 and H57 of the NTE of Prp43 showed a higher oxidation level on the binding of Ntr1(1–120), which indicates that they changed their position during complex formation, probably in co-ordinated movement with the αN1 helix (Figure 4B).
Figure 4.
Relative solvent accessibility determined by hydroxyl radical footprinting. (A) MS2 spectra of modified NIFAHPDGDHITLLNVAYHAFK. Histidine 587 in Prp43 exhibits a typical hydroxyl radical-induced modification, which results in ring-opening and a mass difference of −22. (B) Heatmap shows the residue-based relative quantification of oxidation in Prp43 and Prp43 bound to Ntr1. Glutamic acids in positions 16 and 21 exhibit lower levels of oxidation in the free Prp43 as compared with the bound Prp43, which indicates protection. On the contrary, the remaining residues are more exposed in the complex, with regions in the αN3 and ratchet and C-terminal domains exhibiting the largest levels of oxidation. (C) Domain architecture of Prp43 (PDB-ID 2xau) shows a ring-like structure, with RecA domains in the bottom and C-terminal and Ratchet and winged-helix domains in the upper part. The structure is braced by the N-terminal domain, which connects both ends of the structure with a structured loop. In the bottom, regions that exhibit protection or increased exposure to solvent are indicated.
To take a closer look at the function of the NTE of Prp43, we expressed and purified the variant Prp43(58–767), which lacks the 57 N-terminal amino acids of the helicase. It turned out that this truncation shows no ATPase activity in the absence of Ntr1, but is still able to bind Ntr1(1–120). The highest observed unwinding rate of the Prp43(58–767)/Ntr1(51–110) complex was 29.4 ± 2.4 nM/min, and hence in the same range as for the Prp43/Ntr1(51–110) complex (Figure 2C and Table 2). This result suggests that the observed movement of the N-terminal domain of Prp43 is rather passive in nature and is due to the binding of Ntr1 to the C-terminal domain. As the αN1-helix (19–33) interacts directly with the winged helix domain (457–521), the latter domain is also affected by the rearrangements that occur on binding of Ntr1.
Additionally, differences in surface accessibility could be detected in the C-terminal OB-fold domain, where W693 was not modified in Prp43 alone, but was modified to 90.0 ± 9.5% in the complex, and in the ratchet domain, where H587 was not modified in isolated Prp43, but was modified to 6.0 ± 1.9% in the complex. These changes in oxidation seem to be a result of Ntr1(1–120) binding to the C-terminal domain of Prp43, which induces movements in both domains (Figure 4C).
Taken together, our probing experiments indicate that there are major rearrangements in the OB-fold domain of Prp43 on binding of Ntr1. This is also where we located the binding platform of the G-patch motif on Prp43 according to the protein–protein cross-linking results. Moreover, changes in solvent accessibility are observed for amino acid residues in the NTE of Prp43, and deletion of the NTE affects the intrinsic ATPase activity of Prp43, while additional experiments showed that the NTE of Prp43 is not required for the binding of or the activation by Ntr1.
Protein–RNA cross-linking reveals residue-specific RNA contacts in Prp43 and Ntr1
Ntr1 binding to Prp43 leads to increased unwinding activity, which is why we next set out to investigate the interaction of the helicase with RNA, with and without Ntr1, by UV-induced protein–RNA cross-linking. UV light (254 nm) generates a covalent link between a nucleic acid base and the side chain of an amino acid when they are in proximity. The newly formed covalent bond on the amino-acid residues, a zero-length cross-link, makes it suitable for detection by mass spectrometry, and sequence information about the cross-linked protein region can be obtained (46).
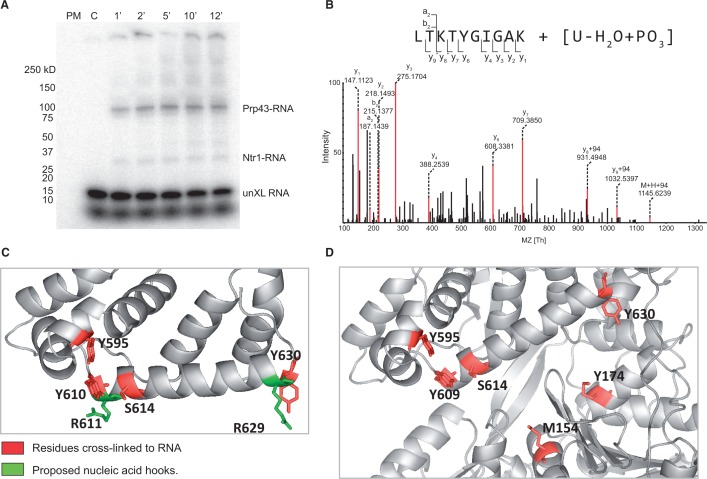
UV-induced cross-linking was initially analysed by SDS-PAGE by using a 21-mer ssRNA labelled with γ-[32P]-ATP. Both Prp43 and Ntr1(1–120) can cross-link to RNA, although Prp43 seems to do so more efficiently (Figure 5A).
Figure 5.
Protein–RNA cross-linking results. (A) SDS-PAGE separation of cross-linked Prp43/Ntr1(1–120) to γ-[32P]-ATP-labelled RNA. Cross-linking was performed at different irradiation times. Both Prp43 and Ntr1 can cross-link RNA even after 1 min of UV irradiation. (B) MS2 spectrum of K60 of Ntr1 cross-linked to U-H2O+PO3. A mass difference of 94 Da corresponding to the uracil nucleobase with a neutral loss of water is observed from y8 onwards, which pinpoints K60 as the cross-linked residue. (C) RNA-cross-linked residues in the long ratchet helix and helix domain. Cross-linked residues in the ratchet domain (red) are localized next to the arginine residues proposed to act as nucleic acid hooks (green). (D) Representation of RNA-cross-linked residues. Residues cross-linked to RNA are oriented towards the nucleic acid binding cavity.
For the identification of the cross-linked protein regions by MS, we digested the cross-linked Prp43–RNA complex with RNAses and trypsin, and enriched for RNA cross-linked peptides by chromatography on TiO2 columns. The mixture was desalted and immediately analysed by MS. Eight different Prp43 peptides cross-linked to RNA were identified. For six of them, the cross-linked amino acid residue could be also identified (Table 4, Supplementary Figure S11). These amino acids are within the RecA-1 (M154, Y174), RecA-2 (K418, Y440) and ratchet (Y595, Y630, Y610, S614) domains, and they are oriented towards the proposed nucleic-acid-binding cavity (36).
Table 4.
Protein RNA cross-links identified in free and bound Prp43
| Domain | Residue | Peptide | RNA | Prp43/Ntr1 | Prp43 | Relative enrichment b/f (%) |
|---|---|---|---|---|---|---|
| RecA-1 | M154 | 151VAAMSVAQR159 | UG | + | + | 33.3 |
| RecA-1 | Y174 | 168LGEEVGYSIR177 | UG | + | + | 24.0 |
| RecA-1 | n.i | 190YM(OX)TDGM(OX)LLREAM(OX)EDHDLSR208 | U | + | + | n.d. |
| RecA-2 | K418 | 408VESLLVSPISKASAQQR424 | U-HPO3 | + | − | 6.3 |
| RecA-2 | Y440 | 439LYTEEAFQK447 | UG | + | + | 3.7 |
| Ratchet | Y595 | 591SDEAYEYGIHK601 | UG | + | + | 132.2 |
| Ratchet | Y630 | 630YNLELNTTDYESPK643 | GU | + | + | 7.3 |
| Ratchet | n.i | 605DHYLNYRSLSAADNIR620 | GU | + | + | n.d |
| Ratchet | Y610 | 605DHYLNYR611 | U | + | + | 339.6 |
| Ratchet | S614 | 612SLSAADNIR620 | UG | + | − | 169.7 |
n.i.: not identified; n.d.: non-determined; (-)cross-link was not sequenced, identification based on MS1; b: bound; f: free. The domain and the cross-linked residue (when identified) are indicated, as well as the RNA oligonucleotide.
To elucidate the functional relevance of residues identified by the UV cross-linking, several mutated variants of the Prp43-Ntr1(51–110) fusion proteins were prepared and analysed. The helicase activity was not affected in case of the K418E and Y630A mutations. In contrast to this, the unwinding activity was significantly decreased in case of the Y595A mutant and the Y610-S614 double mutant, confirming the central role of the long ratchet helix in the unwinding process.
When the peptide–RNA cross-linking results for Prp43–RNA are compared with those for Prp43/Ntr1(1–120)–RNA, no qualitative differences were observed, meaning that the same cross-linking sites to RNA were identified in the presence or absence of Ntr1. Notably, a cross-link of the residue K67 of Ntr1(1–120) to RNA was identified, confirming that this protein is also in direct contact with RNA, in agreement with the SDS-PAGE results. In particular, this shows that the G-patch motif (K67) interacts with RNA. On testing a Prp43-Ntr1(51–110) fusion protein containing the mutation K67E, it turned out that the helicase activity was not significantly affected by this mutation, indicating that this contact is not essential, at least for the unwinding of the used substrate. As the precise target sequence of Prp43 during the disassembly of the intron-lariat spliceosome has not been identified yet, it cannot be ruled out that Ntr1-K67 as well as K418 and Y630 might be essential in vivo.
The two helicase–RNA complexes, with and without Ntr1(1–120), were then compared quantitatively. A remarkable difference was observed in the cross-linking yield of all identified peptide–RNA cross-links, which was found to be more efficient when Prp43 was cross-linked to RNA in the presence of Ntr1. In particular, peptide regions encompassing amino acids 591–601, 605–611 and 612–620 (SDEAYEYGIHK, DHYLNYR and SLSAADNIR, with Y595, Y610 and S614 cross-linked, respectively), which are located in the ratchet helix, were found to be cross-linked with a significantly higher efficiency as compared with Prp43 alone bound to RNA (as calculated from ratio of the intensities of the corresponding non-cross-linked peptide in Prp43 alone and bound to RNA, Table 4). We note that an increase in UV-induced cross-linking yield cannot directly be correlated with RNA affinity at the particular protein region. Nevertheless, it does reflect a shift in equilibrium towards certain conformations that favour RNA cross-linking, in which the RNA is in close spatial proximity with the amino acid residues of this particular domain. Importantly, those regions where we observed a higher cross-linking yield on Ntr1(1–120) binding coincide with those that showed increased solvent accessibility on Ntr1(1–120) binding to Prp43 (Table 4 and Figure 4B). Thus, we propose that the structural rearrangements that occur on Ntr1(1–120) binding allow an increased availability of the nucleic acid-binding cavity, an effect that is evidenced both by an increase in RNA cross-linking yield and by increased solvent accessibility of amino acids in the ratchet domain.
DISCUSSION
We have reported here that the G-patch motif of Ntr1 is intrinsically unstructured, but it can potentially adopt secondary structure elements on binding to Prp43. Ntr1(51–110) is the minimal functional fragment required for the complete activation of Prp43, and it stimulates both, the helicase and the ATPase activity of Prp43. Protein–protein cross-linking in combination with mass spectrometry has revealed that Ntr1 interacts with the C-terminal domains of Prp43. Even more, a protein in which Ntr1(51–110) is directly fused to the C-terminus of Prp43 behaves as a functional complex of both, exhibiting an increased helicase activity. RNA binding contacts were investigated using UV-induced protein–RNA cross-linking, and single amino acid residues were identified that contact RNA directly. The results presented here confirm the crucial participation of the long ratchet helix in RNA binding, and present evidence that Ntr1 is also involved in the interaction with the RNA.
Several structure-function studies for helicases in complex with their activator protein(s) are available, providing different levels of interaction and activation. For example, the activities of the SF1 helicase Upf1 are stimulated by its interaction partner Upf2 (59). Upf2 is an intrinsically unstructured protein that forms a beta-hairpin, and an alpha-helix on interaction with Upf1, decreases the RNA affinity of Upf1 and thereby stimulates its unwinding activity (60,61).
The ATPase and helicase activities of the DEAD-box protein eIF4A are stimulated on binding of eIF4G (62). On binding of eIF4G, the two RecA-like domains of eIF4A become closer to each other, and therefore the affinities for RNA and ATP are increased (63).
Another level of activation has been suggested for the DEAD-box protein Dbp5. The interaction partner Gle1 was shown to decrease the RNA affinity after strand separation, and thus enhances the release of the substrate (64). The structure analysis of a Dbp5/Gle1 complex has revealed that the interaction is mediated by the small molecule inositol hexakisphosphate, which is required for stable complex formation (65).
These examples show the existence of different binding modes of helicase activation by interaction partners based on either direct interactions with the RecA-like domain or on interactions with associated domains like the CH-domain of Upf1. Furthermore, different levels of regulations have evolved, e.g. folding on interaction with a helicase, stabilization of a productive conformation, implication of small molecules in the complex formation and the involvement of several interaction partners.
Although the four spliceosomal DEAH-box helicases in yeast Prp2, Prp16, Prp22 and Prp43 show a high sequence similarity in their helicase core as well as in their C-terminal domains, the N-terminal region that precedes the first RecA-like domain is not conserved.
Prp43 has the shortest NTE (89 residues) compared with Prp2 (220 residues), Prp16 (347 residues) and Prp22 (480 residues). The first 91 amino acids of Prp43 were shown to be dispensable in vivo (58), indicating that they are not involved in the activation of Prp43. We have shown here that the deletion of the N-terminal alpha-helix by which the NTE binds to the winged helix domain affects the intrinsic ATPase activity of Prp43. We furthermore could show that the first 58 residues of Prp43 are not required for the activity in vitro if Ntr1 is present.
The NTE of Prp2 and Prp16 was shown to be functionally dispensable in vivo (66,67). The role of the NTE in Prp22 is more ambiguous: the first 260 are not required, whereas the residues 262–350 are able to enhance the function of Prp22 in vivo (68). Moreover, the deletion of the complete NTE in Prp22 leads to higher ATPase and helicase activities in vitro, but to a defect in its spliceosomal function (68). This suggests that the NTE of Prp22 is required for the recruitment to the spliceosome.
There is a fundamental difference between Prp22 and Prp43: while Prp43 is dependent on the presence of Ntr1 for both, recruitment and activation (31), Prp22 shows intrinsic ATPase and helicase activity in vitro and presumably only requires interaction partners for the recruitment to the spliceosome (69). Comparisons of the C-terminal domains of hPrp22 and yPrp43 with homology models from hPrp2, hPrp16 and hPrp43 reveal that their overall fold is similar, but that patches of different surface charge exist, suggesting that this contributes to the specific interaction with other proteins (37). It can be speculated that differences in the C-terminal domain of Prp22 and Prp43 lead to the different properties of both proteins.
The ATPase activity rate of Prp43 is stimulated by RNA, both in presence and absence of Ntr1 (70,33). We could show here that the extent of stimulation is in the same range in absence and in presence of the truncated Ntr1 variants. Hence, RNA seems to influence the ATP affinity and/or the mechanism of ATP hydrolysis of Prp43.
The crystal structure of the Prp43/ADP complex represents a post-reaction state that cannot accommodate ATP in its active site (35,36). A comparison of the helicase core of Prp43 with the HCV NS3 helicase in its apo=nucleotide-free and in its ATP-bound state suggests that major conformational changes are required within the helicase core of Prp43 to allow ATP binding (Supplementary Figure S9). The overlay of Prp43 with NS3/ATP/RNA shows that major changes occur within the RecA-2 domain, which lead to an enlargement of the nucleotide binding pocket and to a movement of the beta-hairpin that protrudes from this domain. These changes might be induced by a cooperative binding of the RNA substrate as well as of Ntr1.
Using UV-induced protein–RNA cross-linking, we have identified eight residues within Prp43 that interact directly with the RNA. Of these residues, four are conserved in the three other spliceosomal DEAH-box proteins Prp2, Prp16 and Prp22, namely, Y174 of the RecA-1 domain, K418 of the RecA-2 domain and Y595 and Y610 within the ratchet domain. Y595 and Y610 were furthermore identified to be important for the helicase activity, as their mutation leads to a decreased unwinding activity, in agreement with the increased RNA cross-linking yield determined for these residues.
Although it was reported that the ATPase activity of a truncated Prp43 variant lacking the OB-fold domain was not stimulated in the presence of RNA (36), no cross-links were identified within the OB-fold domain of Prp43. This could be due to the fact that a single-stranded RNA substrate was used here, which only included five uridines. In general, most cross-links are found with uridines and rarely with a guanine or a cytidin base. Hence, the bases that potentially were interacting with the OB-fold domain may have been unsuitable for cross-linking, as the uridine nucleotides are located in the center of the used oligonucleotide. Furthermore, it is possible that the OB-fold domain only binds dsRNA, and therefore failed to interact with the ssRNA.
In addition to the cross-links that were found within Prp43, a cross-link between the RNA and the residues K67 of Ntr1 was identified, providing the first experimental evidence of a residue within the G-patch motif directly interacting with RNA. As we could not detect any RNA affinity of isolated Ntr1 in an EMSA (data not shown), the interaction appears to be possible only when Ntr1 is bound to Prp43. Although the mutation K67E did not influence the unwinding activity in vitro, it cannot be ruled out that this residue is essential in vivo. Strikingly, K67 of Ntr1 is conserved within in the G-patch motifs of the second activator of Prp43, Pfa1 and of the human Ntr1 orthologue, TFIP11. Interactions of G-patch motifs with RNA have already been reported for the protein TgDRE from the parasite Toxoplasma gondii using fluorescence anisotropy measurements (71) and for the proteinase from the Mason-Pfizer monkey virus in electrophoretic mobility shift assays (72), but no concrete residues responsible for interaction were reported.
Ntr1 stimulates the helicase activity of Prp43, and the N-terminal fragment Ntr1(1–120) is sufficient to mediate this interaction in vitro (33). These findings were confirmed in this study, and we could demonstrate that the residues 51–110 of Ntr1 are required and sufficient to activate Prp43. Furthermore, we could show that the two fragments Ntr1(51–110) and Ntr1(1–120) are able to stimulate the ATPase activity of Prp43. This result is in contrast to the finding of Tanaka and co-workers who detected no difference in the ATPase activity of Prp43 in absence and presence of full-length Ntr1. Although we used here only a fragment of Ntr1, it seems unlikely that additional parts of Ntr1 can inhibit the stimulatory effect on the ATPase activity. Moreover, the G-patch protein Pfa1 was also shown to stimulate both the helicase and the ATPase activity of Prp43 during ribosome biogenesis (73); therefore, it was suggested that both activities are coupled. The action of Prp43 during the spliceosomal disassembly is most probably regulated in the same manner, meaning that Ntr1 not only stimulates the helicase but also the ATPase activity of Prp43.
The fragment Ntr1(1–122) was shown in yeast two-hybrid studies to be able to interact with Prp43 (74). Additionally, the fragment Ntr1(1–120) was suggested to be sufficient for the stimulation of the helicase activity of Prp43 (33). In vivo studies have revealed that the N-terminal 33 residues of Ntr1 seem to be dispensable (75).
On characterization of the helicase and the ATPase activities, it was shown here that both Ntr1(1–120) and Ntr1(51–110) are able to stimulate Prp43 to the same extent. Moreover, by fusing different fragments of Ntr1 to Prp43, it turned out that for all fragments smaller than Ntr1(51–110), the ATPase activity drops dramatically. Hence, Ntr1(51–110) seems to be both sufficient and required for the activation of Prp43. A sequence alignment of the two different activator proteins of Prp43, Ntr1 and Pfa1, reveals that conserved residues are present in the range of residues 51–103 of Ntr1. This explains why Ntr1(59–110) was not able to completely stimulate Prp43. Admittedly, further studies have to be performed to investigate whether the residues 104–110 of Ntr1 might be dispensable for the complete activation. Furthermore, the cross-linking results as well as the determination of binding constant indicate that also some of the first 50 residues of Ntr1 are involved in interactions with Prp43, although these contacts are not responsible for the activation itself.
In vivo studies have revealed that the C-terminally truncated Prp43(1–732) is still functional, whereas Prp43(1–722) leads to a temperature-sensitive phenotype (58). Furthermore, a truncated Prp43 that lacked the complete OB-fold domain, Prp43(1–657), was shown to be unable to interact with the G-patch containing C-terminal domain of Pfa1 in vitro (36). SAXS studies of the Prp43/Pfa1 complex have proposed that the C-terminal domains of Prp43 are involved in the interaction with Pfa1 (76).These findings are in agreement with the protein–protein cross-linking results reported here, which indicate that the C-terminal domains of Prp43 are responsible for the binding of Ntr1. If the OB-fold domain is removed, apparently the overall structure of this region is disturbed, precluding the binding of Ntr1. Strikingly, it has also been shown for the DEAH-box helicase Prp2 that the interaction with the G-patch protein Spp2 can be disturbed by mutations in the OB-fold domain (30).
Although the used cross-linkers were of medium length and the observed cross-links can correspond to maximal distances of the C-alpha atoms of ∼20 and 24 Å, respectively, only cross-links in a small surface area of Prp43 were identified, proposing that the binding platform of the G-patch motif is located in proximity to this limited region (Supplementary Figure S8).
Although no 3D structure of a G-patch motif has been reported so far, it was suggested that the G-patch motif within the DNA repair protein TgDRE from T. gondii lacks any secondary structure elements (71). The CD spectra of Ntr1(51–110) presented here indicate the same, but also reveal that Ntr1(51–110) has the potential to evolve secondary structure elements on interaction with Prp43. Therefore, Ntr1 could belong to the proteins of ‘dual personality’, which have been proposed to be able to perform transitions between an unstructured and a structured state (77), such as the protein Upf2, which forms an alpha-helix and a beta-sheet upon binding to Upf1.
In summary, the following model for the activation of Prp43 by Ntr1 can be proposed (Supplementary Figure S10): the overhang of a dsRNA could bind within the proposed nucleic binding cavity and interact strongly with the long ratchet helix, as proposed for the Ski2-like helicase Hel308 (34,36). This binding event could induce structural changes, especially within the RecA-2 domain, which lead to the remodelling of the nucleotide binding-pocket to facilitate ATP binding and to a movement of the beta-hairpin. Subsequently, the binding of Ntr1 could adjust the conformation of the three C-terminal domains in such a way that it interacts with the dsRNA substrate as well as with the β-hairpin. The binding of Ntr1 to the OB-fold domain could induce structural changes also in the helicase core domains including the β-harpin, which lead to a higher productivity of Prp43 and thereby to an elevated ATP hydrolysis rate even in absence of RNA.
On hydrolysis of ATP, the conformation of the RecA-2 domain changes and thereby the overhang of the dsRNA could be pulled further into the binding cavity, presumably supported by the long ratchet helix. The beta-hairpin could, eventually in a concerted action with the OB-fold domain, act as steric hindrance for the double-stranded part of RNA and therefore induce the separation of base pairs in this region. The subsequent release of ADP and the binding of ATP could again lead to structural changes, provoking an at least partially processive unwinding of the dsRNA as proposed in the backbone stepping motor model for the HCV NS3 helicase (78). The fine details of this activation will be revealed when additional crystallographic and functional studies succeed and further describe the molecular details of the interaction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The DFG [DFG Grant GSC 226/1 to H.C., SFB860 TP A2 to R.F. and SFB860 TP A10 to H.U.]. Funding for open access charge: Deutsche Forschungsgemeinschaft [DFG, SFB860].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Monika Raabe and Uwe Plessmann for expert technical assistance in MS, Katharina Kramer for fruitful discussions in protein–RNA cross-linking experiments and Uzma Zaman for assistance in radiolabelling. They are also thankful to Petra Hummel (IT&Elektronik) for help in footprinting data analysis, Claudia Höbartner for use of the phosphoimager and Sebastián Díaz for language editing. Furthermore, they are grateful to Achim Dickmanns for critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Hocine S, Singer R, Grünwald D. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010;2:a000752. doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore M, Proudfoot N. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Will C, Lührmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:pii: a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahl M, Will C, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Tarn W, Steitz J. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 6.Turunen J, Niemelä E, Verma B, Frilander M. The significant other: splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA. 2012;4:61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 8.Staley J, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 9.Cordin O, Hahn D, Beggs J. Structure, function and regulation of spliceosomal RNA helicases. Curr. Opin. Cell Biol. 2012;24:431–438. doi: 10.1016/j.ceb.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Kistler A, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, Query C. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghunathan P, Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 13.Small E, Leggett S, Winans A, Staley J. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng C, Liu H, Cheng S. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011;17:145–154. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umen J, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3’ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 16.Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 2009;16:1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 17.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 18.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol. Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arenas J, Abelson J. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl Acad. Sci. USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourmann J, Schmitzová J, Christian H, Urlaub H, Ficner R, Boon K, Fabrizio P, Lührmann R. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013;27:413–428. doi: 10.1101/gad.207779.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebaron S, Froment C, Fromont-Racine M, Rain J, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohnsack M, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess S, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 24.Semlow D, Staley J. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem. Sci. 2012;37: 263–273. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F, Wang X-Y, Zhang Z-M, Pu J, Fan Y-J, Zhou J, Query CC, Xu Y-Z. Splicing proofreading at 5’ splice sites by ATPase Prp28p. Nucleic Acids Res. 2013;41:4660–4670. doi: 10.1093/nar/gkt149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fairman-Williams M, Guenther U, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanya H, Bird L, Brannigan J, Wigley D. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 28.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 29.Pyle A. Translocation and unwinding mechanisms of RNA and DNA helicases. Ann. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 30.Silverman E, Maeda A, Wei J, Smith P, Beggs J, Lin R. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 2004;24:10101–10110. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai R, Tseng C, Lee P, Chen H, Fu R, Chang K, Yeh F, Cheng S. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell. Biol. 2007;27:8027–8037. doi: 10.1128/MCB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aravind L, Koonin E. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 1999;24:342–344. doi: 10.1016/s0968-0004(99)01437-1. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Büttner K, Nehring S, Hopfner K. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 35.He Y, Andersen G, Nielsen K. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–186. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–2204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudlinzki D, Schmitt A, Christian H, Ficner R. Structural analysis of the C-terminal domain of the spliceosomal helicase Prp22. Biol. Chem. 2012;393:1131–1140. doi: 10.1515/hsz-2012-0158. [DOI] [PubMed] [Google Scholar]
- 38.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl F-U, Ban N, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 39.Schmitzová J, Rasche N, Dybkov O, Kramer K, Fabrizio P, Urlaub H, Lührmann R, Pena V. Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein. EMBO J. 2012;31:2222–2234. doi: 10.1038/emboj.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Chance M. Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting. Anal. Chem. 2011;83:7234–7241. doi: 10.1021/ac200567u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belon C, Frick D. Monitoring helicase activity with molecular beacons. BioTechniques. 2008;45:433–440, 442. doi: 10.2144/000112834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 43.Mészáros B, Simon I, Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharp JS, Becker JM, Hettich RL. Protein surface mapping by chemical oxidation: Structural analysis by mass spectrometry. Anal. Biochem. 2003;313:216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- 45.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 46.Kramer K, Hummel P, Hsiao H, Luo X, Wahl M, Urlaub H. Mass-spectrometric analysis of proteins cross-linked to 4-thio-uracil- and 5-bromo-uracil-substituted RNA. Int. J. Mass Spectrom. 2011;304:184–194. [Google Scholar]
- 47.Schmidt C, Urlaub H. iTRAQ-labeling of in-gel digested proteins for relative quantification. Methods Mol. Biol. 2009;564:207–226. doi: 10.1007/978-1-60761-157-8_12. [DOI] [PubMed] [Google Scholar]
- 48.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 49.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 50.Charvátová O, Foley BL, Bern MW, Sharp JS, Orlando R, Woods RJ. Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to galectin-1. J. Am. Soc. Mass Spectrom. 2008;19:1692–1705. doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sturm M, Bertsch A, Gröpl C, Hildebrandt A, Hussong R, Lange E, Pfeifer N, Schulz-Trieglaff O, Zerck A, Reinert K, et al. OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertsch A, Gröpl C, Reinert K, Kohlbacher O. OpenMS and TOPP: open source software for LC-MS data analysis. Methods Mol. Biol. 2011;696:353–367. doi: 10.1007/978-1-60761-987-1_23. [DOI] [PubMed] [Google Scholar]
- 53.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 54.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu H, Freitas MA. MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine research articles. J. Proteome Res. 2008;7:138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo P, Baldwin R. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 58.Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 59.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 60.Clerici M, Mourão A, Gutsche I, Gehring N, Hentze M, Kulozik A, Kadlec J, Sattler M, Cusack S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009;28:2293–2306. doi: 10.1038/emboj.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 62.Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schütz P, Bumann M, Oberholzer A, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl Acad. Sci. USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weirich C, Erzberger J, Flick J, Berger J, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 65.Montpetit B, Thomsen N, Helmke K, Seeliger M, Berger J, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwalds-Gilbert G, Kim D, Silverman E, Lin R. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA. 2004;10:210–220. doi: 10.1261/rna.5151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hotz H, Schwer B. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics. 1998;149:807–815. doi: 10.1093/genetics/149.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider S, Schwer B. Functional domains of the yeast splicing factor Prp22p. J. Biol. Chem. 2001;276:21184–21191. doi: 10.1074/jbc.M101964200. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka N, Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- 70.Tanaka N, Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6510–6521. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frénal K, Callebaut I, Wecker K, Prochnicka-Chalufour A, Dendouga N, Zinn-Justin S, Delepierre M, Tomavo S, Wolff N. Structural and functional characterization of the TgDRE multidomain protein, a DNA repair enzyme from Toxoplasma gondii. Biochemistry. 2006;45:4867–4874. doi: 10.1021/bi051948e. [DOI] [PubMed] [Google Scholar]
- 72.Svec M, Bauerová H, Pichová I, Konvalinka J, Strísovský K. Proteinases of betaretroviruses bind single-stranded nucleic acids through a novel interaction module, the G-patch. FEBS Lett. 2004;576:271–276. doi: 10.1016/j.febslet.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 73.Lebaron S, Papin C, Capeyrou R, Chen Y, Froment C, Monsarrat B, Caizergues-Ferrer M, Grigoriev M, Henry Y. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009;28:3808–3819. doi: 10.1038/emboj.2009.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsai R, Fu R, Yeh F, Tseng C, Lin Y, Huang Y, Cheng S. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pandit S, Lynn B, Rymond B. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl Acad. Sci. USA. 2006;103:13700–13705. doi: 10.1073/pnas.0603188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Behrens M, He Y, Oliveira C, Andersen G, Pedersen J, Nielsen K. Structural Analysis of RNA Helicases with Small-Angle X-ray Scattering. Methods Enzymol. 2012;511:191–212. doi: 10.1016/B978-0-12-396546-2.00031-0. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, Stec B, Godzik A. Between order and disorder in protein structures: analysis of “dual personality” fragments in proteins. Structure. 2007;15:1141–1147. doi: 10.1016/j.str.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Appleby T, Anderson R, Fedorova O, Pyle A, Wang R, Liu X, Brendza K, Somoza J. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J. Mol. Biol. 2011;405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.