Abstract
Glycogen storage disease type II is a lysosomal storage disorder due to mutations of the GAA gene, which causes lysosomal alpha-glucosidase deficiency. Clinically, glycogen storage disease type II has been classified in infantile and late-onset forms. Most late-onset patients share the leaky splicing mutation c.-32-13T>G. To date, the mechanism by which the c.-32-13T>G mutation affects the GAA mRNA splicing is not fully known. In this study, we demonstrate that the c.-32-13T>G mutation abrogates the binding of the splicing factor U2AF65 to the polypyrimidine tract of exon 2 and that several splicing factors affect exon 2 inclusion, although the only factor capable of acting in the c.-32-13 T>G context is the SR protein family member, SRSF4 (SRp75). Most importantly, a preliminary screening using small molecules described to be able to affect splicing profiles, showed that resveratrol treatment resulted in a significant increase of normal spliced GAA mRNA, GAA protein content and activity in cells transfected with a mutant minigene and in fibroblasts from patients carrying the c-32-13T>G mutation. In conclusion, this work provides an in-depth functional characterization of the c.-32-13T>G mutation and, most importantly, an in vitro proof of principle for the use of small molecules to rescue normal splicing of c.-32-13T>G mutant alleles.
INTRODUCTION
Glycogen storage disease type II (GSDII- MIM number 232300, Pompe disease) is an autosomal recessive inherited lysosomal storage disorder arising from a deficiency of acid alpha-glucosidase (GAA; E.C.3.2.1.20) that results in impaired glycogen degradation and accumulation within the lysosomes (1).
Clinically, GSDII encompasses a continuous spectrum of phenotypes, ranging from a rapidly progressive infantile form to a slowly progressive late-onset (LO) form that affects mobility and respiratory function. Classic infantile GSDII manifests soon after birth and is characterized by absent or nearly absent enzyme activity, severe muscle weakness, cardiomegaly/cardiomyopathy and respiratory insufficiency, that typically lead to death within the first year of life (1–4). LO GSDII comprises all milder subtypes; partial enzyme deficiency manifests in children and adults as a slowly progressive skeletal muscle weakness without cardiac involvement. Respiratory muscle weakness, particularly of the diaphragm, is the leading cause of death in the LO cases (1,2,4–6). In nearly all patients, the disease progression results in severe physical handicap that heavily affects health status and quality of life.
The GAA gene (MIM number 606800) has been localized to human chromosome 17q25.2-25.3. Almost 300 different mutations of the GAA gene have been identified to date and are listed in the Pompe center database (http://www2.eur.nl/fgg/ch1/pompe). Although some mutations are common in different ethnic groups (p.R854X among African–Americans, p.D645E among Asians and del525T among Dutch people), the mutation profile is very heterogeneous and most mutations are present in single individuals or small number of families. The only exception is represented by the intronic mutation c.-32-13T>G that is present in 40–70% of the alleles in patients affected with the LO form of the disease (7–12).
The c.-32-13T>G mutation has been described for the first time by Huie et al. (13) in a patient affected by the adult onset form of GSDII. Studies performed in cells from patients carrying the c.-32-13T>G mutation have demonstrated that this mutation leads to the synthesis of three different aberrant splicing variants: SV1, which retains the first 36 nt of intron 1 and lacks exon 2, SV2 in which exon 2 is completely spliced out and SV3 in which exon 2 is partially spliced out (Figure 1A), as well as a small amount of normally spliced GAA mRNA. The same aberrant splicing variants were detected in low quantities in cells from normal controls (14), and these data were then further confirmed in vitro using a minigene assay (15). Using this system, the results suggested that the major effect of the mutation was to affect the overall splicing efficiency of the pre-mRNA transcript to yield the protein-coding normal isoform rather than its processing quality. At the clinical level, therefore, these data suggest that the residual expression of a certain amount of ‘normal spliced’ mRNA may be the general mechanisms that define the delayed phenotypic expression in patients carrying the c.-32-13T>G mutation (15).
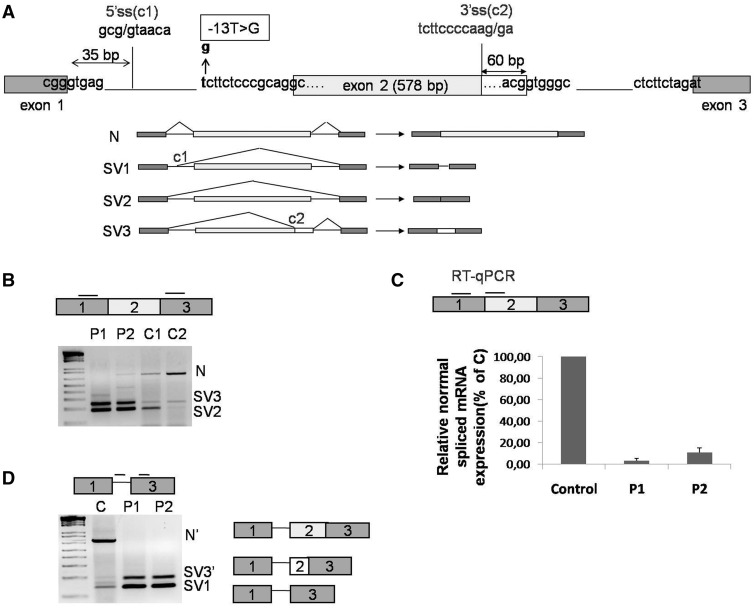
Figure 1.
GAA mRNA splicing variants expressed in cultured fibroblasts. (A) Schematic representation of the 5′ region of the GAA gene (exons 1–3). The position of the c.-32-13T>G mutation is highlighted in bold. The cryptic splice sites located at position +35 from the normal donor splice site of exon 1 and at −60 nt from the donor site of exon 2 are shown as C1 and C2, respectively. Lower panel, schematic diagram of the GAA mRNA spliced variants already described in human fibroblasts: N, SV1, SV2 and SV3. (B) RT-PCR analysis of the region encompassing exons 1–3 of GAA mRNA in fibroblasts from patients carrying the c.-32-13T>G mutation (P1 and P2) and normal controls (C1 and C2). Three fragments corresponding to normal (N) and SV2 and SV3 variants were detected in fibroblasts both from patients and controls. (C) real time PCR quantification of the normal spliced GAA mRNA (N) in fibroblasts from patients P1 and P2 and a normal control. Normal spliced GAA mRNA in P1 and P2 fibroblasts is expressed as percentage of the normal spliced mRNA detected in the normal control. Data represent the means ± SD of three independent experiments. (D) RT-PCR analysis of GAA mRNA using primers to specifically amplify SV1 variant in fibroblast from patients P1 and P2 and from a normal control. In addition to the SV1 variant, other two PCR fragments identical to N and SV3 but retaining the first 35 nt of intron 1 were detected (N′ and SV3′).
At the molecular level, it was therefore hypothesized that the presence of the c.-32-13T>G mutation may function by altering the binding of trans-acting factors to the polypyrimidine tract of exon 2. However, until now, the exact mechanism(s) by which this mutation could affect the splicing process has never been studied in detail.
In this work, we have now further characterized the splicing variants generated by wild type and c.-32-13T>G alleles in human fibroblasts and investigated the molecular mechanism by which the c.-32-13T>G mutation affects the pre-mRNA splicing of GAA gene.
In addition, several strategies have been recently developed to rescue/increase normal splicing of transcripts carrying mutations that affect the splicing process (16). However, to our knowledge, none of these approaches have yet been tested in GSDII. In this respect, therefore, we now also provide an in vitro proof-of-principle for the use of small molecules to partially rescue normal splicing of c.-32-13T>G mutant alleles.
MATERIALS AND METHODS
Reverse transcription-polymerase chain reaction and sequencing of GAA mRNA from patient’s fibroblasts
Total RNA was isolated from cultured skin fibroblasts from two patients affected by GSDII, genotype (c.-32-13T>G: unknown) and two healthy controls, using RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. Demographic and clinical characteristics of these patients are shown in Supplementary Table S1.
For reverse trasncription and polymerase chain reaction (RT-PCR) analysis, the first strand complementary DNA (cDNA) was synthesized using random hexamer primers. The amplification of the splice variants generated in fibroblasts by the allele carrying the c.-32-13T>G mutation was analyzed using primers WTF/SKIP2R reported in Table 1. The RT-PCR products were cloned (Topocloning, Invitrogen) and analyzed by automated sequencing (ABI Prism 3500×l genetic analyzer). The specific amplification of normal spliced GAA mRNA and SV1 variant was performed using primers WTF/WTR and SV1F/SKIP2R, respectively (Table 1). The relative abundance of the normal spliced GAA mRNA was assessed by real time PCR as described later.
Table 1.
Primers used for GAA cDNA amplification
| Primer | Sequence |
|---|---|
| WT F | 5′-CCACCTCTAGGTTCTCCTCGT |
| SKIP2R | 5′-CGGAGAACTCCACGCTGTA |
| WT R | 5′-GGCCTGGACAGCTCCTACA |
| SV1F | 5′-AGCGGGTGAGACACCTGAC |
| Bra2 | 5′-TAGGATCCGGTCACCAGGAAGTTGGTTAAATCA |
| a2-3 | 5′-CAACTTCAAGCTCCTAAGCCACTGC |
| F3 | 5′-CCCGGCCTGGAGTACAATG |
| R3 | 5′-CAGGAGTGCAGCGGTTGC |
| HPRT FW: | 5′-GACCAGTCAACAGGGGACAT |
| HPRT RV: | 5′-GTGTCAATTATATCTTCCACAATCAAG |
| GAPDH FW | 5′-ACCCACTCCTCCACCTTTGACG |
| GAPDH RV | 5′-CTCTCTTCCTCTTGTGCTCTTGC |
Pulldown assay
The pulldown assay was performed as previously described (17). Briefly, ∼500 pmoles of the target RNA transcribed from the wild type and mutated sequences were placed in a 400-µl reaction mixture containing 100 mM NaOAC, pH 5.0, and 5 mM sodium m-periodate (Sigma), incubated for 1 h in the dark at room temperature, ethanol precipitated and resuspended in 100 µl of 0.1 M NaOAC, pH 5.0. To this RNA, 300 µl of adipic acid dihydrazide agarose bead 50% slurry (Sigma) equilibrated in 100 mM NaOAC, pH 5.0, was added, and the mix was incubated for 12 h at 4°C on a rotator. The beads with the bound RNA were then pelleted, washed 3 times with 1 ml of 2 M NaCl and equilibrated in washing buffer (5 mM HEPES, pH 7.9, 1 mM MgCl2, 0.8 mM magnesium acetate). They were then incubated on a rotator with 1 mg of HeLa cell nuclear extract for 30 min at room temperature. Heparin was added to a final concentration of 5 mg/ml. The beads were then pelleted and washed 4 times with 1.5 ml of washing buffer, before addition of sodium dodecyl sulphate (SDS) sample buffer and loading on a 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis.
Minigene construction and transfection in culture cells
The Human wild type GAA minigene constructs (WT) has been prepared by amplifying human exon 2 and 50 nt of flanking intronic sequences using the following forward 5′-aattaaacatatggagctctttgagagccccgtgagt-3′ and reverse 5′-aattaaacatatgggatccgtcgatgtccacgcgcac-3′ primers and cloned inside the NdeI restriction site of the pTB plasmid. Further modification of the upstream intron to introduce the −13T/G substitution (MUT) was performed by site-directed mutagenesis using specific primers. Primer sequences for this MUT can be provided on request. This GAA sequence was also cloned in Bls KS+ plasmid using the SacI-BamH1 restriction sites in the primers to transcribe the RNA sequence for pulldown analysis.
In vitro splicing
In vitro splicing assays were essentially performed as previously described (18). The substrates for the in vitro reaction were prepared using RT-PCR by designing a primer that carried the T7 sequence together with the last portion of the tropomyosin exon 2 as contained in the PY7 plasmid (5′-taatacgactcactataggccgacgagaccgccgccaag-3′, T7PY7dir1) and two reverse primers that carried the 3′ss of GAA exon 2 either in its WT form (5′-cctggacagctcctacaggcctgcgggagaagaaagcgggcctcgaggcccaaga-3′) or mutated form (5′-cctggacagctcctacaggcctgcgggagaagcaagcgggcctcgaggcccaaga-3′). These primers were used to amplify wild type and mutated substrates from the PY7 plasmid for the in vitro splicing reaction as schematically shown in Figure 2D. The spliced products were then amplified using the forward T7PY7dir1 primer and the reverse primer that only contained the exonic portions of GAA exon 2 (5′-cctggacagctcctacaggc-3′) and run on a standard agarose gel.
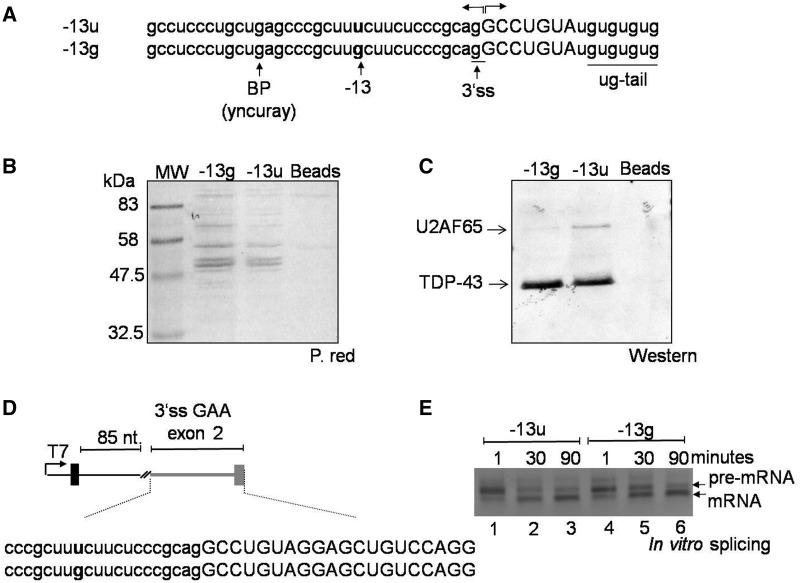
Figure 2.
Effect of the c.-32-13T>G mutation on U2AF65 binding. (A) Sequence of RNA oligonucleotides used in a pulldown assay. Each RNA oligo carried the polypyrimidine tract, putative branch point and 3′ss sequence of exon 2 either in its wild type or in its mutated form. Each also carried a ug-tail to control for equal pulldown efficiencies by measuring the amounts of TDP-43 protein. (B) Analysis of total proteins pulled down by the −13g and −13u RNA oligonucleotide as detected using Red Ponceau before western blot analysis. (C) Western blot analysis of U2AF65 and TDP-43 in the RNA–protein complex. The presence of the c.-32-13T>G mutation almost completely abrogated the binding of the U2AF65 to the RNA. The pulldown efficiency of wild type or mutated RNA was comparable as demonstrated by the intensity of the TDP-43 signals. (D) Schematic representation of the small in vitro splicing substrate used to test for the effects of the −13T>G substitution on the 3′ss of GAA exon 2. Gray line and boxes represent GAA sequence (reported in full below, small letters intron and capital letters exon), whereas black line and boxes represent tropomyosin sequences. The position of the −13T>G substitution is highlighted in bold. (E) In vitro splicing analysis at different time points of the constructs shown in Figure 2D carrying either the GAA wild type (-13u) or mutated sequence (-13g). The result shown here is representative of three independent experiments.
Cell culture and transient transfection
Human fibroblasts were cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal calf serum and 50 mg/ml penicillin/streptomycin (Gibco, Paisley, UK). HeLa cells were grown in Dulbecco’s modified Eagle’s medium high glucose medium (Gibco, Paisley, UK) supplemented with 10% (v/v) fetal bovine serum, 50 mg/ml penicillin/streptomycin and 2 mM l-glutamine (Gibco, Paisley, UK). Cells were maintained at 37°C in a humidified atmosphere enriched with 5% (v/v) CO2 and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) using 0.8–4 µg of total plasmid DNA Endofree purified (Qiagen GmbH, Hilden, Germany) following manufacturer’s instructions. For minigene transfection, 0.8 µg of WT or MUT plasmid was transfected into 3 × 105 Hela cells in serum-free medium with 8 µl of Lipofectamine reagent (Invitrogen). After 24 h, total RNA was extracted using RNeasy Mini Kit (QIAGEN) following manufacturer’s instructions. Then, 1–3 µg of total RNA was reverse transcribed using random hexamer primers and cDNA was amplified by PCR in a total volume of 20 µl using primers specifically designed to amplify processed transcripts derived from the minigene, and not the endogenous, GAA mRNA (Bra2 and a2-3). Each transfection experiment was performed at least three times, and representative gels are shown in each case. PCR products were resolved in a 1% agarose gel and sequenced. For overexpression experiments, 1 µg of each plasmid expressing either SR or hnRNP proteins was co-transfected with either the WT or MUT minigene and the resulting RNA analyzed as described earlier.
Inhibition of SRSF4 expression by small interfering RNA
Small interfering RNA (siRNA) duplex derived from the human SRSF4 sequence was purchased from Invitrogen (Pre-designed siRNA, HSS109661, against human SRSF4 gene). Cultures of fibroblasts were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions and 72 h later were harvested and analyzed for SRSF4 expression and for the relative abundance of the normal spliced GAA mRNA by western blot and real time-PCR, respectively.
Cell treatments
HeLa cells transfected with the MUT minigene or human fibroblasts were treated with resveratrol (50–200 µM), amiloride (150–300 µM), tannic acid (50–100 µM), kinetin (100–200 µM), C6 pyridinium ceramide (5 µM), sodium butyrate (0.5–5 µg/ml), phenylbutyric acid (0.5–2 mM) and SAHA (3–20 µM). Cells were harvested after 18–24-h treatment for the analysis of splicing factor and GAA mRNA expression and after 48 h for the analysis of GAA protein and enzymatic activity.
Real time-PCR
The abundance of the endogenous normal spliced GAA mRNA generated in fibroblasts of patients carrying the c.-32-13T>G mutation and the abundance of the normal spliced GAA mRNA generated by the MUT minigene were analyzed by quantitative real time-PCR using the primers WTF/WTR and F3/R3, respectively (Table 1).
Amplification of HPRT and GAPDH genes was used as an internal standard gene.
Primers were designed from available human sequences using the primer analysis software Primer3. Real time-PCR was performed using a Roche LightCycler 480 Real-Time PCR System and the LightCycler 480 SYBR Green I Master (Roche), following manufacturer’s instructions. Standard curves using a ‘calibrator’ cDNA (chosen among the cDNA samples) were prepared for each target and internal standard gene. To verify the specificity of the amplification, a melt-curve analysis was performed immediately after the amplification protocol. Non-specific PCR products were not found in any case. The relative quantification was made using the Pfaffl modification of the ΔΔCt equation, taking into account the efficiencies of individual genes. The results were normalized to HPRT and GAPDH, and the initial amount of normal spliced mRNA of each sample was determined as relative expression versus its reference sample, which in each case was considered the 1× sample. At least three independent determinations for each gene were performed.
Western blot
Samples were dissolved in standard 3× SDS loading buffer and were heated for 5 min in boiling water before loading on a 10% SDS-polyacrylamide denaturing gel. The gel was then electroblotted onto a Hybond-C Extra membrane (Amersham) according to standard protocols (Amersham Biosciences) and blocked with 10% skimmed milk (non-fat dry milk in 1× phosphate-buffered saline, 0.1% Tween 20). Membranes targeted for SR protein recognition were blocked using western blocking reagent (Roche) according to the manufacturer’s instructions. Proteins were probed with different antibodies and detected with a chemiluminescence kit (ECL; Pierce Biotechnology) according to manufacturer’s instructions. Anti-SR protein antibodies were purchased from Invitrogen Histo-Line Laboratories. Polyclonal rabbit antibodies against U2AF65 and DAZAP-1 were home-made in ICGEB using standard immunization protocols and used in western blot at a dilution of 1:1000 and 1:1500, respectively. The YB-1 antibody was obtained from Dr T.A. Cooper and was used at a dilution of 1:1000. Membranes targeted from GAA protein were probed with an antiserum against GAA as described elsewhere (19). An anti-rabbit horseradish peroxidase-conjugated antibody (DAKO, Glostrup, Denmark) was used as a second antibody and developed by enhanced chemiluminescence using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific). The signals were normalized to those obtained for actin, using a polyclonal anti actin antibody (Sigma, St Louis, MO, USA).
Enzymatic activity
GAA activity was measured using the fluorogenic substrate 4-methylumbelliferyl-α-d-glucopyranoside (Glycosynth, Cheshire, England) (20). Protein concentration of the samples was determined by the Lowry method. Enzymatic activity was expressed as nanomoles of substrate hydrolyzed per milligram of total protein per hour. All assays were done in triplicate.
RESULTS
Characterizing the patient’s mRNA splicing profile in the presence of the c-32-13T>G mutation
The GAA gene (Ensembl number ENST00000302262) consists of 20 exons that must be joined together by the splicing process to form the mature mRNA. One unusual feature of exon 2 (where the starting ATG is located) is represented by its length of 578 nt, which is considerably longer than the average length of human exons (∼150 nt) (Figure 1A).
The only previous data describing in some detail the effects of the c.-32-13 T>G mutation on the GAA mRNA splicing process in vivo have been published >15 years ago (14). In this previous study, as summarized in Figure 1A, following the introduction of the c-32-13T>G mutation, four splicing isoforms could be detected in the mRNA extracted from patient’s cells. These included some residual normal splicing (N) together with three aberrantly spliced transcripts that used a cryptic donor site in intron 1, 5′ss(c1) at position +35 with respect to the normal exon 1 donor site, and a cryptic acceptor sequence, 3′ss(c2), located in exon 2 at −60 nt from exon 2 natural donor site (Figure 1A, lower panel).
To confirm these observations, it was decided to analyze the GAA mRNA variants transcribed from the c.-32-13T>G allele in fibroblasts obtained from 2 LO-GSDII patients (P1-P2) previously characterized in our laboratory (7). These two patients were chosen because they both carried the c.-32-13T>G mutation in heterozygosis with an unknown mutation that abrogated the expression of their allele (thus avoiding the need to take into account its eventual splicing profile). This was demonstrated by the analysis of a c.324C>T polymorphism present in heterozygosis in exon 2 of the GAA gene. PCR amplification of exon 2 and its 5′ flanking region followed by subcloning and sequencing of the amplified products showed that the c.-32-13T>G mutation was in cis with the T at position 324. Because in both cases only the allele carrying the T at position 324 was detected in the cDNA, this indicated that only this allele was expressed in the patient’s fibroblasts (Supplementary Figure S1).
It was then decided to use primers WTF and SKIP2R (Table 1) to amplify a region encompassing exons 1–3 of the GAA cDNA from these two patients (Figure 1B, P1 and P2). As controls, we also used these primers to look at the splicing profile of two samples from healthy individuals (Figure 1B and C1 and C2). As shown in Figure 1B, normal spliced GAA mRNA (N) and the two different splicing variants already described, SV2 and SV3, were detected both in fibroblasts from patients carrying the c.-32-13T>G mutation and in normal controls (Figure 1A and B). In the patients versus controls samples, however, the relative preponderance of the various splicing isoforms differed greatly. For example, significant amounts of normal spliced GAA mRNAs were present in the controls, but were only slightly present in patient 2 and absent in patient 1 (Figure 1B). In this respect, however, the subsequent cloning and sequencing of the PCR products from the patient samples showed that patient 1 also expressed low amounts of normal mRNA even if not visible in the agarose gel of Figure 1B. Therefore, to better quantify these data, we then designed primers to specifically amplify only the normal spliced mRNA and estimated its relative expression in P1 and P2 by quantitative real time PCR. As shown in Figure 1C, the abundance of normal (N) mRNA in fibroblasts from patients 1 and 2 was low, representing 3 and 10% of the amount detected in normal fibroblasts, respectively.
Finally, the original report from Boerkoel et al. (14) also described the presence of an isoform called SV1 in which the activation of a cryptic intron 1 donor site, 5′ss(c1), was described. However, an effect on the 5′ donor site of intron 1 (leading to the synthesis of SV1) is quite unexpected, especially considering that the c.-32-13T>G mutation is located 2651 bp of the normal intron 5′ splice site. To further characterize this possible effect of the c.-32-13T>G mutation in our patients, we designed two primers to specifically amplify the SV1 variant (SV1F/SKIP2R-Table 1). Using this strategy, we were able to demonstrate that both normal (C) and patient’s fibroblasts (P1-P2) express the spliced variants SV1, albeit to a low extent. In addition, an extra variant containing exons 1, 2 and 3, but retaining the 36 nt of intron 1 (called N’ here), was present in normal cells, and an extra variant containing both the 36 nt of intron 1 and the 60 nt of exon 2 (called SV3’) was present both in normal and patient’s cells (Figure 1D). These two novel splicing variants have not been reported in the dbEST of NCBI (http://www.ncbi.nlm.nih.gov/dbEST/) and its functionality, if any, remains unknown. It should be noted, however, that these variants are expressed at low levels and can be detected only through the use of dedicated primers (see earlier).
Taken together, these results indicate that two versions of splicing variants N, SV2 and SV3 exist both in patients and controls: one in which intron 1 is completely spliced out (N and SV2 and SV3 variants) and the other in which 36 nt of intron 1 are retained (N’, SV1 and SV3’).
In conclusion, our results suggest that the use of these two donor sites, the 5′ss of intron 1 and 5′ss(c1), is independent of the presence of the c.-32-13T>G mutation and should therefore be considered as part of the normal physiological mRNA processing of this region.
Effect of the c.-32-13T>G mutation on the binding of U2AF65
Because the c.-32-13T>G mutation is located within the polypyrimidine tract of exon 2, it is reasonable to assume that it will affect the binding of regulatory proteins to this region. Therefore, to experimentally address this hypothesis, the binding of normal and mutated mRNA to U2AF65 was analyzed by a pulldown assay using short RNA oligos that carried the polypyrimidine tract, putative branch point and 3′ss sequence of exon 2 either in its wild type (-13u) or mutated (-13g) form. These oligos also carried a short ug-tail needed to control for equal pulldown efficiencies using the well-known ug-repeat binding factor, TDP-43 (Figure 2A). As shown in Figure 2B, Red Ponceau staining of the total protein profiles pulled down by the −13g and −13u oligos did not display significant qualitative differences in terms of specific protein signatures (that could have been gained or lost following the introduction of the u-to-g substitution). It should be noted, however, that Red Ponceau staining does not provide a high sensitivity with regard to many splicing factors. Therefore, we decided to further investigate this issue using the more sensitive western blot assay.
In particular, we focused on the basic splicing factors that are well known to recognize 3′ splice sites that in general are U2AF proteins, 35 and 65 and branch-point binding protein. Considering that the −13T>G mutation falls within the polypyrimidine tract, the U2AF65 factor represented the most likely protein to be affected by this nucleotide change. Therefore, to test this hypothesis, we performed the pulldown under splicing-competent conditions and tested directly for U2AF65 binding using a specific antibody. As shown in Figure 2C, the presence of the c.-32-13T>G mutation completely abrogated the binding of the U2AF65 to the mutated RNA as opposed to the wild type.
Second, to provide some additional mechanistic insight with regard to the effects of this mutation, we set up an in vitro splicing system to investigate the effects of the −13T>G substitution in the absence of any other GAA regulatory motifs. To achieve this, we used a small in vitro splicing system based on the PY7 plasmid that carries a short tropomyosin intronic sequence (21) (Figure 2D). In this system, a short intronic sequence from the tropomyosin gene (85 nt) separates a viable 5′ss sequence from either the wild type or mutated GAA exon 2 acceptor site. As shown in Figure 2E, our results demonstrate that the introduction of the −13G substitution in an isolated GAA exon 2 acceptor sequence can substantially hinder, but not completely abolish, 3′ss recognition. In particular, the presence of the −13g substitution considerably slows down the processing of the pre-mRNA at early reaction time points (30 min, compare lanes 2 and 5 in Figure 2E). Besides mimicking the results obtained in the native GAA gene context, this result further confirms that from a molecular point of view the −13 substitution is affecting the basic recognition of the acceptor site and does not depend on additional regulatory elements specific of the GAA pre-mRNA.
Mimicking the effects of the c-32-13T>G mutation in a minigene context
To further characterize the molecular mechanisms by which the c.-32-13T>G mutation affects GAA pre-mRNA splicing, we constructed a minigene system in which the entire exon 2 and 50 nt of the upstream and downstream intronic regions were inserted in the NdeI site of the pTB plasmid (Figure 3A, upper diagram). WT and MUT minigenes were then transfected in HeLa cells and the processed GAA mRNA species were analyzed by RT-PCR using minigene-specific primers (a2-3, Bra2, Figure 3A, Table 1). As shown in Figure 3A, lower panel, only the WT construct that does not carry the c.-32-13T>G mutation displayed inclusion of the full-length exon 2 (N isoform), while this recognition seems to be abolished in the MUT construct. In both cases, the SV2 and SV3 isoforms were present. In the case of the WT construct, they are present in higher amounts than in the RT-PCR from the human control samples (compare Figure 3A, WT with Figure 1B, lanes C1–C2).
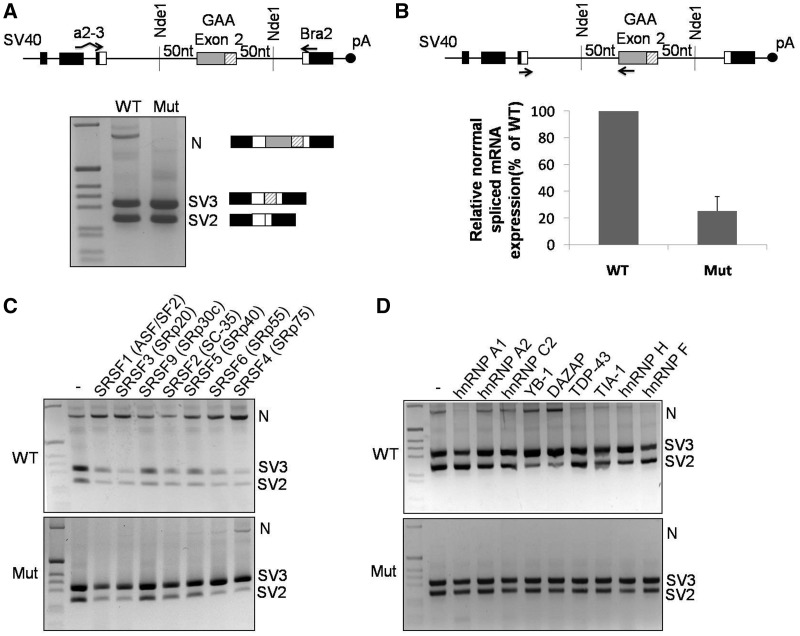
Figure 3.
GAA mRNA splicing variants expressed in cells transfected with the WT and MUT minigenes. (A) Schematic diagram of the minigene system to test the effect of the c.-32-13T>G mutation on exon 2 inclusion. The α-globin, fibronectin EDB and human GAA exon 2 are shown as black, white and gray boxes, respectively. The shaded box represents the segment of exon 2 that becomes included following the activation of the cryptic (c2) acceptor site. Minigenes carrying either WT or MUT GAA sequence were transfected in HeLa cells, and the splicing variants generated were analyzed by RT-PCR using minigene-specific primers (a2-3, Bra2). Cells transfected with the WT construct displayed inclusion of full-length exon 2 (N isoform) as well as total or partial exclusion of exon 2 (SV2 and SV3, respectively). Cells transfected with the MUT minigene expressed SV2 and SV3 variants, while the N variant was undetectable. (B) Relative quantification on the N isoform in cells transfected with WT or MUT minigenes using minigene specific primers designed to amplify only this variant. The abundance of the N isoform generated in cells transfected with the MUT minigene was expressed as percentage of the N isoform detected in cells transfected with the WT minigene. Data represent means ± SD of three independent experiments. (C) The effect of various SR protein overexpressions on exon 2 inclusion was assessed in HeLa cells co-transfected with the WT (upper panel) or MUT (lower panel) minigenes. (D) The effect of various hnRNP protein overexpression on exon 2 inclusion was assessed in HeLa cells co-transfected with the WT (upper panel) or MUT (lower panel) minigenes.
In keeping with this experimental bias, and just like it was observed in the patients, when we performed a quantitative real time-PCR using primers to specifically amplify only the normal spliced mRNA, we observed that a fair amount of normal spliced GAA mRNA is also produced in cells transfected with the MUT minigene (∼20% of WT) (Figure 3B) and that the WT GAA minigene is including the full-length exon 2 with much higher efficiency than that apparently shown in the RT-PCR assay shown in Figure 3A.
In conclusion, therefore, the mRNA variants expressed by the minigenes seemed to qualitatively reflect the variants and splicing efficiencies present in human control and patient fibroblasts. For this reason, we decided to further use these constructs to analyze the effect of auxiliary splicing factors on GAA exon 2 expression.
Effect of various SR and hnRNP splicing factors on exon 2 recognition
Because the presence of the c.-32-13T>G mutation completely abrogated the binding of U2AF65 to the polypyrimidine tract (Figure 2), the simple overexpression of this protein was not a viable strategy to rescue correct exon 2 splicing. In fact, as expected, overexpression of this factor failed to rescue exon 2 inclusion of the MUT minigene. Furthermore, it also failed to have an effect on the WT minigene (Supplementary Figure S2A), suggesting that its endogenous levels are quite effective to guarantee maximum splicing efficiency of this region. However, because of the unusual length of exon 2, it was likely that many auxiliary splicing factors may help to promote its physiological inclusion in the mature mRNA. To help answer this question, HeLa cells transfected with both the WT and MUT minigenes were co-transfected with constructs expressing some of the major splicing factors involved in the regulation of pre-mRNA splicing.
In particular, because of their well-known activity to promote exon inclusion in both alternative and constitutive systems, we first analyzed whether the overexpression of different SR proteins could favor the processing of normal spliced GAA mRNA. As shown in Figure 3C, the expression of various SR family members had a generally positive effect on WT exon 2 inclusion, with SRSF4 (SRp75) being the strongest enhancer of exon 2 inclusion (N isoform). Interestingly, co-transfection of these factors with the MUT minigene had very little effect on increasing the amount of correctly spliced mRNA, with SRSF4 representing the only SR protein capable of rescuing the presence of the N isoform. In the MUT context, in fact, overexpression of the various SR proteins generally causes just a shift of isoforms from SV2 to SV3 with respect to the normal situation. This is an interesting result, as it suggests that in the presence of the c.-32-13T>G mutation, the effect of SR protein overexpression generally results in promoting the use of the internal cryptic 3′ acceptor sequence, 3'ss(c2), with the consequent higher inclusion of the last 60 nt of exon 2. Finally, with regard to SRSF4, no further improvements in N isoform production could be detected when it was overexpressed together with U2AF65 either in the WT or MUT minigenes (Supplementay Figure S2A), suggesting that these factors may act in an independent manner (see Discussion).
Next, we also investigated the effects of overexpressing several hnRNP factors in the presence of the WT or MUT minigenes (Figure 3D). In this assay, we overexpressed a variety of hnRNP factors that have been previously reported to have either enhancer or silencer ability with regard to pre-mRNA splicing process. In the WT context, overexpression of hnRNP A1, TDP-43, TIA-1 and hnRNP H/F had a generally inhibitory action on inclusion of the full-length exon 2, while hnRNP A2 and hnRNP C2 had no detectable effect. Of all the tested factors, only the YB-1 and DAZAP proteins possessed a moderate enhancer activity in the WT context. Unlike SRSF4, however, the enhancing effect of all these hnRNP factors was completely absent when they were overexpressed in the presence of the MUT minigene. In addition, no effect on splicing of the N isoform could also be observed when overexpressing PTB, another well-known splicing affecting alternative splicing processes (Supplementary Figure S2B).
In conclusion, from the point of view of rescuing the correct processing of GAA exon 2 in the presence of the c.-32-13T>G mutation, the only auxiliary splicing factor capable of promoting full-length exon 2 inclusion, both in the WT and in the MUT context, was the SR protein family member, SRSF4 (Figure 3C).
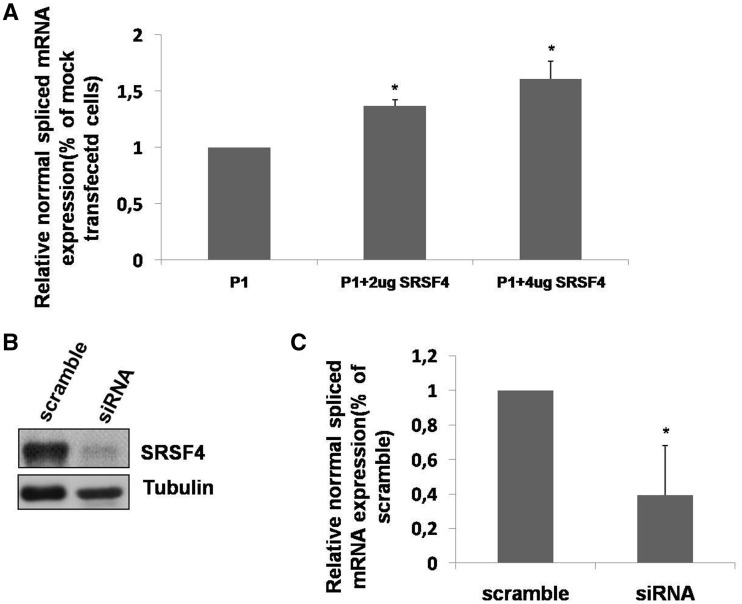
The effect of SRSF4 on exon 2 GAA inclusion was further confirmed in vivo. In fact, the overexpression of SRSF4 in patient’s fibroblasts (P1) led to a dose-dependent increase of normal spliced GAA mRNA (Figure 4A). Furthermore, when the expression of SRSF4 was knocked down in normal fibroblasts using siRNA, the level of normal spliced GAA mRNA was significantly reduced (Figure 4B–C).
Figure 4.
Effect of SRSF4 on normal spliced GAA mRNA in fibroblasts. (A) P1 fibroblasts were transfected with 2–4 ug of SRSF4, and the relative amount of normal spliced GAA mRNA was assessed by real time PCR. Results are expressed as percentage of the normal variant detected in fibroblasts transfected with the empty vector (mock). Data are means ± SD of three independent experiments. *P < 0.05. (B) Western blot analysis of endogenous SRSF4 protein expression in primary cultured fibroblasts from a healthy donor after knocking down the expression of SRSF4 by siRNA. (C) Relative abundance of the normal GAA variant assessed by real time PCR in cultured fibroblasts from a healthy donor after knocking down the expression of SRSF4 by siRNA. Results are expressed as percentage of the normal variant detected in fibroblasts transfected with the scramble siRNA. Data are means ± SD of three independent experiments. *P < 0.05.
Rescuing normal spliced GAA mRNA using small molecules
Based on these results, we hypothesized that it may be possible to rescue/increase normal spliced GAA mRNA of transcripts carrying the c.-32-13T>G mutation by using small molecules described in literature capable to increase the expression/activity of splicing factors. Therefore, using our MUT minigene system, we tested the ability to rescue the expression of normal spliced GAA mRNA (N) of the following small molecules: resveratrol (22), amiloride (23,24), tannic acid (25), kinetin (26) and C6 pyridinium ceramide (27). Because of its greater sensitivity to detect the presence of the normal N isoform, the eventual increase in correct exon 2 inclusion was measured using quantitative real time PCR.
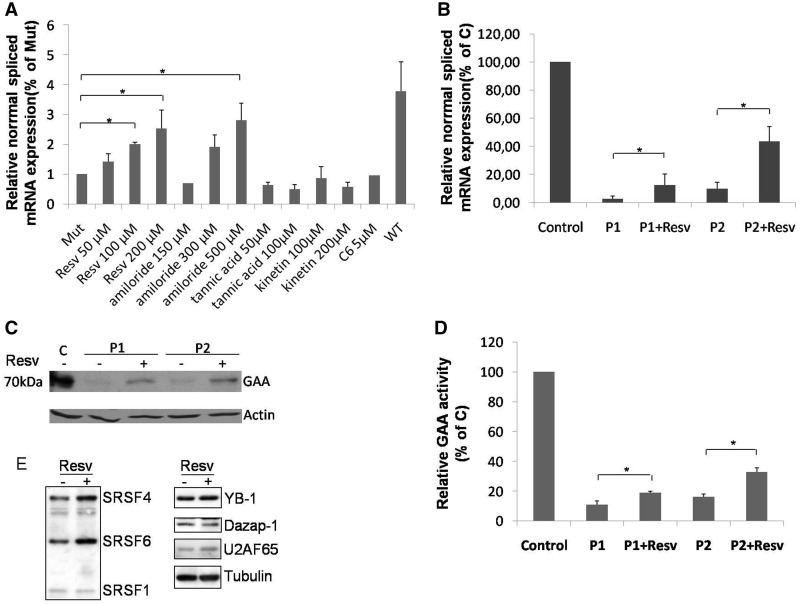
As shown in Figure 5A, a significant increase in the normal spliced mRNA produced by the MUT minigene carrying the c.-32-13T>G mutation was observed only in cells treated with resveratrol (100 and 200 µM) and amiloride (500 µM). However, the concentrations of amiloride required to obtain a significant rescue of normal splicing were toxic and resulted in a significant reduction of cell viability (data not shown). Therefore, we decided to further test only the effects of resveratrol on GAA mRNA splicing and protein expression in vivo using fibroblasts from P1 and P2. The results in Figure 5B showed that even if under basal conditions P1 and P2 express different levels normal spliced mRNA (3 and 10%, respectively) resveratrol treatment resulted in 5-fold increase of normal GAA mRNA expression for both of them. Furthermore, this increase was associated with an increase of GAA protein expression and a 2-fold increase of GAA enzymatic activity (Figures 5Cand D, respectively). With regard to the mechanism of action of resveratrol, it should be noted that all these compounds cause a change in a wide number of splicing factors, thus making it difficult to ascribe their effects on a single protein. Nonetheless, with regard to the splicing regulators tested in this work, treatment of HeLa cells with this compound displayed a significant increase in the expression of SRSF4 (∼1.6-fold, as measured by densitometric analysis using the ImageJ software), the most effective splicing factor capable of promoting normal exon 2 processing both in the presence or absence of the c.-32-13T>G mutation (Figure 5E). These effects were already observed after treating the cells with a concentration of 50 μM of resveratrol for 18 h as previously described (22). In addition to SRSF4, treatment with resveratrol was also associated with an increase in SRSF6 expression (1.7-fold) that was also observed to increase the efficiency of GAA normal splicing but only in a WT context. Finally, no changes were detected for the hnRNP proteins capable of increasing GAA normal splicing in the WT context, Dazap-1 and YB-1 (Figure 5E, right). Regarding previous results obtained by Markus et al., 2011, following resveratrol treatment using identical conditions in HEK293 cells, it should be noted that in our HeLa cells we did not observe any increase in SRSF1 expression levels. Besides possible technical differences, these results also suggest that resveratrol may have a cell-specific effect in influencing SR protein and hnRNP expression levels.
Figure 5.
Effect of small molecules on GAA splicing. (A) Cells transfected with the MUT minigene were treated for 24 h with resveratrol (Resv), amiloride, tannic acid, kinetin or C6 pyridinium ceramide (C6). The relative expression of the N isoform was analyzed by real time PCR, using minigene-specific primers designed to amplify only this variant. The results are expressed as fold of N isoform detected in untreated cells. The abundance of the N isoform produced by cells transfected with the WT minigene is also shown for comparison. Data represent the means ± SD of three independent experiments. *P < 0.05. (B) Cultured fibroblasts from P1 and P2 patients were treated for 24 h with 200 µM of resveratrol and the relative abundance of the N variant was assessed by real time PCR. Results are expressed as percentage of the N variant detected in fibroblasts from a normal control. Data are means ± SD of three independent experiments. *P < 0.05. (C) Western blot analysis of the GAA mature protein (70 kD) in cultured fibroblast from a normal control (C) and the two P1 and P2 patients treated or not with resveratrol. (D) GAA enzymatic activity in cultured fibroblast from a normal control and P1 and P2 patients treated or not with resveratrol. Results are expressed as percentage of the activity detected in the normal control. Data represent the means ± SD of three independent experiments. *P < 0.05. (E) On the left, western blot analysis of endogenous SRSF proteins in HeLa cells treated or not with resveratrol. The positions of SRSF4, SRSF6 and SRSF1 are indicated. On the right, western blot analyses of DAZAP, YB-1 and U2AF65 factors. A western blot against tubulin was added to show loading control.
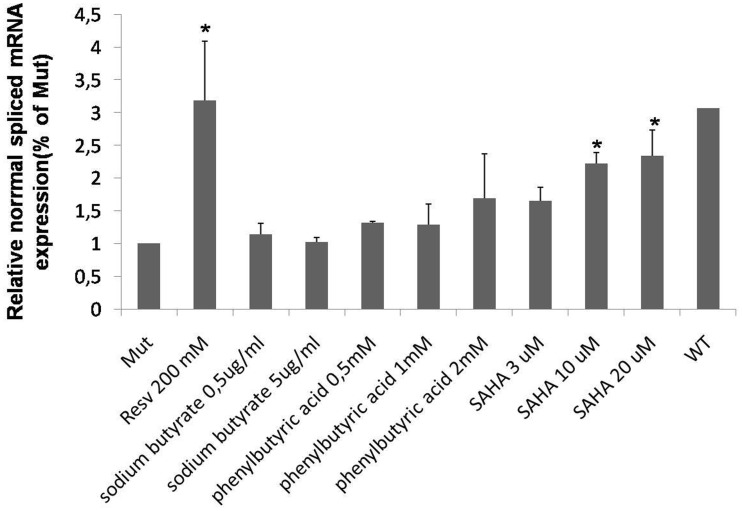
Finally, it was considered that resveratrol is a well-known inhibitor of the histone deacetylase (HDAC) activity. In this respect, it has been reported that other HDAC inhibitors, such us sodium butyrate (28), phenylbutyric acid (29) and SAHA (30), are also able to modify the mRNA splicing patterns. Therefore, we tested the effect of these molecules on the GAA mRNA splicing using our MUT minigene system. Although none of these molecules was as effective as resveratrol in rescuing normal GAA mRNA splicing, a dose-dependent increase in the normal spliced mRNA produced by the MUT minigene could be observed in cells treated with SAHA (Figure 6).
Figure 6.
Effect of HDAC inhibitors on GAA splicing. Cells transfected with the MUT minigene were treated for 24 h with sodium butyrate, phenylbutyric acid and SAHA. The relative expression of the N isoform was analyzed by real time PCR, using minigene-specific primers designed to amplify only this variant. The results are expressed as fold of N isoform detected in untreated cells. The abundance of the N isoform produced by cells transfected with the WT minigene is also shown for comparison. Data represent the means ± SD of three independent experiments. *P < 0.05.
DISCUSSION
The intronic GAA mutation c.-32-13T>G is the most frequent mutation in patients affected by the LO form of GSDII accounting for ∼40–70% of the LO alleles (7–12). Although this mutation was first reported many years ago, the mechanism by which it affects GAA pre-mRNA splicing has not been studied in detail to date.
The initial studies performed in cells from patients carrying this mutation have shown that it leads to the generation of three aberrant splice variants in which GAA exon 2 was partially or completely spliced out in addition to retaining small amounts of normal spliced mRNA (14). As a consequence, patients carrying this mutation expressed some residual GAA activity, which might be enough to prevent the onset of severe phenotype and could explain the LO origin of the disease. Therefore, the purpose of this study was to better characterize this splicing event and suggest possible ways to rescue correct splicing efficiency able to revert the pathologic phenotype.
Here, using fibroblasts from two patients who only express the c.-32-13T>G allele and normal controls, we found that in the absence of the c.-32-13T>G mutation small amounts of splice variants SV2 and SV3 are produced, as previously reported. It is worth noting that an analysis in silico of the 3′ acceptor splice site of exon 2 showed that even in the absence of the mutation, these sequences would still represent a viable splice site. Such a characteristic, together with the unusual length of the exon, may explain the generation of small amounts of the aberrant splice variants even in the absence of the c.-32-13T>G mutation.
In addition to the variants already reported, two additional mRNA isoforms that resulted from the usage of the cryptic donor site located 36-nt downstream of the canonic 5′ splice site of intron 1 were detected (N’ and SV3’) suggesting that, even in the absence of the mutation, the normal processing of this GAA region could lead to the generation of six different splice variants. Taken together, our results show that in this already very complex situation, the presence of the c.-32-13-T>G mutation leads to a very significant increase in all these aberrant splice variants at the expense of the one leading to full exon 2 inclusion and the production of an active GAA enzyme.
In this work, we have demonstrated that the c.-32-13T>G mutation can abrogate the binding of basic splicing factor U2AF65 to the polypyrimidine tract of exon 2 and that, as expected, several auxiliary splicing factors can affect exon 2 inclusion. Most importantly, the only factor capable of acting both in the wild type and c.-32-13 T>G context was represented by the SR protein SRSF4 (SRp75). At present, we do not know why only SRSF4 was able to display this effect, and its mechanism of action, which seems to be independent of U2AF65, has not been yet clarified. However, the observation that this protein can directly interact with SF3a subunit (31) could suggest that binding of SRSF4 to GAA exon 2 might be able to recruit directly additional U2 snRNP factors to the 3′ splice site.
Because the c.-32-13 T>G mutation is extremely common among LO GSDII patients and taking into account our results, it was then decided to test whether altering splicing factors levels could represent a viable alternative or complementary solution to existing therapeutic approaches for GSDII.
To this date, in fact, the only treatment currently approved for GSDII is represented by enzyme replacement therapy with recombinant human GAA (rhGAA). Although most of the studies on enzyme replacement therapy in GSDII support its efficacy in improving survival or in stabilizing the disease course, limitations of this approach are becoming evident (32) and innovative, and more effective therapies are highly desirable.
Recently, several strategies have been developed to explore therapeutic approaches that may rescue/increase normal splicing of transcripts carrying mutations that affect the mRNA splicing process, including the use of small molecules that increase the expression or activity of splicing factors (33). Of all the compounds tested, only resveratrol treatment of fibroblasts from patients carrying the c-32-13T>G mutation resulted in a significant increase of normal spliced GAA mRNA. Most importantly, this rescue of normal splicing was associated with a 2-fold increase in GAA enzymatic activity and protein content expressed by these fibroblasts. It is worth noting that the effect of resveratrol was similar in both patient's cell lines, independently of the different level of basal normal spliced mRNA and protein expression and activity. Considering that some patients carrying the c.-32-13T>G mutation express up to 20% of normal GAA activity (34,35), this increase might be enough to achieve a beneficial effect in clinical settings.
Beneficial effects of resveratrol on many biological processes have been reported. It has been shown to protect against cardiovascular diseases, type II diabetes and neurological diseases. In addition, several effects on skeletal muscle, such as an inhibition of protein catabolism, an enhancement of glucose transport and protection against oxidative stress, injury and cells death have also been reported (36). However, it is worth noting that resveratrol is also an inductor of autophagy, a critical player in the pathogenesis of GSDII (37). Therefore, even if resveratrol is able to partially rescue normal GAA splicing and activity of alleles carrying the c.-32-13T>G mutation, it might also increase the intracellular autophagic build up. These potential effects need to be further investigated. Nonetheless, the observation that another inhibitor of HDAC activity, SAHA, displayed a lower, but still beneficial, effect on the rescue of correct GAA splicing suggests that this class of molecules may represent a priority research area to find even more effective effectors.
In conclusion, our results have provided the first in vitro proof of principle for the use of small molecules to partially rescue normal splicing of c.-32-13T>G mutant alleles. As such, these results provide the justification to perform future high-throughput screening assays of small molecules that could efficiently rescue normal GAA mRNA expression and could eventually represent an alternative strategy to delay or rescue disease progression in almost all LO GSDII patients that are characterized by the presence of the c.-32-13T>G mutation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Italian Health Ministry ‘Svillupo e utilizzo dei percorsi assistenziali diagnostico-terapeutici per malattie rare’. Funding for open access charge: Italian Health Ministry.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Hirschhorn R, Reuser AJ. Glycogen storage disease type II: acid α-glucosidase (acid maltase) deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. Vol. III. New York: McGraw-Hill; 2001. pp. 3389–3420. [Google Scholar]
- 2.Raben N, Plotz P, Byrne BJ. Acid alpha-glucosidase deficiency (glycogenosis type II, Pompe disease) Curr. Mol. Med. 2002;2:145–166. doi: 10.2174/1566524024605789. [DOI] [PubMed] [Google Scholar]
- 3.Van den Hout HM, Hop W, Van Diggelen OP, Smeitink JA, Smit GP, Poll-The BT, Bakker HD, Loonen MC, De Klerk JB, Reuser AJ, et al. The natural course of infantile Pompe's disease: 20 original cases compared with 133 cases from the literature. Pediatrics. 2003;112:332–340. doi: 10.1542/peds.112.2.332. [DOI] [PubMed] [Google Scholar]
- 4.Kishnani PS, Howell RR. Pompe disease in infants and children. J. Pediatr. 2004;144:S35–S43. doi: 10.1016/j.jpeds.2004.01.053. [DOI] [PubMed] [Google Scholar]
- 5.Hagemans ML, Janssens AC, Winkel LP, Sieradzan KA, Reuser AJ, Van Doorn PA, Van der Ploeg AT. Late-onset Pompe disease primarily affects quality of life in physical health domains. Neurology. 2004;63:1688–1692. doi: 10.1212/01.wnl.0000142597.69707.78. [DOI] [PubMed] [Google Scholar]
- 6.Hagemans ML, Winkel LP, Van Doorn PA, Hop WJ, Loonen MC, Reuser AJ, Van der Ploeg AT. Clinical manifestation and natural course of late-onset Pompe's disease in 54 Dutch patients. Brain. 2005;128:671–677. doi: 10.1093/brain/awh384. [DOI] [PubMed] [Google Scholar]
- 7.Montalvo AL, Bembi B, Donnarumma M, Filocamo M, Parenti G, Rossi M, Merlini L, Buratti E, De Filippi P, Dardis A, et al. Mutation profile of the GAA gene in 40 Italian patients with late onset glycogen storage disease type II. Hum. Mutat. 2006;27:999–1006. doi: 10.1002/humu.20374. [DOI] [PubMed] [Google Scholar]
- 8.Gort L, Coll MJ, Chabás A. Glycogen storage disease type II in Spanish patients: high frequency of c.1076-1G>C mutation. Mol. Genet. Metab. 2007;92:183–187. doi: 10.1016/j.ymgme.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Nascimbeni AC, Fanin M, Tasca E, Angelini C. Molecular pathology and enzyme processing in various phenotypes of acid maltase deficiency. Neurology. 2008;19:617–626. doi: 10.1212/01.wnl.0000299892.81127.8e. [DOI] [PubMed] [Google Scholar]
- 10.Joshi PR, Gläser D, Schmidt S, Vorgerd M, Winterholler M, Eger K, Zierz S, Deschauer M. Molecular diagnosis of German patients with late-onset glycogen storage disease type II.J. Inherit. Metab. Dis. 2008;31:S261–S265. doi: 10.1007/s10545-008-0820-2. [DOI] [PubMed] [Google Scholar]
- 11.Wan L, Lee CC, Hsu CM, Hwu WL, Yang CC, Tsai CH, Tsai FJ. Identification of eight novel mutations of the acid alpha-glucosidase gene causing the infantile or juvenile form of glycogen storage disease type II.J. Neurol. 2008;255:831–838. doi: 10.1007/s00415-008-0714-0. [DOI] [PubMed] [Google Scholar]
- 12.Herzog A, Hartung R, Reuser AJ, Hermanns P, Runz H, Karabul N, Gökce S, Pohlenz J, Kampmann C, Lampe C, et al. A cross-sectional single-centre study on the spectrum of Pompe disease, German patients: molecular analysis of the GAA gene, manifestation and genotype-phenotype correlations. Orphanet J. Rare Dis. 2012;7:35–48. doi: 10.1186/1750-1172-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huie ML, Chen AS, Tsujino S, Shanske S, DiMauro S, Engel AG, Hirschhorn R. Aberrant splicing in adult onset glycogen storage disease type II (GSDII): molecular identification of an IVS1 (-13T—>G) mutation in a majority of patients and a novel IVS10 (+1GT—>CT) mutation. Hum. Mol. Genet. 1994;3:2231–2236. doi: 10.1093/hmg/3.12.2231. [DOI] [PubMed] [Google Scholar]
- 14.Boerkoel CF, Exelbert R, Nicastri C, Nichols RC, Miller FW, Plotz PH, Raben N. Leaky splicing mutation in the acid maltase gene is associated with delayed onset of glycogenosis type II. Am. J. Hum. Genet. 1995;56:887–897. [PMC free article] [PubMed] [Google Scholar]
- 15.Raben N, Nichols RC, Martiniuk F, Plotz PH. A model of mRNA splicing in adult lysosomal storage disease (glycogenosis type II) Hum. Mol. Genet. 1996;5:995–1000. doi: 10.1093/hmg/5.7.995. [DOI] [PubMed] [Google Scholar]
- 16.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;20:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prudencio M, Jansen-West KR, Lee WC, Gendron TF, Zhang YJ, Xu YF, Gass J, Stuani C, Stetler C, Rademakers R, et al. Misregulation of human sortilin splicing leads to the generation of a nonfunctional progranulin receptor. Proc. Natl Acad. Sci. USA. 2012;109:21510–21515. doi: 10.1073/pnas.1211577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passoni M, De Conti L, Baralle M, Buratti E. UG repeats/TDP-43 interactions near 5′ splice sites exert unpredictable effects on splicing modulation. J. Mol. Biol. 2012;6:46–60. doi: 10.1016/j.jmb.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Montalvo AL, Cariati R, Deganuto M, Guerci V, Garcia R, Ciana G, Bembi B, Pittis MG. Glycogenosis type II: identification and expression of three novel mutations in the acid alpha-glucosidase gene causing the infantile form of the disease. Mol. Genet. Metab. 2004;81:203–208. doi: 10.1016/j.ymgme.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 20.Hermans MM, Kroos MA, Van Beeumen J, Oostra BA, Reuser AJ. Human lysosomal alpha-glucosidase. Characterization of the catalytic site. J. Biol. Chem. 1991;266:13507–13512. [PubMed] [Google Scholar]
- 21.Deirdre A, Scadden J, Smith CW. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;3:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markus MA, Marques FZ, Morris BJ. Resveratrol, by modulating RNA processing factor levels, can influence the alternative splicing of pre-mRNAs. PLoS One. 2011;6:e28926. doi: 10.1371/journal.pone.0028926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang WH, Liu TC, Yang WK, Lee CC, Lin YH, Chen TY, Chang JG. Amiloride modulates alternative splicing in leukemic cells and resensitizes Bcr-AblT315I mutant cells to imatinib. Cancer Res. 2011;71:383–392. doi: 10.1158/0008-5472.CAN-10-1037. [DOI] [PubMed] [Google Scholar]
- 24.Chang JG, Yang DM, Chang WH, Chow LP, Chan WL, Lin HH, Huang HD, Chang YS, Hung CH, Yang WK. Small molecule amiloride modulates oncogenic RNA alternative splicing to devitalize human cancer cells. PLoS One. 2011;6:e8643. doi: 10.1371/journal.pone.0018643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bian Y, Masuda A, Matsuura T, Ito M, Okushin K, Engel AG, Ohno K. Tannic acid facilitates expression of the polypyrimidine tract binding protein and alleviates deleterious inclusion of CHRNA1 exon P3A due to an hnRNP H-disrupting mutation in congenital myasthenic syndrome. Hum. Mol. Genet. 2009;18:1229–1237. doi: 10.1093/hmg/ddp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slaugenhaupt SA, Mull J, Leyne M, Cuajungco MP, Gill SP, Hims MM, Quintero F, Axelrod FB, Gusella JF. Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 2004;13:429–436. doi: 10.1093/hmg/ddh046. [DOI] [PubMed] [Google Scholar]
- 27.Sumanasekera C, Kelemen O, Beullens M, Aubol BE, Adams JA, Sunkara M, Morris A, Bollen M, Andreadis A, Stamm S. C6 pyridinium ceramide influences alternative pre-mRNA splicing by inhibiting protein phosphatase-1. Nucleic Acids Res. 2012;40:4025–4039. doi: 10.1093/nar/gkr1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl Acad. Sci. USA. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreassi C, Angelozzi C, Tiziano FD, Vitali T, De Vincenzi E, Boninsegna A, Villanova M, Bertini E, Pini A, Neri G, et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 30.Hahnen E, Eyüpoglu IY, Brichta L, Haastert K, Tränkle C, Siebzehnrübl FA, Riessland M, Hölker I, Claus P, Romstöck J, et al. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J. Neurochem. 2006;98:193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- 31.Van der Ploeg AT, Reuser AJ. Pompe's disease. Lancet. 2008;11:1342–1353. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 32.Tazi J, Durand S, Jeanteur P. The spliceosome: a novel multi-faceted target for therapy. Trends Biochem. Sci. 2005;30:469–478. doi: 10.1016/j.tibs.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Kroos MA, Pomponio RJ, Hagemans ML, Keulemans JL, Phipps M, DeRiso M, Palmer RE, Ausems MG, Van der Beek NA, Van Diggelen OP, et al. Broad spectrum of Pompe disease in patients with the same c.-32-13T->G haplotype. Neurology. 2007;68:110–115. doi: 10.1212/01.wnl.0000252798.25690.76. [DOI] [PubMed] [Google Scholar]
- 34.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroos M, Hoogeveen-Westerveld M, Van der Ploeg A, Reuser AJ. The genotype-phenotype correlation in Pompe disease. Am. J. Med. Genet. C Semin. Med. Genet. 2012;160:59–68. doi: 10.1002/ajmg.c.31318. [DOI] [PubMed] [Google Scholar]
- 36.Dirks Naylor AJ. Cellular effects of resveratrol in skeletal muscle. Life Sci. 2009;84:637–640. doi: 10.1016/j.lfs.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda T, Roberts A, Ahearn M, Zaal K, Ralston E, Plotz PH, Raben N. Autophagy and lysosomes in Pompe disease. Autophagy. 2006;2:318–320. doi: 10.4161/auto.2984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.